UniProt Tools: BLAST, Align, Peptide Search, and ID Mapping
Sandra Orchard, Sandra Orchard, The UniProt Consortium, The UniProt Consortium, Rossana Zaru, Rossana Zaru
Abstract
The Universal Protein Resource (UniProt) is a comprehensive resource for protein sequence and annotation data (UniProt Consortium, 2023). The UniProt website receives about 800,000 unique visitors per month and is the primary means to access UniProt. Along with various datasets that you can search, UniProt provides four main tools. These are the “BLAST” tool for sequence similarity searching, the “Align” tool for multiple sequence alignment, the “Peptide Search” tool for retrieving proteins containing a short peptide sequence, and the “Retrieve/ID Mapping” tool for using a list of identifiers to retrieve UniProt Knowledgebase (UniProtKB) proteins and to convert database identifiers from UniProt to external databases or vice versa. This article provides four basic protocols and seven alternate protocols for using UniProt tools. © 2023 The Authors. Current Protocols published by Wiley Periodicals LLC.
Basic Protocol 1 : Basic local alignment search tool (BLAST) in UniProt
Alternate Protocol 1 : BLAST through UniProt text search results pages
Alternate Protocol 2 : BLAST through UniProt basket
Basic Protocol 2 : Multiple sequence alignment in UniProt
Alternate Protocol 3 : Align tool through UniProt results pages and entry pages
Alternate Protocol 4 : Align tool through UniProt basket
Basic Protocol 3 : Peptide search in UniProt
Basic Protocol 4 : Batch retrieval and ID mapping in UniProt
Alternate Protocol 5 : Retrieve/ID Mapping tool through UniProt text search results pages and BLAST and Align results pages
Alternate Protocol 6 : Retrieve/ID Mapping tool through UniProt basket
Alternate Protocol 7 : Retrieve/ID Mapping tool through UniProt search box
INTRODUCTION
UniProt, or the Universal Protein Resource, provides an up-to-date, comprehensive body of protein information at a single site (UniProt Consortium, 2023). The UniProt Knowledgebase (UniProtKB) delivers high-quality functional information for a selected set of protein sequences. To build upon these protein data and to aid analysis, UniProt provides four main tools: “BLAST” (Basic Local Alignment Search Tool), the “Align” multiple sequence alignment tool, “Peptide Search,” and “Retrieve/ID Mapping” for batch retrievals of UniProt entries and ID mapping between UniProt and external databases. These tools are available on their own dedicated pages on the UniProt website and are also accessible directly from other parts of the website, such as the basket, search/tool results pages, and protein entry pages. Having these tools in the UniProt website creates an integrated hub of data and analysis tools, allowing both to leverage each other. For example, if you come across sequences while browsing UniProt databases that you would like to BLAST or align, you can select your sequence and submit it to the relevant tool directly. Consequently, results from tools provide links directly to all relevant data from UniProt and allow you to filter by attributes, like whether you are looking for reviewed (UniProtKB/Swiss-Prot) or unreviewed (UniProtKB/TrEMBL) UniProtKB entries, entries with 3D structures, or entries that are part of a proteome, among others. All data in UniProtKB are freely available to users.
The UniProt website can be accessed at http://www.uniprot.org/. The following protocols describe how you can access and navigate the “BLAST” (Basic Protocol 1 and Alternate Protocols 1 and 2), “Align” (Basic Protocol 2 and Alternate Protocols 3 and 4), “Peptide Search” (Basic Protocol 3), and “Retrieve/ID Mapping” (Basic Protocol 4 and Alternate Protocols 5 to 7) tools on the UniProt website and use them in your analysis.
Basic Protocol 1: BASIC LOCAL ALIGNMENT SEARCH TOOL (BLAST) IN UniProt
The UniProt website provides BLAST (UniProt Consortium, 2023), which finds regions of local similarity between sequences. This can be used to infer functional and evolutionary relationships between sequences as well as to help identify members of gene families. The BLAST tool page can be reached from a link in the header on all pages of the UniProt website.
Necessary Resources
- An up-to-date web browser
1.Go the UniProt home page at http://www.uniprot.org/ using an up-to-date web browser.
2.Click on the “BLAST” link.


3.To run a BLAST search, enter either a UniProt identifier into the first input box or a protein or nucleotide sequence into the second input box provided.
| Advanced option | Significance |
|---|---|
| Database | Database against which the search is performed; UniProtKB or clusters of sequences with 100%, 90%, or 50% identity. |
| Restrict by taxonomy | Select a specific taxon against which the search is performed. |
| E-threshold | The expectation value (E) threshold is a statistical measure of the number of expected matches in a random database. The lower the E-value, the more likely the match is to be significant. E-values between 0.1 and 10 are generally dubious and over 10 are unlikely to have biological significance. In all cases, those matches need to be verified manually. You may need to increase the E-value threshold if you have a very short query sequence, if you need to detect very weak similarities or similarities in a short region, or if your sequence has a low-complexity region and you use the “filter” option. |
| Matrix | The matrix assigns a score for each position in an alignment. The BLOSUM matrix assigns a score based on the frequency at which that substitution is known to occur among consensus blocks within related proteins. BLOSUM62 is among the best of the available matrices for detecting weak protein similarities. The PAM set of matrices is also available. If “Auto” is set, the matrix will be selected depending on the query sequence length. |
| Filtering | Low-complexity regions (e.g., stretches of cysteine in Q03751 or, in some cases, hydrophobic regions in membrane proteins) tend to produce spurious, insignificant matches with sequences in the database that have the same kind of low-complexity regions but are unrelated biologically. If “Filter low complexity regions” is selected, the query sequence will be run through the program SEG, and all amino acids in low-complexity regions will be replaced by an X. |
| Gapped | This will allow gaps to be introduced in the sequences when the comparison is done. |
| Hits | This limits the number of returned alignments. The user can limit the number of returned alignments to 50, 100, 250, 500, 750, or 1000. By default, the limit is set to 250. |
| HSPs per hit | This limits the number of high-scoring segment pairs (HSPs), each of which has local alignment with no gaps that achieves one of the highest alignment scores in a given search. |
4.Click on the “Run BLAST” button to execute the query.

Alternate Protocol 1: BLAST THROUGH UniProt TEXT SEARCH RESULTS PAGES
As an alternative to Basic Protocol 1, queries can be submitted to the BLAST tool directly through UniProt search results pages and when you come across a sequence you would like to analyze using a sequence similarity search. This allows for a flexible workflow between browsing data and analyzing data.
Necessary Resources
- An up-to-date web browser
1.Go the UniProt home page at http://www.uniprot.org/ using an up-to-date web browser.
2.Choose the search dataset using the drop-down menu to the left of the search box. Select “UniProtKB,” “UniRef,” or “UniParc.”
3.Enter a query in the search box, for example “insulin,” and click on the “Search” button.
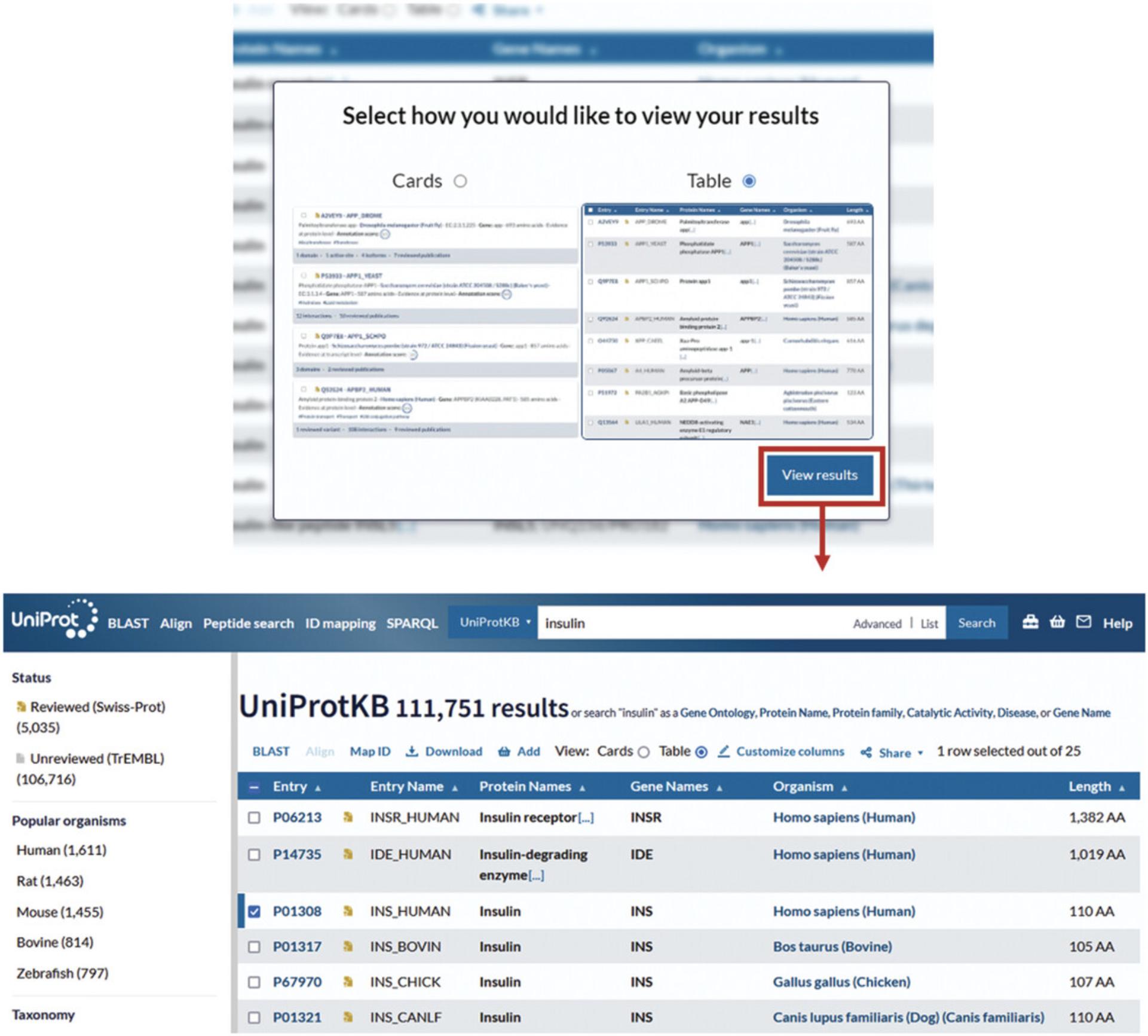
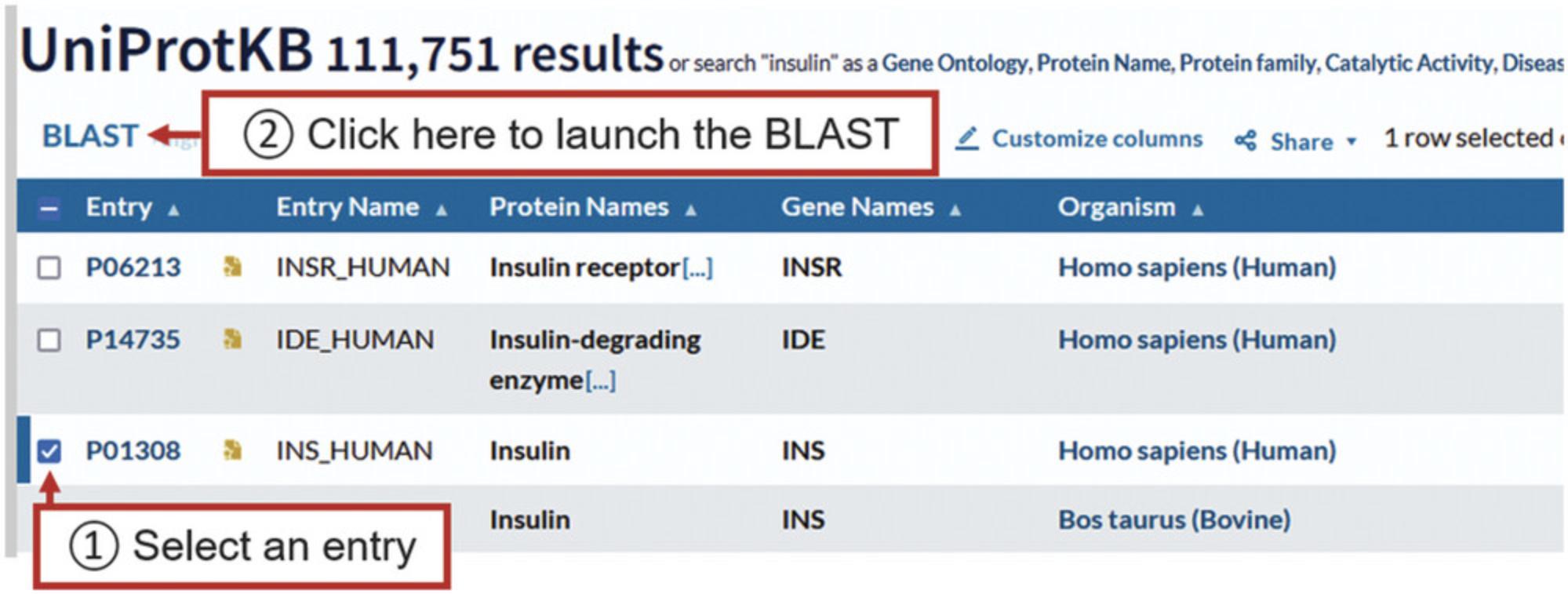
4.To run a BLAST search for a protein in the search results, click on the checkbox in the left-hand column for that protein row.
5.Click on the “BLAST” button just above the search results table, as shown in Figure 6, which will open the BLAST page, from where the BLAST can be launched as described in Basic Protocol 1, steps 3 and 4.

Alternate Protocol 2: BLAST THROUGH UniProt BASKET
As an alternative to Basic Protocol 1, queries can be submitted to the BLAST tool directly through the UniProt basket feature. The UniProt basket allows you to store entries from UniProtKB, UniRef, or UniParc. You can use the basket to build a set of your proteins across different searches. The basket then allows you to download your dataset to access analysis tools (i.e., BLAST, Align, and Retrieve/ID Mapping). Your basket is saved as long as you do not clear your cookies.
Necessary Resources
- An up-to-date web browser
1.Go the UniProt home page at http://www.uniprot.org/ using an up-to-date web browser.
2.Choose the search dataset using the drop-down menu to the left of the search box. Select “UniProtKB,” “UniRef,” or “UniParc.”
3.Enter a query in the search box, for example “insulin,” and click on the “Search” button.
4.For the entry of interest, click on the checkbox to the left of its accession and then click on “Add” next to the basket icon button at the top of the results table, as shown in Figure 8.

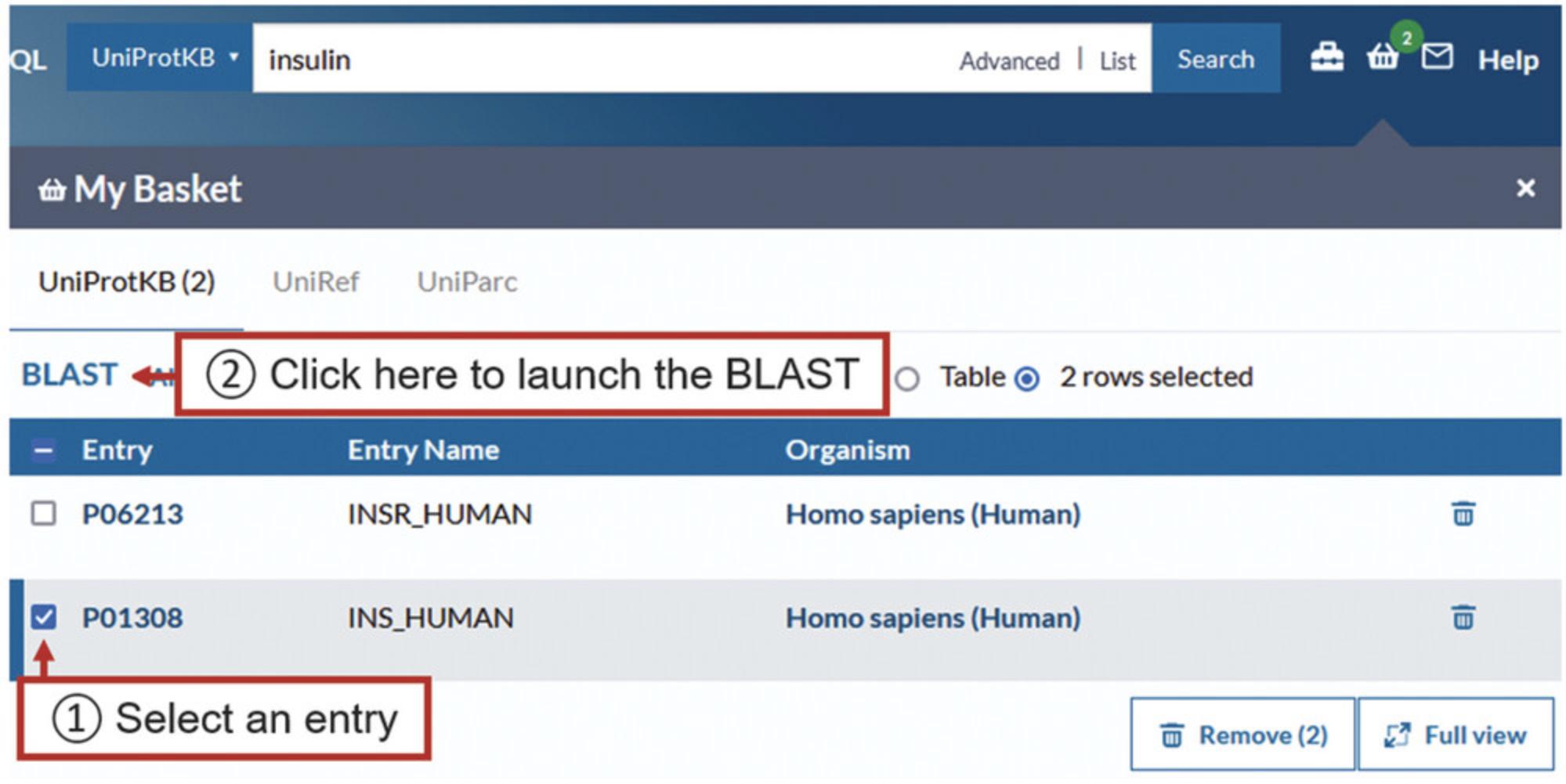
5.When ready to analyze the entries in the basket, click on the basket to open it.
6.Click on the checkbox to the left of the entry of interest and then click on “BLAST,” as shown in Figure 9.
Basic Protocol 2: MULTIPLE SEQUENCE ALIGNMENT IN UniProt
The UniProt website provides a multiple sequence alignment tool for proteins called “Align.” This tool runs the Clustal Omega algorithm to find areas of similarity in the entries being aligned. This can be used to find conserved residues and regions that can help infer evolutionary and functional relationships (see Current Protocols article: Simossis, Kleinjung, & Heringa, 2003).
Necessary Resources
- An up-to-date web browser
1.Go the UniProt home page at http://www.uniprot.org/ using an up-to-date web browser.
2.Click on the “Align” link.

3.To execute the multiple sequence alignment, enter either the UniProt identifiers into the first input box or the protein sequences in FASTA format into the second sequence input box provided and click “Align sequences.”
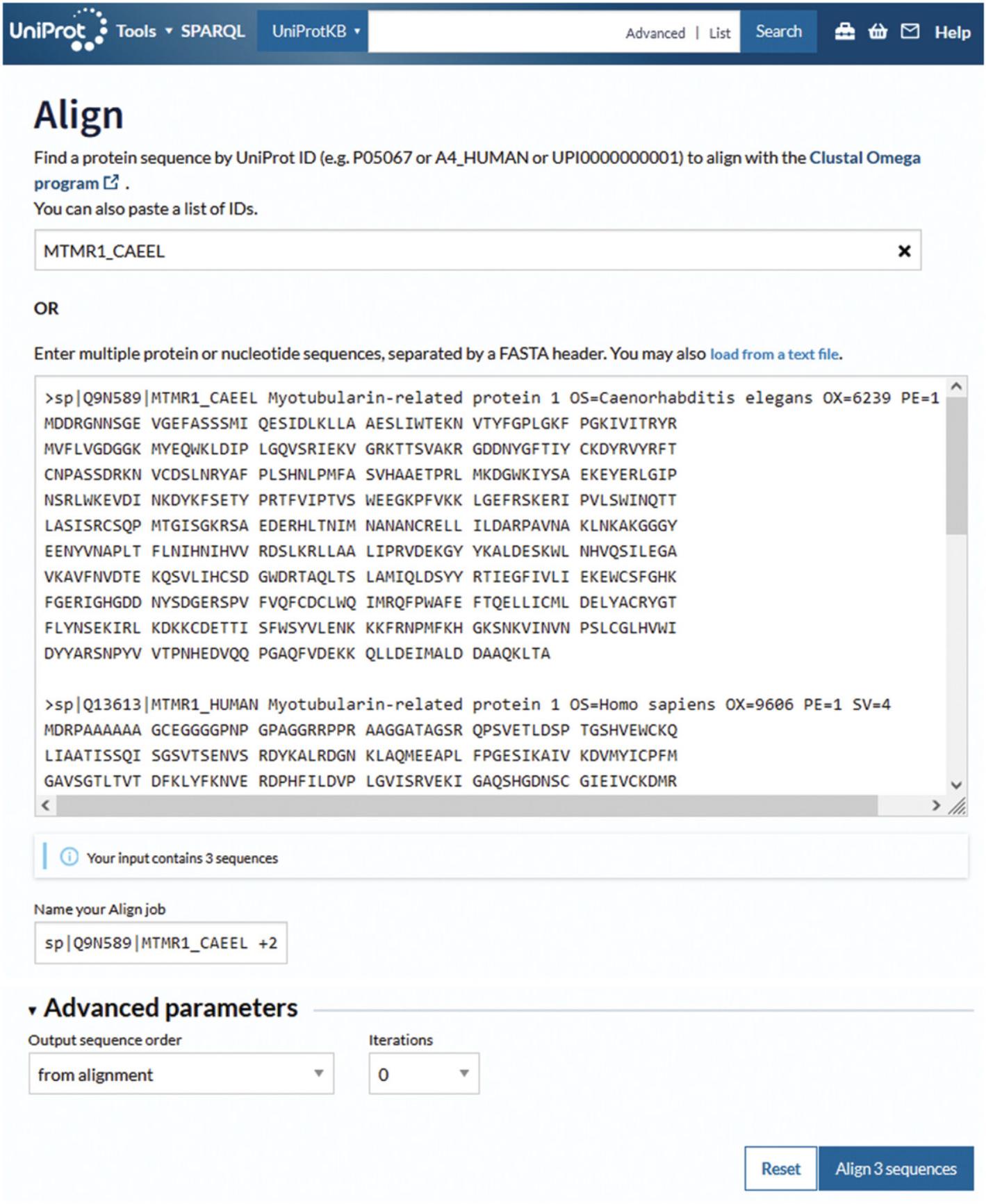

4.Once the alignment is finished, click on “completed” to open the Align results page, which is shown in Figure 13.
Alternate Protocol 3: ALIGN TOOL THROUGH UniProt RESULTS PAGES AND ENTRY PAGES
As an alternative to Basic Protocol 2, queries can be submitted to the Align tool directly through UniProt search results pages.
Necessary Resources
- An up-to-date web browser
1.Go the UniProt home page at http://www.uniprot.org/ using an up-to-date web browser.
2.Select “UniProtKB” as the search dataset using the drop-down menu to the left of the search box.
3.Enter a query in the search box, for example “insulin,” and click on the “Search” button.
4.Click on two or more checkboxes to align these protein entries, as shown in Figure 14.

5.Click on the “Align” button just above the search results table.
6.Click the “Align sequence” button.
7.Once the alignment is finished, click on “completed” to open the Align results page.
Alternate Protocol 4: ALIGN TOOL THROUGH UniProt BASKET
As an alternative to Basic Protocol 2, queries can also be submitted to the Align tool directly through the UniProt basket feature.
Necessary Resources
- An up-to-date web browser
1.Go the UniProt home page at http://www.uniprot.org/ using an up-to-date web browser.
2.Select “UniProtKB” as the search dataset using the drop-down menu to the left of the search box.
3.Enter a query in the search box, for example “insulin,” and click on the “Search” button.
4.For entries of interest, click on the checkboxes to the left of their accession numbers in the results table and then click on “Add” next to the basket icon at the top of the results table.
5.When ready to analyze the entries in the basket, click on the basket to open it, as shown in Figure 15.

6.Click on the checkboxes to the left of the entries to be aligned and then click on “Align.”
7.Click the "Align sequence" button.
8.Once the alignment is finished, click on “completed” to open the Align results page.
Basic Protocol 3: PEPTIDE SEARCH IN UniProt
The UniProt website provides a tool that allows you to upload short peptide sequences of at least three residues and find all UniProtKB sequences that have an exact match to the query sequence. These peptide sequences can come from proteomics experiments or from the design of peptides for antibody production, for example.
Necessary Resources
- An up-to-date web browser
1.Go the UniProt home page at http://www.uniprot.org/ using an up-to-date web browser.
2.Click on the “Peptide search” link.

3.Enter the peptide(s) of interest into the input box as shown in Figure 17 and click on the “Run Peptide search” button.


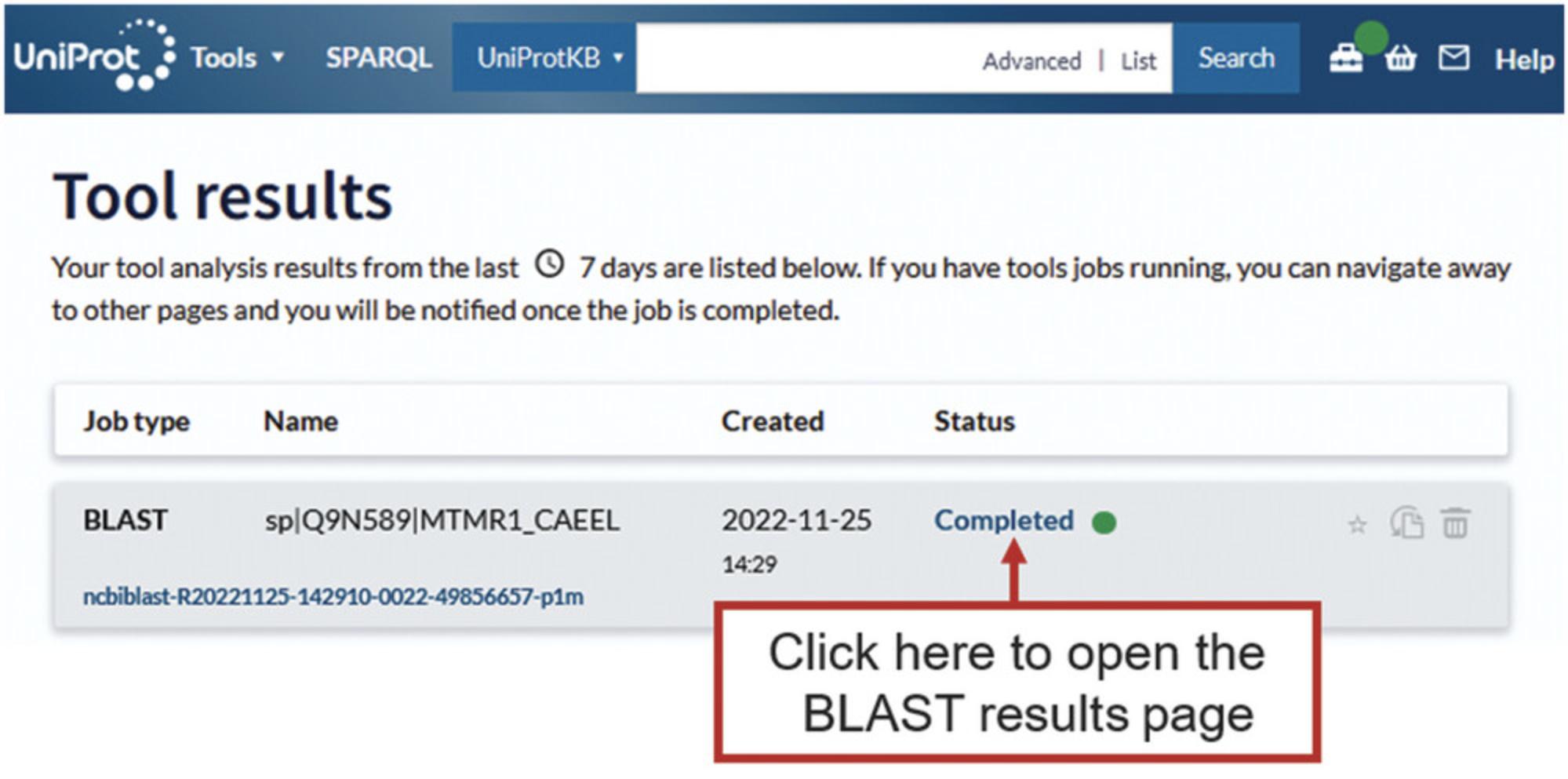
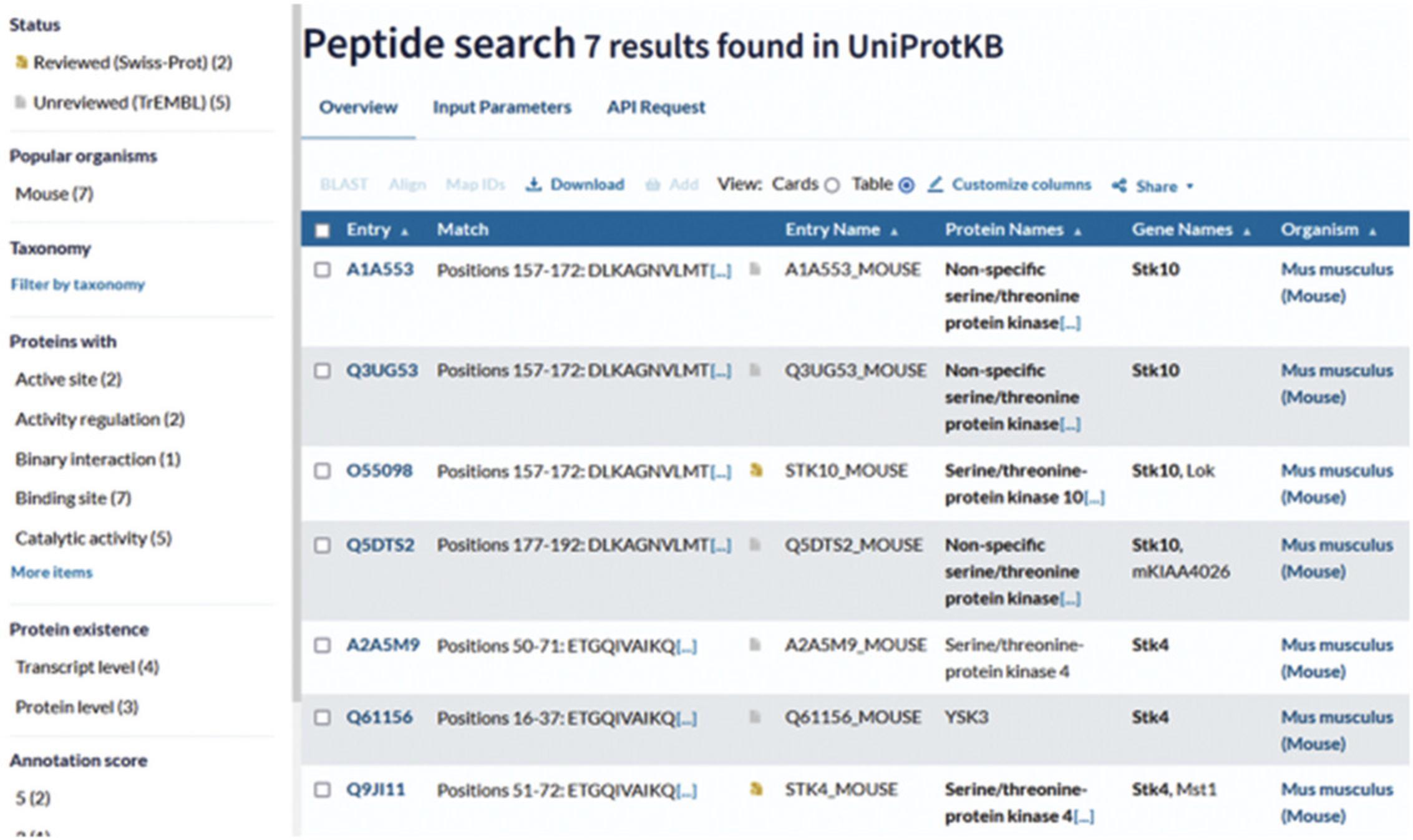
4.Once the search is finished, click on “completed” to open the results page, which is shown in Figure 19.
Basic Protocol 4: BATCH RETRIEVAL AND ID MAPPING IN UniProt
The UniProt website provides a tool that allows you to upload a list of UniProt identifiers and batch-retrieve all the corresponding UniProt entries. It allows you to convert or “map” your identifiers from UniProtKB to over 100 external databases that UniProt is cross-referenced to and vice versa (e.g., Ensembl, PDB, RefSeq; Huang et al., 2011). This covers a number of databases from different categories, including databases covering sequence, 3D structure, protein-protein interaction, protein family and group, chemistry, post-translational modification, and genome annotation, among others. This tool is called “Retrieve/ID Mapping.”
Necessary Resources
- An up-to-date web browser
1.Go the UniProt home page at http://www.uniprot.org/ using an up-to-date web browser.
2.Click on the “ID Mapping” link.

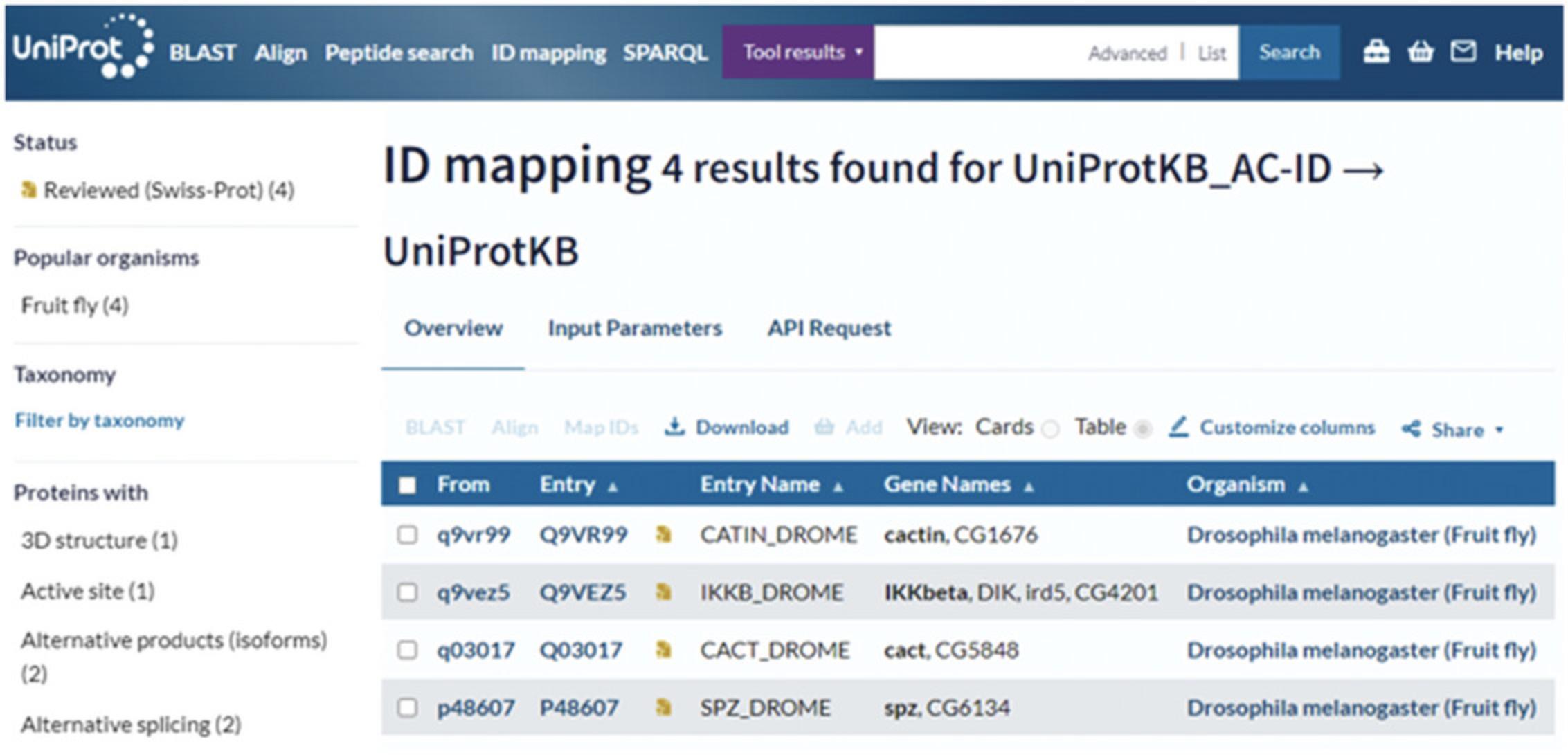
3.To retrieve functional information or look at the sequences for a list of UniProt IDs (for example, Q9VR99, Q9VEZ5, Q03017, and P48607), paste the list into the input box provided or upload a file.
4.Leave “From” and “To” as “UniProtKB” and click the “Map IDs” button.

5.Once the mapping is finished, click on “completed” to open the results page as shown in Figure 22.
6.To map a list of UniProt IDs to IDs from an external database such as Ensembl, RefSeq, or one of the model organism databases (or vice versa), upload or paste in the IDs. Then, select the source database in the “From” drop-down menu and the target database in the “To” drop-down menu.
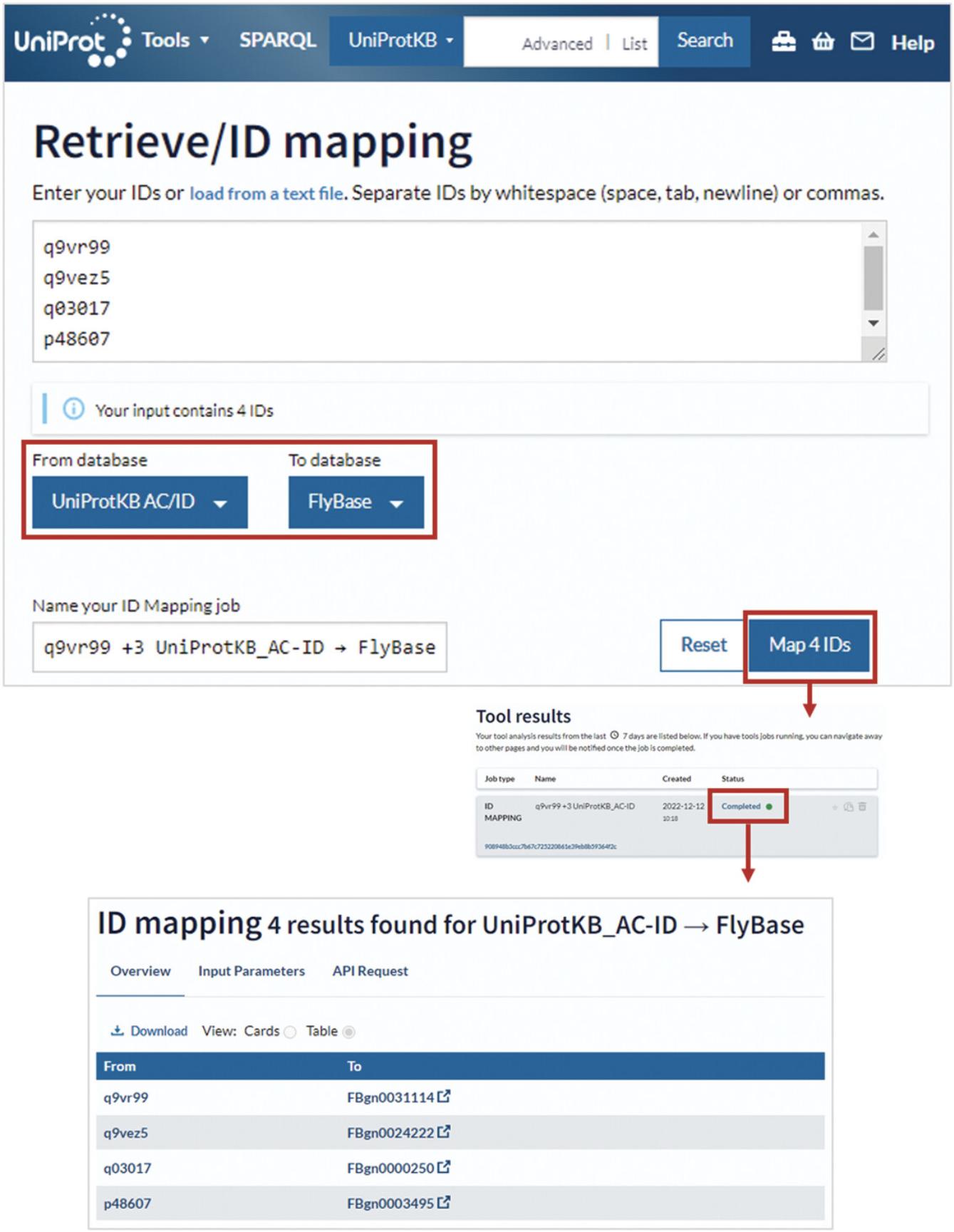
7.Click the “Map IDs” button as shown in Figure 23.
Alternate Protocol 5: RETRIEVE/ID MAPPING TOOL THROUGH UniProt TEXT SEARCH RESULTS PAGES AND BLAST AND ALIGN RESULTS PAGES
As an alternative to Basic Protocol 4, queries can be submitted to the “Retrieve/ID Mapping” tool directly through UniProt search results pages, BLAST results pages, or Align results pages.
Necessary Resources
- An up-to-date web browser
1.Go the UniProt home page at http://www.uniprot.org/ using an up-to-date web browser.
2.Select “UniProtKB” as the search dataset using the drop-down menu to the left of the search box.
3.Enter a query in the search box, for example “insulin,” and click on the “Search” button.
4.Click on two or more checkboxes as shown in Figure 24.

5.Click on the “MapIDs” button just above the search results table.
Alternate Protocol 6: RETRIEVE/ID MAPPING TOOL THROUGH UniProt BASKET
Queries can be submitted to the “Retrieve/ID Mapping” tool directly through the UniProt basket. The UniProt basket allows you to store entries from UniProtKB, UniRef, or UniParc. You can use the basket to build a set of your proteins across different searches. The basket then allows you to download your dataset to access analysis tools (i.e., BLAST, Align, and Retrieve/ID Mapping). Your basket is saved as long as you do not clear your cookies.
Necessary Resources
- An up-to-date web browser
1.Go the UniProt home page at http://www.uniprot.org/ using an up-to-date web browser.
2.Choose “UniProtKB” for the search dataset using the drop-down menu to the left of the search box.
3.Enter a query in the search box, for example “insulin,” and click on the “Search” button.
4.For entries of interest, click on the checkboxes to the left of their accession numbers in the results table and then click on “Add” next to the basket icon at the top of the results table, as shown in Figure 8.
5.When ready to analyze the entries in the basket, click the basket to open it.
6.To map the UniProt IDs to an external database from the basket, click on the checkboxes to the left and click on the “Map IDs” button in the basket, as shown in Figure 25.

Alternate Protocol 7: RETRIEVE/ID MAPPING TOOL THROUGH UniProt SEARCH BOX
As an alternative to Basic Protocol 4, queries can be submitted to the “Retrieve/ID Mapping” tool directly through the UniProt search box.
Necessary Resources
- An up-to-date web browser
1.Go the UniProt home page at http://www.uniprot.org/ using an up-to-date web browser.
2.Click on “List” on the left-hand side of the general search box as shown in Figure 26.

GUIDELINES FOR UNDERSTANDING RESULTS
UniProt BLAST Results
The UniProt BLAST results appear as shown in Figure 4. The results page provides (1) a left-hand side panel with filters and (2) a results table that contains the protein entries (hits) that match your query. The left-hand side panel allows you to filter the results based on the percent identity, the score, or the E-value. You can also filter for reviewed entries (UniProtKB/Swiss-Prot), unreviewed entries (UniProtKB/TrEMBL), entries with specific features such as proteins with a 3D structure, or entries from a specific organism. For example, Figure 27 shows how you can use the filter by taxonomy to display only mouse entries.

In the results table, the protein entries are sorted according to their score in descending order. For each entry, various information is displayed, including the name, the organism, and the protein length. The last column on the left provides a basic graphical view of the sequence alignment showing the percent identity, the score, and the E-value in small boxes. Clicking on the sequence box opens a window at the bottom of the page where you can see a detailed view of the alignment between the query and the matched entry, as shown in Figure 28. The alignment can be explored as described in the UniProt Align Results section below.

On the top of the page, various tabs allow you to view the results according to their “Taxonomy,” or “Hit Distribution,” or as a “Text Output.”
You can download your alignment in various formats by clicking on the “Download” button. You can also select one or multiple entries by clicking the checkboxes next to them to run a BLAST or an alignment or to store the entries in the basket for further analysis. Using the “Customize columns” button above the table allows you to change the type of information displayed. For example, in Figure 29, the subcellular location is shown.

The “API request” tab provides you with the code to run the current job with the same input on the command line using curl.
You can save the URL of your sequence alignment to access it at any time for up to 7 days from when you first ran the query. The “Input Parameters” tab provides detailed information about the alignment parameters, including the job identifier, which can be used to access your sequence alignment at any time for up to 7 days from when you first ran the query.
UniProt Align Results
Multiple sequence alignments can help understand evolutionary conservation of structurally and functionally important regions of protein sequences (see Current Protocols article: Simossis et al., 2003). To obtain meaningful results and minimize errors in the alignment, it is necessary to align sequences that are likely to be related to each other.
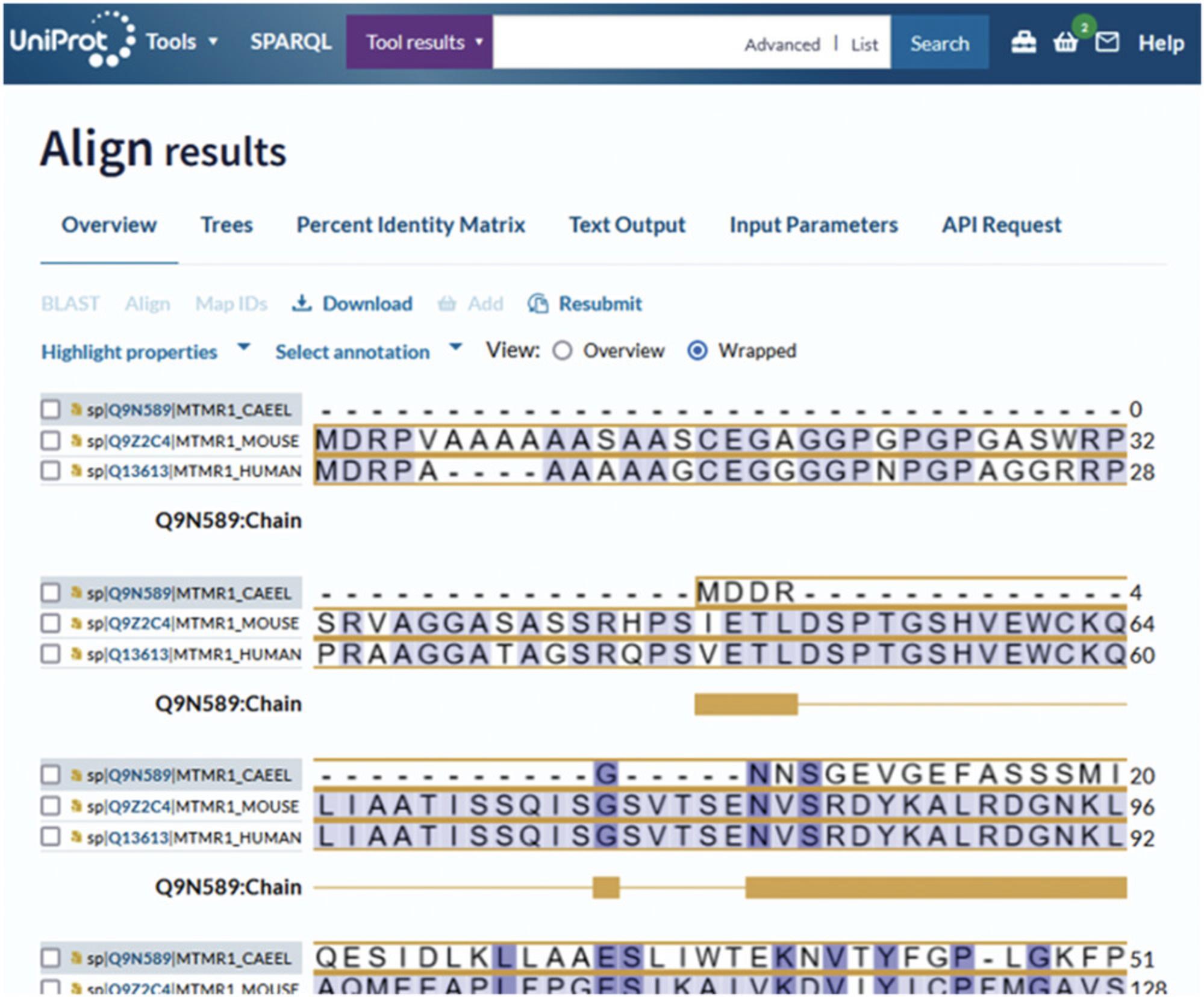
The UniProt Align results appear as shown in Figure 13. The results page displays the full sequence alignment by default (“wrapped” view). You can change how the alignment is displayed by selecting one of the “view” options above the sequence on the right. The “overview” mode allows you to zoom in using the gray sequence toggles and explore a specific region of the alignment, as shown in Figure 30.
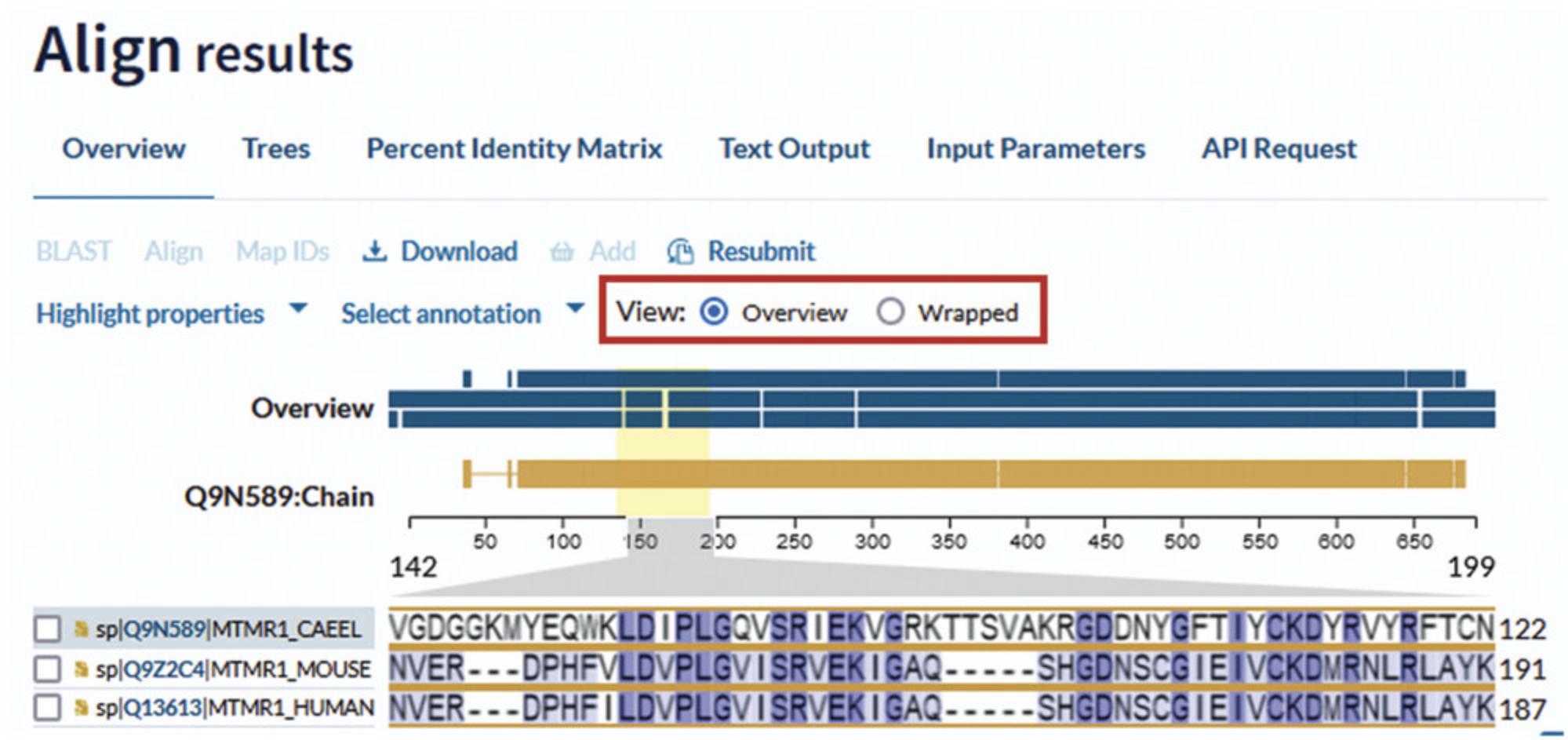
Above the aligned sequence on the left, there are two drop-down menus that allow you to select sequence annotations (i.e., domains, sites) to view them highlighted across the aligned sequences. For example, in Figure 31, the active site has been selected using “Select annotation” and is shown highlighted in the sequence. Clicking on the active-site pictogram opens a box where you can find additional information about the site. You can also highlight sequence features by amino acid properties (i.e., hydrophobicity). Both menus are available in the “overview” and “wrapped” views.

On the top of the results page, there are various tabs that give you the option to change how the alignment is visualized. The alignment can be viewed as “Trees,” as shown in Figure 32; as “Percent Identity Matrix;” or as “Text Output.”

You can download your alignment in various formats by clicking on the “Download” button. You can also select one or multiple entries by clicking the checkbox next to them to run a BLAST or a new alignment or to store the entries in the basket for further analysis.
The “API request” tab provides you with the code to run the current job with the same input on the command line using curl.
You can save the URL of your sequence alignment to access it at any time for up to 7 days from when you first ran the query. The “Input Parameters” tab provides detailed information about the alignment parameters, including the job identifier, which can be used to access your sequence alignment at any time for up to 7 days from when you first ran the query.
UniProt Peptide Search Results
The “Peptide Search” results provide UniProtKB entries with sequences that contain an exact match for each peptide that you searched for.
The results page for the peptide search is shown in Figure 19. For each protein entry row, the position of the peptide in the sequence and the sequence of the peptide are provided in the “Match” column. The filters on the left-hand side can be used to restrict the search to specific features. For example, you can display only UniProtKB/Swiss-Prot/reviewed entries.
Protein entries can be selected by clicking the checkboxes and then can be downloaded or stored in the basket for analysis at a later time point.
The “API request” tab at the top of the results page provides you with the code to run the current job with the same input on the command line using curl.
You can also save the URL of your peptide search to access it at any time for up to 7 days from when you first ran the query. The “Input Parameters” tab provides detailed information about the parameters, including the job identifier, which can be used to access your peptide results table at any time for up to 7 days from when you first ran the query.
UniProt Retrieve/ID Mapping Results
If you retrieve a batch of UniProt entries for a list of IDs using this tool, you will get a results page as shown in Figure 22. This results page provides filters in the left-hand side panel and a main results table. The results table starts with a column titled “From” that shows your input identifiers. The next columns in the results table show information from the corresponding UniProt entries that were found. You can edit these columns by clicking on the “Customize Columns” button above the results table. You can also run tools such BLAST and Align and add entries to your basket by selecting the corresponding checkboxes and then clicking on the buttons available at the top of the results page. You can download the full results table or just the list of identifiers using the “Download” button.
If you use the tool to map UniProt IDs to external database IDs (or vice versa; Huang et al., 2011), you will get a results table with two columns showing your input IDs and the corresponding mapped IDs, as shown in Figure 23. You can use the “Download” button to download your results.
The “API request” tab provides you with the code to run the current job with the same input on the command line using curl.
You can save the URL of your ID mapping to access it at any time for up to 7 days from when you first ran the query. The “Input Parameters” tab provides detailed information about the parameters, including the job identifier, which can be used to access your mapping results table at any time for up to 7 days from when you first ran the query.
COMMENTARY
Background Information
UniProt aids scientific discovery by collecting, interpreting, and organizing information so that it is easy to access and use. In addition to providing data through various datasets, UniProt also provides tools to help researchers analyze these data. UniProtKB is the central hub for the collection of functional information and other rich annotations on proteins. It is further divided into the Reviewed (UniProtKB/Swiss-Prot), expertly annotated section and the Unreviewed (UniProtKB/TrEMBL), automatically annotated section. UniParc is a non-redundant archive containing all the publicly available protein sequences in the world. UniRef provides clustered sets of sequences from UniProtKB (including isoforms) and selected UniParc entries. UniRef reduces redundancy and provides complete coverage of the sequence space at three levels of sequence identity (i.e., 100%, 90%, and 50% identity). The Proteomes dataset provides protein sets for organisms with completely sequenced genomes. Supporting datasets are a collection of meta-information about proteins in UniProtKB entries, such as literature citations, taxonomy, subcellular locations, keywords, cross-referenced databases, and diseases. The tools UniProt provides are BLAST, Align, Peptide Search, and Retrieve/ID Mapping. The UniProt website was designed following a user-centered design process and is flexible, powerful, and user friendly. It provides many ways of accessing these tools. Using tools within UniProt, you can easily chain activities by (1) searching for data, (2) running a BLAST search for a sequence in your results, (3) running a multiple sequence alignment on sequences in your BLAST results, and then (4) mapping the IDs of these sequences to an external database.
UniProt provides training material through the European Bioinformatics Institute (EMBL-EBI) online training portal, including a quick tour (https://www.ebi.ac.uk/training/online/courses/uniprot-quick-tour/) and a detailed course (https://www.ebi.ac.uk/training/online/courses/uniprot-exploring-protein-sequence-and-functional-info/). UniProt also provides short video tutorials embedded in the website, and they are available on a YouTube channel at https://www.youtube.com/uniprotvideos.
Critical Parameters
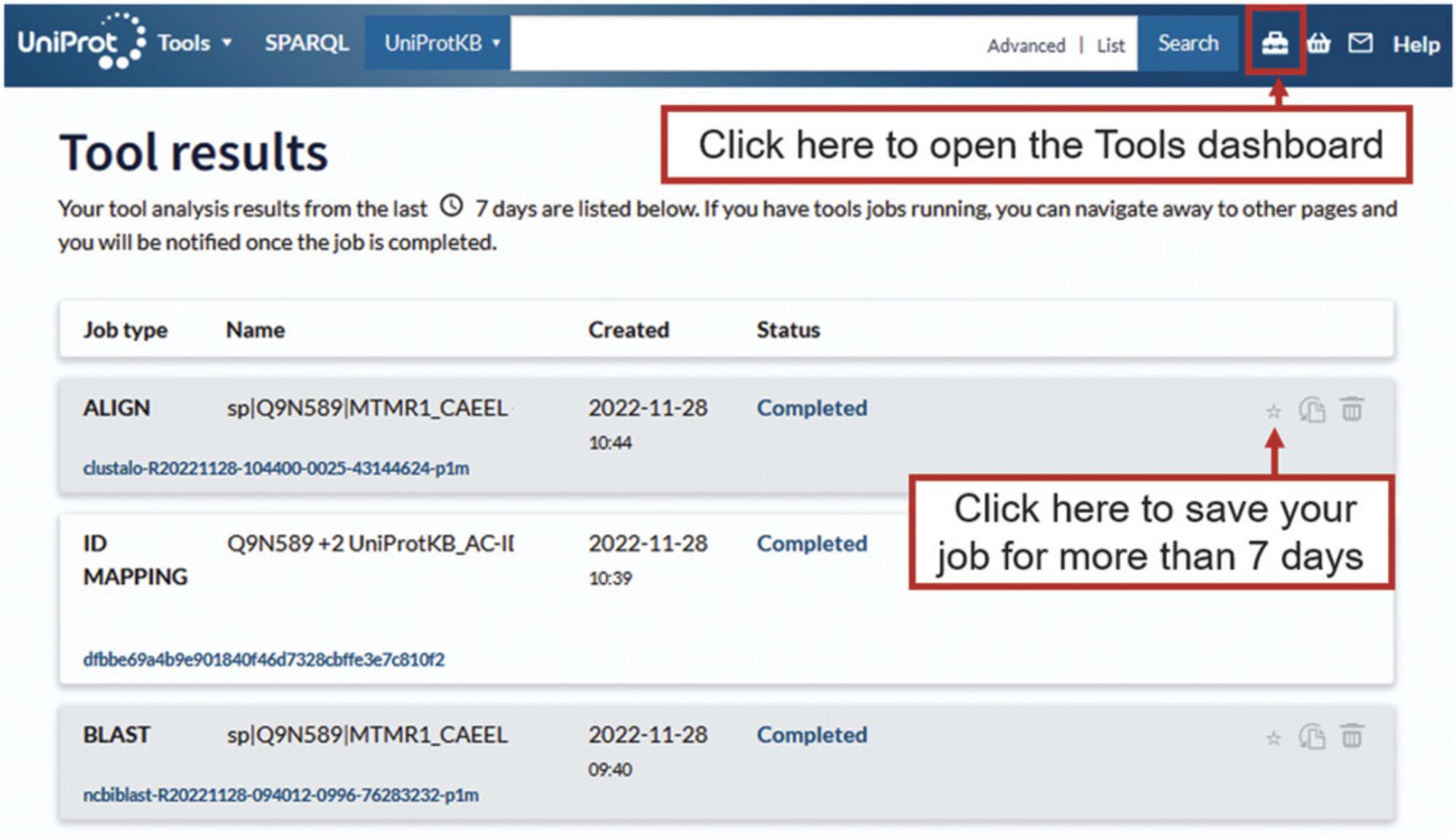
The Tools dashboard is where all the results of the Tools queries that you have run are stored for 7 days from when you first ran the query. You can access the dashboard by clicking the toolbox icon on the right-hand side of the UniProt head banner as shown in Figure 33. For each job, you have the option to resubmit the query, with the possibility to modify the parameters, store the job for more than 7 days (this requires the user to not clear UniProt website cookies), or delete the job.
Acknowledgments
This work was supported by the National Human Genome Research Institute (NHGRI), Office of the Director (OD/DPCPSI/ODSS), National Institute of Allergy and Infectious Diseases (NIAID), National Institute on Aging (NIA), National Institute of General Medical Sciences (NIGMS), National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK), National Eye Institute (NEI); National Cancer Institute (NCI), and National Heart, Lung, and Blood Institute (NHLBI) of the National Institutes of Health under Award Number [U24HG007822].
Open access funding enabled and organized by Projekt DEAL.
Author Contributions
Rossana Zaru : Writing – original draft, writing – review and editing; Sandra Orchard : Funding acquisition, writing – review and editing.
Conflict of Interest
The authors declare no conflict of interest.
Open Research
Data Availability Statement
The data in UniProt are freely available and can be accessed at https://www.uniprot.org.
Literature Cited
- Huang, H., McGarvey, P. B., Suzek, B. E., Mazumder, R., Zhang, J., Chen, Y., & Wu, C. H. (2011). A comprehensive protein-centric ID mapping service for molecular data integration. Bioinformatics , 27, 1190–1191. doi: 10.1093/bioinformatics/btr101
- Ladunga, I. (2017). Finding homologs in amino acid sequences using network BLAST searches. Current Protocols in Bioinformatics , 59(1), 3.4.1–3.4.24. doi: 10.1002/cpbi.34
- Simossis, V., Kleinjung, J., & Heringa, J. (2003). An overview of multiple sequence alignment. Current Protocols in Bioinformatics , 3(1), 3.7.1–3.7.26. doi: 10.1002/0471250953.bi0307s03
- Suzek, B. E., Wang, Y., Huang, H., McGarvey, P. B., Wu, C. H., & UniProt Consortium. (2015). UniRef clusters: A comprehensive and scalable alternative for improving sequence similarity searches. Bioinformatics , 31(6), 926–932. doi: 10.1093/bioinformatics/btu739
- UniProt Consortium. (2023). UniProt: The universal protein knowledgebase in 2023. Nucleic Acids Research , 51(D1), D523–D531. doi: 10.1093/nar/gkac1052
Citing Literature
Number of times cited according to CrossRef: 29
- Eman Fikry, Raha Orfali, Nora Tawfeek, Shagufta Perveen, Safina Ghafar, Maher M. El-Domiaty, Azza M. El-Shafae, Unveiling the Bioactive Efficacy of Cupressus sempervirens ‘Stricta’ Essential Oil: Composition, In Vitro Activities, and In Silico Analyses, Pharmaceuticals, 10.3390/ph17081019, 17 , 8, (1019), (2024).
- Jovito Cesar Santos-Álvarez, Juan Manuel Velázquez-Enríquez, Rafael Baltiérrez-Hoyos, Evaluation of the Molecular Mechanism of Chlorogenic Acid in the Treatment of Pulmonary Arterial Hypertension Based on Analysis Network Pharmacology and Molecular Docking, Journal of Vascular Diseases, 10.3390/jvd3010002, 3 , 1, (11-33), (2024).
- Helena Lucena-Padros, Nereida Bravo-Gil, Cristina Tous, Elena Rojano, Pedro Seoane-Zonjic, Raquel María Fernández, Juan A. G. Ranea, Guillermo Antiñolo, Salud Borrego, Bioinformatics Prediction for Network-Based Integrative Multi-Omics Expression Data Analysis in Hirschsprung Disease, Biomolecules, 10.3390/biom14020164, 14 , 2, (164), (2024).
- Jia Yang, Yu-Hong Jiang, Xin Zhou, Jia-Qi Yao, Yang-Yang Wang, Jian-Qin Liu, Peng-Cheng Zhang, Wen-Fu Tang, Zhi Li, Material basis and molecular mechanisms of Chaihuang Qingyi Huoxue Granule in the treatment of acute pancreatitis based on network pharmacology and molecular docking-based strategy, Frontiers in Immunology, 10.3389/fimmu.2024.1353695, 15 , (2024).
- Jianwei Li, Lianwei Sun, Lingbo Liu, Ziyu Li, MIFAM-DTI: a drug-target interactions predicting model based on multi-source information fusion and attention mechanism, Frontiers in Genetics, 10.3389/fgene.2024.1381997, 15 , (2024).
- Kaare Lund Rasmussen, Bodil Bundgaard Rasmussen, Thomas Delbey, Ilaria Bonaduce, Frank Kjeldsen, Vladimir Gorshkov, Analyses of the brown stain on the Parthenon Centaur head in Denmark, Heritage Science, 10.1186/s40494-023-01126-9, 12 , 1, (2024).
- Hannah L. Morgan, Nader Eid, Nadine Holmes, Sonal Henson, Victoria Wright, Clare Coveney, Catherine Winder, Donna M. O’Neil, Warwick B. Dunn, David J. Boocock, Adam J. Watkins, Paternal undernutrition and overnutrition modify semen composition and preimplantation embryo developmental kinetics in mice, BMC Biology, 10.1186/s12915-024-01992-0, 22 , 1, (2024).
- Mahmood Kalemati, Mojtaba Zamani Emani, Somayyeh Koohi, DCGAN-DTA: Predicting drug-target binding affinity with deep convolutional generative adversarial networks, BMC Genomics, 10.1186/s12864-024-10326-x, 25 , 1, (2024).
- Jacob B. Geri, William Pao, Elucidating the Cell Surfaceome to Accelerate Cancer Drug Development, Cancer Discovery, 10.1158/2159-8290.CD-24-0088, 14 , 4, (639-642), (2024).
- Lavanya Rumandla, Mounika Badineni, Ramesh Malikanti, Rajender Vadija, Kiran Kumar Mustyala, Vasavi Malkhed, Virtual Screening Technique to Identify Inhibitors of Mycobacterium tuberculosis Rv3032 Protein Involved in MGLP Biosynthesis, Russian Journal of Bioorganic Chemistry, 10.1134/S1068162024030300, 50 , 3, (1067-1081), (2024).
- Léo C. Caulat, Aurélie Lotoux, Maria C. Martins, Nicolas Kint, Cyril Anjou, Miguel Teixeira, Filipe Folgosa, Claire Morvan, Isabelle Martin-Verstraete, Physiological role and complex regulation of O 2 -reducing enzymes in the obligate anaerobe Clostridioides difficile , mBio, 10.1128/mbio.01591-24, 15 , 10, (2024).
- Rémy Luthringer, Morgane Raphalen, Carla Guerra, Sébastien Colin, Claudia Martinho, Min Zheng, Masakazu Hoshino, Yacine Badis, Agnieszka P. Lipinska, Fabian B. Haas, Josué Barrera-Redondo, Vikram Alva, Susana M. Coelho, Repeated co-option of HMG-box genes for sex determination in brown algae and animals, Science, 10.1126/science.adk5466, 383 , 6689, (2024).
- Daniel W A Buchan, Lewis Moffat, Andy Lau, Shaun M Kandathil, David T Jones, Deep learning for the PSIPRED Protein Analysis Workbench, Nucleic Acids Research, 10.1093/nar/gkae328, 52 , W1, (W287-W293), (2024).
- Haiping Zhang, Hongjie Fan, Jixia Wang, Tao Hou, Konda Mani Saravanan, Wei Xia, Hei Wun Kan, Junxin Li, John Z H Zhang, Xinmiao Liang, Yang Chen, Revolutionizing GPCR–ligand predictions: DeepGPCR with experimental validation for high-precision drug discovery, Briefings in Bioinformatics, 10.1093/bib/bbae281, 25 , 4, (2024).
- Ru Wang, Yongjian Luo, Zheng Lan, Daoshou Qiu, Insights into structure, codon usage, repeats, and RNA editing of the complete mitochondrial genome of Perilla frutescens (Lamiaceae), Scientific Reports, 10.1038/s41598-024-64509-3, 14 , 1, (2024).
- Yuqin Wu, Ashish Foollee, Andrea Y. Chan, Susanne Hille, Jana Hauke, Matthew P. Challis, Jared L. Johnson, Tomer M. Yaron, Victoria Mynard, Okka H. Aung, Maria Almira S. Cleofe, Cheng Huang, Terry C. C. Lim Kam Sian, Mohammad Rahbari, Suchira Gallage, Mathias Heikenwalder, Lewis C. Cantley, Ralf B. Schittenhelm, Luke E. Formosa, Greg C. Smith, Jürgen G. Okun, Oliver J. Müller, Patricia M. Rusu, Adam J. Rose, Phosphoproteomics-directed manipulation reveals SEC22B as a hepatocellular signaling node governing metabolic actions of glucagon, Nature Communications, 10.1038/s41467-024-52703-w, 15 , 1, (2024).
- Yashan Yang, Qianqian Shao, Mingcheng Guo, Lin Han, Xinyue Zhao, Aohan Wang, Xiangyun Li, Bo Wang, Ji-An Pan, Zhenguo Chen, Andrei Fokine, Lei Sun, Qianglin Fang, Capsid structure of bacteriophage ΦKZ provides insights into assembly and stabilization of jumbo phages, Nature Communications, 10.1038/s41467-024-50811-1, 15 , 1, (2024).
- Zhen-Ru Zhou, Fen Liu, Shan Li, Chang-Zhi Dong, Lei Zhang, A fungal P450 enzyme from Fusarium equiseti HG18 with 7β-hydroxylase activity in biosynthesis of ursodeoxycholic acid, The Journal of Steroid Biochemistry and Molecular Biology, 10.1016/j.jsbmb.2024.106507, 240 , (106507), (2024).
- Jerine Peter Simon, Shouliang Dong, In-silico screening of missense nsSNPs in Delta-opioid receptor protein and their restoring tendency on MCRT interaction; focusing on dynamic nature, International Journal of Biological Macromolecules, 10.1016/j.ijbiomac.2024.133710, 275 , (133710), (2024).
- Hale İnci Öztürk, Aysun Oraç, Harvesting bioactive peptides from sustainable protein sources: Unveiling technological and functional properties through in silico analyses, Food and Humanity, 10.1016/j.foohum.2024.100294, 2 , (100294), (2024).
- Aleksandra Karolak, Konstancja Urbaniak, Andrii Monastyrskyi, Derek R. Duckett, Sergio Branciamore, Paul A. Stewart, Structure-independent machine-learning predictions of the CDK12 interactome, Biophysical Journal, 10.1016/j.bpj.2024.05.017, 123 , 17, (2910-2920), (2024).
- Mengmeng Wei, Jingdian Liu, Suoming Wang, Xiyong Wang, Haisuang Liu, Qing Ma, Jiancheng Wang, Wei Shi, Genetic Diversity and Phylogenetic Analysis of Zygophyllum loczyi in Northwest China’s Deserts Based on the Resequencing of the Genome, Genes, 10.3390/genes14122152, 14 , 12, (2152), (2023).
- Guillermo Mateos, Adrián Martínez-Bonilla, José M. Martínez, Ricardo Amils, Vitamin B12 Auxotrophy in Isolates from the Deep Subsurface of the Iberian Pyrite Belt, Genes, 10.3390/genes14071339, 14 , 7, (1339), (2023).
- Eman Fikry, Raha Orfali, Shaimaa S. El-Sayed, Shagufta Perveen, Safina Ghafar, Azza M. El-Shafae, Maher M. El-Domiaty, Nora Tawfeek, Potential Hepatoprotective Effects of Chamaecyparis lawsoniana against Methotrexate-Induced Liver Injury: Integrated Phytochemical Profiling, Target Network Analysis, and Experimental Validation, Antioxidants, 10.3390/antiox12122118, 12 , 12, (2118), (2023).
- Silvia Villaró-Cos, Tomas Lafarga, Online tools to support teaching and training activities in chemical engineering: enzymatic proteolysis, Frontiers in Education, 10.3389/feduc.2023.1290287, 8 , (2023).
- Stephanie E Mohr, Ah-Ram Kim, Yanhui Hu, Norbert Perrimon, Finding information about uncharacterized Drosophila melanogaster genes , GENETICS, 10.1093/genetics/iyad187, 225 , 4, (2023).
- Dao Feng Xiang, Maggie Xu, Manas K. Ghosh, Frank M. Raushel, Metabolic Pathways for the Biosynthesis of Heptoses Used in the Construction of Capsular Polysaccharides in the Human Pathogen Campylobacter jejuni , Biochemistry, 10.1021/acs.biochem.3c00390, 62 , 21, (3145-3158), (2023).
- Lu-Dan Zhang, Ling-Yu Song, Ming-Jin Dai, Jin-Yu Liu, Jing Li, Chao-Qun Xu, Ze-Jun Guo, Shi-Wei Song, Jing-Wen Liu, Xue-Yi Zhu, Hai-Lei Zheng, Inventory of cadmium-transporter genes in the root of mangrove plant Avicennia marina under cadmium stress, Journal of Hazardous Materials, 10.1016/j.jhazmat.2023.132321, 459 , (132321), (2023).
- Yung‐Chen Sun, Tsung‐Lun Hsieh, Chia‐I Lin, Wan‐Yu Shao, Yu‐Hao Lin, Jie‐rong Huang, A Few Charged Residues in Galectin‐3′s Folded and Disordered Regions Regulate Phase Separation, Advanced Science, 10.1002/advs.202402570.

