Structure and Stability of Ago2 MID-Nucleotide Complexes: All-in-One (Drop) His6-SUMO Tag Removal, Nucleotide Binding, and Crystal Growth
Li Lei, Li Lei, Joel M. Harp, Joel M. Harp, John C. Chaput, John C. Chaput, Kelly Wassarman, Kelly Wassarman, Mark K. Schlegel, Mark K. Schlegel, Muthiah Manoharan, Muthiah Manoharan, Martin Egli, Martin Egli
Abstract
The middle (MID) domain of eukaryotic Argonaute (Ago) proteins and archaeal and bacterial homologues mediates the interaction with the 5′-terminal nucleotide of miRNA and siRNA guide strands. The MID domain of human Ago2 (hAgo2) is comprised of 139 amino acids with a molecular weight of 15.56 kDa. MID adopts a Rossman-like beta1-alpha1-beta2-alpha2-beta3-alpha3-beta4-alpha4 fold with a nucleotide specificity loop between beta3 and alpha3. Multiple crystal structures of nucleotides bound to hAgo2 MID have been reported, whereby complexes were obtained by soaking ligands into crystals of MID domain alone. This protocol describes a simplified one-step approach to grow well-diffracting crystals of hAgo2 MID-nucleotide complexes by mixing purified His6-SUMO-MID fusion protein, Ulp1 protease, and excess nucleotide in the presence of buffer and precipitant. The crystal structures of MID complexes with UMP, UTP and 2′-3′ linked α-L-threofuranosyl thymidine-3′-triphosphate (tTTP) are presented. This article also describes fluorescence-based assays to measure dissociation constants (Kd) of MID-nucleotide interactions for nucleoside 5′-monophosphates and nucleoside 3′,5′-bisphosphates. © 2024 The Authors. Current Protocols published by Wiley Periodicals LLC.
Basic Protocol 1 : Crystallization of Ago2 MID-nucleotide complexes
Basic Protocol 2 : Measurement of dissociation constant Kd between Ago2 MID and nucleotides
INTRODUCTION
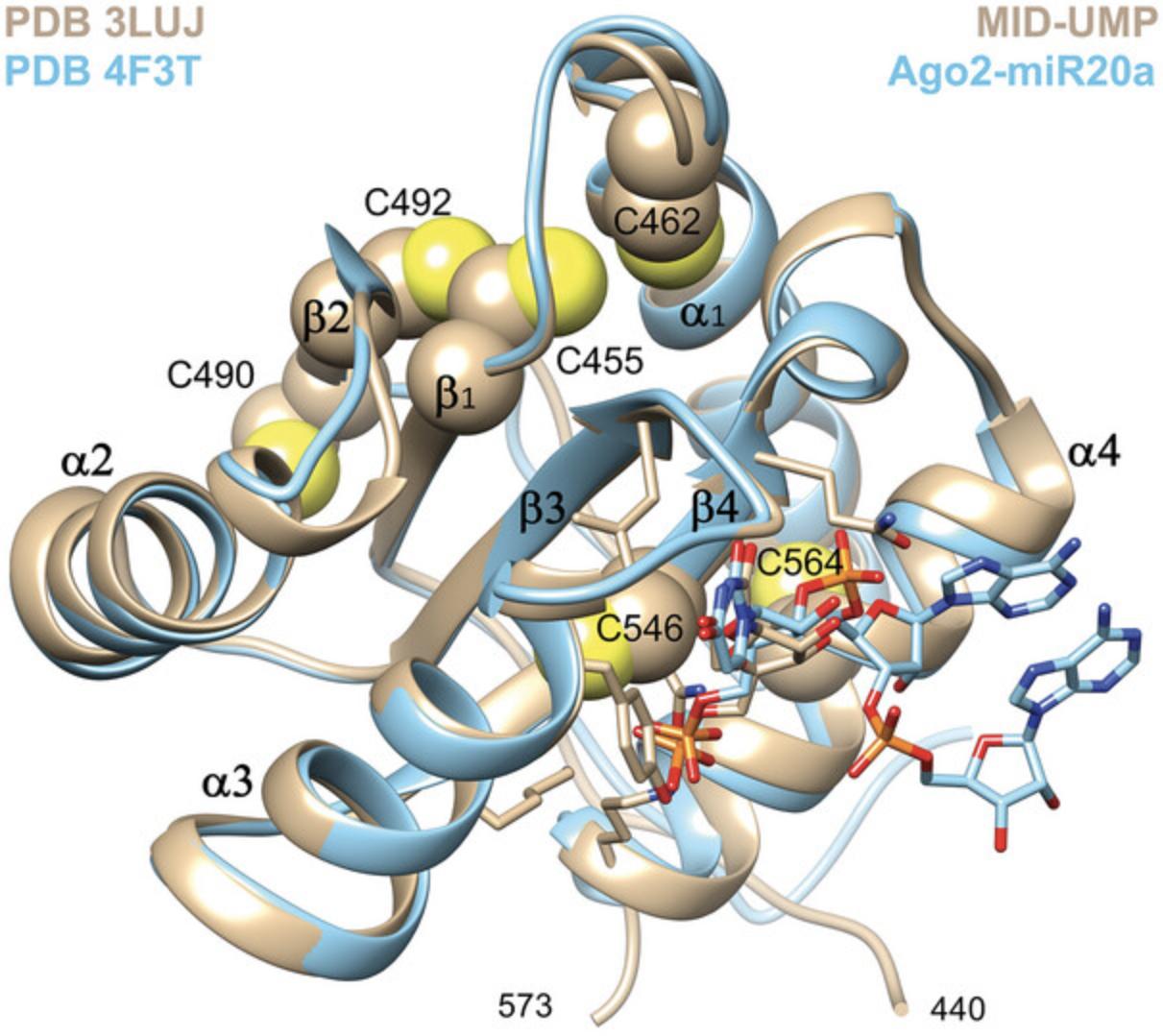
The human Ago2 MID domain is a compact protein of 139 amino acids in length (N439 to L578) that forms the binding site for the 5′-terminal nucleotide of miRNA and siRNA guide strands. Other important Ago2 domains are PIWI that harbors the endonuclease for cleaving the target strand and PAZ that binds to the 3′-terminal dinucleotide overhang of the siRNA guide (Elkayam et al., 2012; Schirle & MacRae, 2012). MID adopts an alpha-beta sandwich fold and contains six cysteines that do not engage in disulfide bridges in crystal structures of full-length Ago2 and MID alone (Fig. 1).
The MID domain forms a relatively shallow depression that harbors the binding site for the 5′-terminal phosphate of the guide strand residue (AS1, Fig. 2). The electrostatic surface potential (ESP) inside and around the depression is strongly positive, thereby promoting not just binding of 5′-monophosphorylated species but also ATP (Fig. 2A). Viewed from the side, the ribose 2′- and 3′-oxygen atoms and a portion of the six-membered ring of adenine are visible, consistent with the shallow nature of the binding pocket that only buries the 5′-phosphate (Fig. 2B). An overlay of the MID domain complex and the structure of the complex between full-length Ago2 and miR-20a demonstrates that the second guide strand residue (AS2) is exposed and not buried inside the pocket (Fig. 2C).
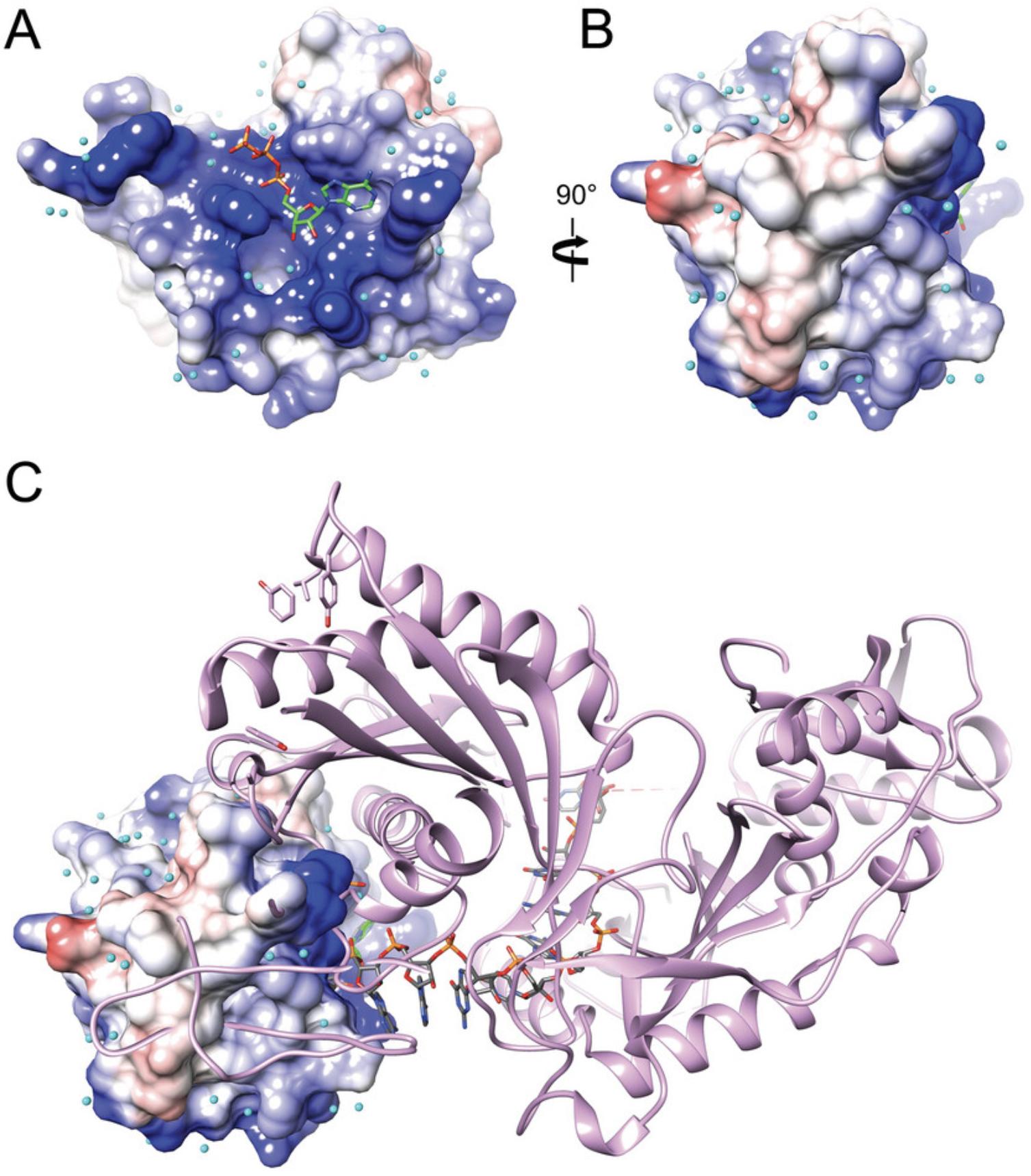
The positive ESP of the MID binding pocket and the area around it also permit binding of m7GpppG dinucleotide and trapping the complex in a crystal (Fig. 3). The 5′-phosphate of m7G is located roughly at the site where the 5′-phosphate of NMPs and the α-phosphate of ATP are bound. However, the additional phosphates then curl around and emerge from the binding pocket such that there is no interaction between G and any MID side chains. The observation that both ATP and m7GpppG produce complex crystals attests to the importance of the electrostatic contribution to the binding affinity. However, no structures of more standard di- or trinucleotides, e.g., 5′-pUpU or 5′-pUpUpU, respectively, bound to MID domain alone have been reported to date.
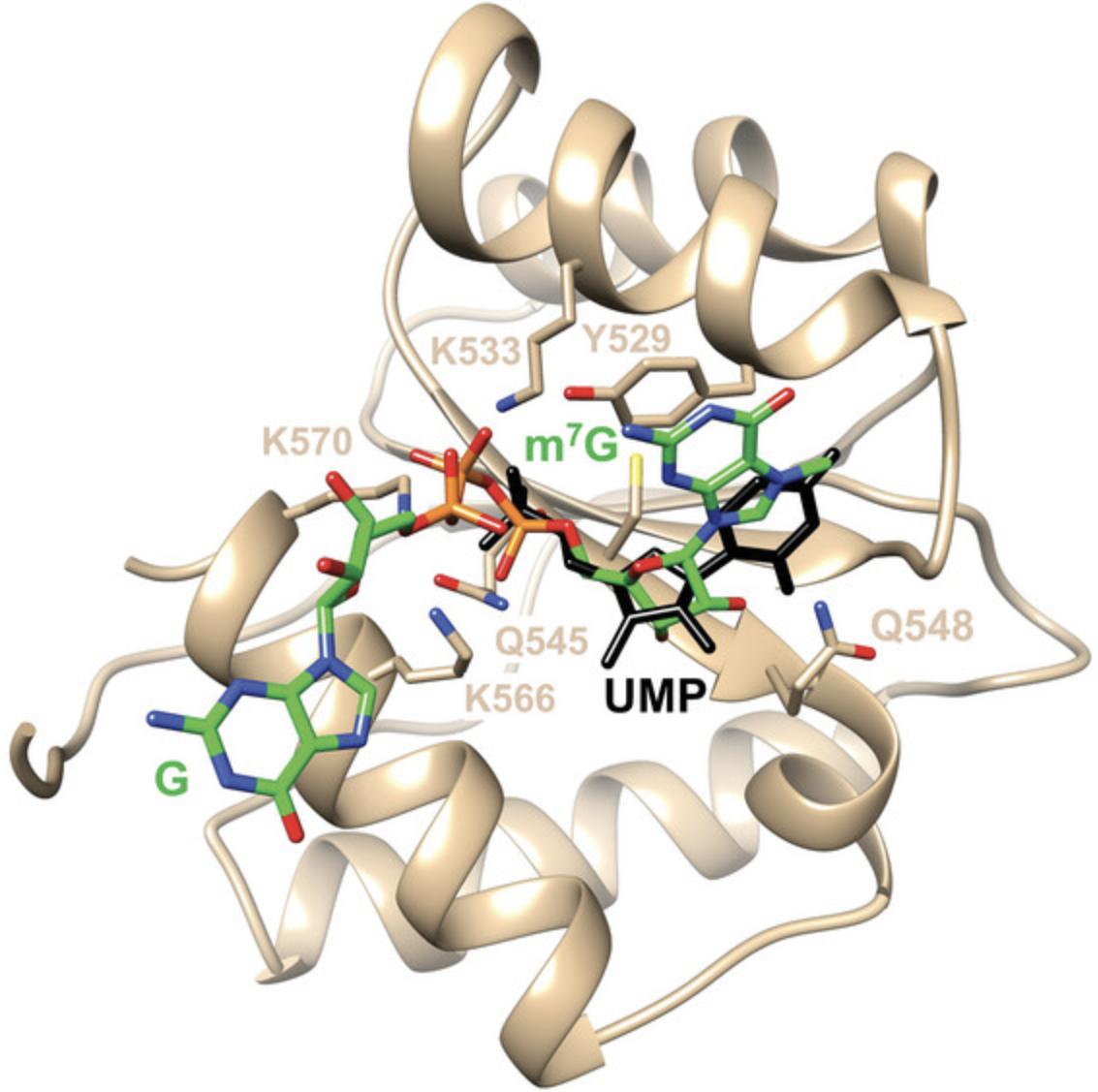
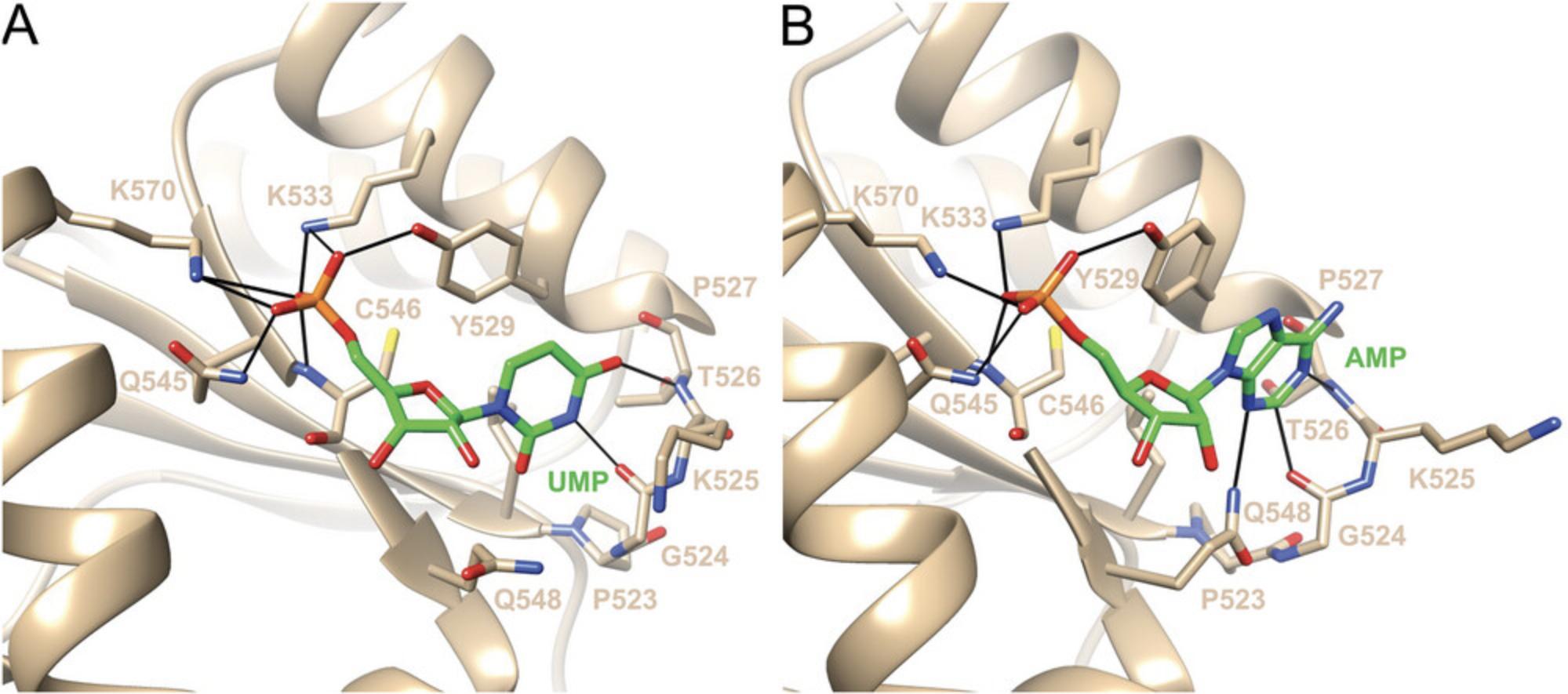
Frank et al. (2010) reported crystal structures of MID complexes with UMP and AMP. MID crystals were grown from solutions containing 10 to 15 mg/ml protein that were mixed 1:1 with a solution containing 0.1 M imidazole (pH 8), 0.2 M NaCl, 0.46 M NaH2PO4, and 1.84 M K2HPO4. Native crystals were then soaked in a drop containing 0.2 M (NH4)2SO4, 0.1 M Na cacodylate buffer (pH 6.5), 15% PEG 8000, 20% glycerol, and 20 mM NMP. However, unlike the complexes with UMP and AMP obtained in this fashion, the occupancy of CMP and GMP in the MID structures was insufficient to place these nucleotides into the binding pocket and refine the structures of the corresponding complexes (Frank et al., 2010). One reason for this limitation may be related to the soaking approach, i.e., CMP and GMP could not displace phosphate bound to MID in crystals that were grown in the presence of high concentrations of phosphate buffer. We also know from stability data that the MID complexes with UMP (Kd = 0.12 mM) or AMP (Kd = 0.26 mM) are more stable than those with CMP (Kd = 3.6 mM) or GMP (Kd = 3.3 mM) (Frank et al., 2010). Both uracil and adenine use their Watson-Crick edges to engage in H-bond interactions with residues from the P523 to P527 loop (G524 and T526, Fig. 4). In the complex with AMP, there is an additional interaction between Q548 and the N3 of adenine. By comparison, neither cytosine nor guanine can establish these interactions; thus, structures of these ‘complexes’ deposited in the Protein Data Bank (PDB) just contain phosphate in the MID binding pocket. It is possible that GMP is in a conformational equilibrium and binds as a mixture of the nucleoside in the syn and anti conformations (see the syn conformation of m7G in Fig. 3).
As pointed out above, the complex with m7GpppG shows that dinucleotides can in principle be trapped in MID crystal structures. Moreover, comparison of MID in complex with UMP and full-length Ago2 in complex with miR-20a reveals that MID residues can potentially form stabilizing interactions not just with AS1 but also AS2: Q548, K550, and N551 with P2, and Q558 and N562 with the nucleobase (Fig. 5). However, the nucleoside portion of AS3 appears outside the reach of any MID side chain, although K566 can potentially interact with the second phosphodiester moiety (P3, Fig. 5). MID also lacks R812 that interacts with the 5′-phosphate directly and via water (P1) and R792 that interacts with P3 in the Ago2 complex; both are part of the PIWI domain.
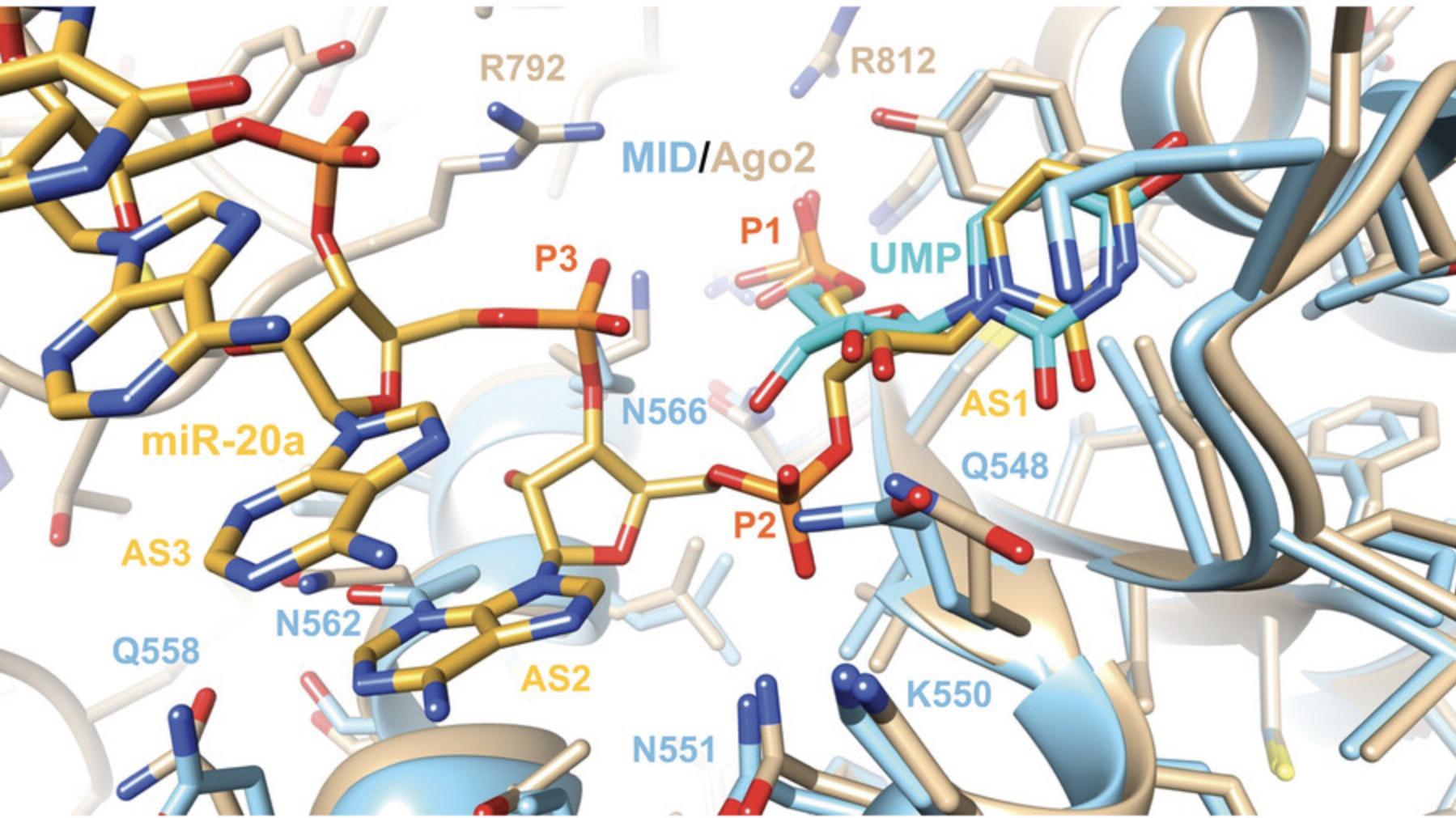
An interesting observation concerns the ribose conformation of UMP in the MID complex and pU (AS1) at the 5′-end of the guide strand in complex with full-length Ago2 (Fig. 5). Thus, the pucker of these residues is in the C2′-endo or South range of the pseudorotation phase cycle. A closer examination of more structures reveals that this is a recurring feature of Ago2 complexes, and that the 5′-terminal nucleotide adopts either a C2′-endo or the neighboring C1′-exo pucker (Egli & Manoharan, 2019). The metabolic stability of the guide strand and stable phosphorylation of its 5′-terminus are critical for siRNA activity. RNA, DNA, 2′-OMe-RNA, and 2′-F RNA are all tolerated at the AS1 position and the (E)-vinylphosphonate moiety precludes dephosphorylation and rephosphorylation of the guide strand (Elkayam et al., 2017). Alternative modifications at the 5′-end of the guide strand continue to be evaluated.
In the recently published crystal structures of MID in complex with either 8-Br-AMP or 6-mCEPh-purine (Shinohara et al., 2021), the conformations of the ribose do not match those in the complexes with UMP and AMP or the puckers of AS1 in the structures of miRNA or siRNAs with full-length Ago2, i.e., C2′-endo or C1′-exo (Elkayam et al., 2012; Elkayam et al., 2017; Frank et al., 2010; Schirle & MacRae, 2012). The pucker of the ribose in 8-Br-AMP is C4′-exo , possibly due to the bromine at the C8 position. The pucker of the ribose in 6-mCEPh-purine is C3′-endo and possibly the result of additional interactions of the modified base with residues of the abovementioned P523 to P527 loop (Fig. 4).
To potentially determine structures of new 5′-guide strand modifications as well as di- and trinucleotides bound to MID, we developed an alternative approach for growing crystals of complexes that precludes removal of the MID fusion partner (His6-SUMO) prior to crystallization by relying on in situ proteolysis. Also, our procedure skips the soaking step of MID-phosphate crystals by equilibrating an all-in-one crystallization drop containing His6-SUMO-MID, Ulp1 protease, nucleotide, buffer, and precipitant against the well solution (Basic Protocol 1). In addition, we present a fluorescence-based assay to measure the dissociation constant Kd o f MID-nucleotide complexes (Basic Protocol 2).
Basic Protocol 1: CRYSTALLIZATION OF Ago2 MID-NUCLEOTIDE COMPLEXES
This protocol describes the crystallization of Ago2 MID domain complexes with nucleotides from a mixture of His6-SUMO-MID fusion protein, Ulp1 protease, nucleotide and crystallization buffer and precipitant. DNA encoding residues 439-578 of hAgo2 MID was cloned into the pSMT3 vector (Mossessova & Lima, 2000) at the BamHI and NotI sites; the vector contains an N-terminal, Ulp1 cleavable His6-SUMO fusion (Frank, 2011; Frank et al., 2010). The plasmid was a gift from Prof. Bhushan Nagar (McGill University, Montreal, Quebec, Canada). His6-SUMO-MID fusion protein was expressed in BL21(DE)3 cells, and purified by Ni-affinity chromatography, ion-exchange chromatography (Q and S columns), and Superdex-75 size exclusion chromatography. Briefly, after expression suspended cells were lysed and sonicated with Buffer A consisting of 50 mM Tris·HCl pH 7.8, 20% glycerol, 800 mM NaCl, 3 mM beta-mercaptoethanol (BME), 0.5 mM tris(2-carboxyethyl)phosphine (TCEP), and 10 mM imidazole. Following centrifugation, cat. no. 355884, the supernatant was loaded onto the Ni column, washed with Buffer A, and then eluted with 400 mM imidazole. After diluting NaCl to a 50 to 80 mM concentration, the sample was passed over a Q column and then loaded onto an S column. Using a gradient of Buffer B [50 mM Tris·HCl pH 7.8, 1 mM dithiothreitol (DTT) and up to 1M NaCl], the protein was eluted when NaCl reached a concentration of 400 mM. Finally, the sample was run on a Superdex-75 size exclusion column with Buffer C consisting of 25 mM Tris·HCl pH 8.0, 150 mM NaCl, and 3 mM DTT. The purified His6-SUMO-MID protein was concentrated to 15 mg/ml. The protein was >95% pure based on SDS-PAGE. The identity of the MID domain fusion protein was established by tryptic digestion in combination with MS analysis.
Materials
-
15 mg/ml His6-SUMO-MID protein (see above)
-
UMP (Sigma, CAS no. 3387-36-8)
-
UTP (Millipore Sigma, CAS no. 19817-92-6)
-
tTTP (2′-3′ linked α-L-threofuranosyl thymidine-3′-triphosphate) (Zhang et al., 2013)
-
H2O, nuclease free (Promega, cat. no. P119E-C)
-
Ulp1 protease (His-tagged SUMO protease, Vanderbilt Center for Structural Biology https://static.yanyin.tech/literature/current_protocol/10.1002/cpz1.1088/attachments/Purif.Ulp1.pdf; or ThermoFisher Scientific, cat. no. 12588018)
-
Index screen, 96-condition crystallization kit (Hampton Research, cat. no. HR2-144)
-
Paratone-N oil, Parabar 10312 (Hampton Research, cat. no. HR2-643)
-
Columns for His6-SUMO-MID protein purification (see above):
- Q sepharose fast flow (GE Healthcare, cat. no. 17-0510-01)
- SP sepharose fast flow (GE Healthcare, cat. no. 17-0720-01)
- Superdex 75 10/30 GL (GE Healthcare, cat. no. 17-5174-01)
-
1.5-ml Eppendorf tubes
-
Ice
-
MRC 2 crystallization plates (Hampton Research, cat. no. HR3-082)
-
Microscope
-
75-μm MicroMount (MiTeGen, cat. no. M2-)
-
Liquid N2
-
Crystal imager (Formulatrix imager with UV fluorescence detector)
1.In an Eppendorf tube mix His6-SUMO-MID protein with nucleotide ligand, e.g., UMP, UTP, tTTP (in nuclease-free water), in a 1:700 to 1:1000 molar ratio and store the tube on ice for 2 hr.
2.Add Ulp1 protease in a 1:1000 (w/w) ratio of Ulp1:His6-SUMO-MID.
3.Using the Index crystallization screen, set up sitting drops in a 96-well MRC 2 crystallization plate by mixing 200 nl of the His6-SUMO-MID/nucleotide/Ulp1 solution from step 2 with 200 nl of the individual screen condition. Equilibrate each drop against 80 μl of the particular screen solution.
4.Check the plate once every 24 hr using a microscope; crystals typically appeared after 3 to 7 days (Fig. 6) with Index screen conditions 20 (0.1 M HEPES pH 7.5, 1.4 M sodium citrate tribasic dihydrate), 21 (1.8 M ammonium citrate tribasic pH 7.0), and 23 (2.1 M DL-malic acid pH 7.0).

5.Mount crystal in a 75-μm MiTeGen MicroMount following cryo-protection with Paratone-N taking care to remove excess oil before plunging in liquid nitrogen. Store in liquid nitrogen prior to X-ray diffraction data collection (Table 1).
| Parameter | MID-UMP complex | MID-UTP complex | MID-tTTP complex |
|---|---|---|---|
| Space group | P1 | P1 | P1 |
| Complexes/unit cell | 3 | 3 | 3 |
|
Unit cell constants a, b, c (Å); α, β, γ (°) |
a = 39.608 b = 46.339 c = 64.920 α = 87.58 β = 74.68 γ = 85.87 |
a = 40.106 b = 46.511 c = 65.240 α = 87.78 β = 74.53 γ = 85.41 |
a = 37.549 b = 46.899 c = 66.209 α = 86.74 β = 74.02 γ = 84.76 |
| Data collection | In-house | In-house | In-house |
| X-ray instrument | Bruker MetalJet | Bruker MetalJet | Bruker MetalJet |
| Wavelength (Å) | 1.342 | 1.342 | 1.342 |
| Resolution (Å) |
21.58-1.595 (1.652-1.595) |
20.53-1.78 (1.844-1.78) |
20.5-1.90 (1.93-1.90) |
| Total reflections | 856,515 (59,080) | 337,798 (25,492) | 186,059 (4801) |
| Unique reflections | 118,253 (11,241) | 86,315 (8459) | 41,796 (1612) |
| Multiplicity | 7.2 (5.0) | 3.9 (3.0) | 4.5 (3.0) |
| Completeness (%) | 99.17 (94.79) | 98.88 (97.57) | 99.04 (99.21) |
| Mean I/σ (I) | 15.20 (1.98) | 5.72 (0.68) | 7.48 (0.11) |
| R-merge | 0.082 (0.835) | 0.120 (2.263) | 0.061 (7.834) |
| R-meas | 0.088 (0.937) | 0.138 (2.729) | 0.068 (9.535) |
| R-pim | 0.031 (0.418) | 0.066 (1.501) | 0.029 (5.354) |
| CC1/2 | 0.999 (0.587) | 0.994 (0.273) | 0.999 (0.247) |
| CC | 1 (0.86) | 0.999 (0.655) | 1 (0.629) |
| Refl. used in ref. | 117,364 (11,235) | 85,817 (8448) | 33,833 (1385) |
| Refl. used for R-free | 5,665 (509) | 4221 (429) | 1710 (49) |
| R-work | 0.1645 (0.2667) | 0.1902 (0.3733) | 0.2504 (0.4082) |
| R-free | 0.1892 (0.2885) | 0.2346 (0.4262) | 0.2885 (0.4300) |
| CC (work) | 0.972 (0.824) | 0.970 (0.589) | – |
| CC (free) | 0.959 (0.758) | 0.961 (0.579) | – |
| No. non-H atoms | 3607 | 3530 | 3268 |
| No. macromol. atoms | 3132 | 3084 | 3118 |
| No. ligand atoms | 0 | 87 | 84 |
| No. of solvent mols. | 475 | 359 | 68 |
| Protein residues | 394 | 396 | 400 |
| R.m.s.d. bonds (Å) | 0.011 | 0.009 | 0.023 |
| R.m.s.d. angles (°) | 0.95 | 0.88 | 1.74 |
| Ramachandran favored (%) | 99.74 | 99.74 | 99.49 |
| Ramachandran allowed (%) | 0.26 | 0.26 | 0.25 |
| Ramachandran outliers (%) | 0.00 | 0.00 | 0.25 |
| Rotamer outliers (%) | 0.00 | 0.29 | 2.89 |
| Clashscore | 3.77 | 4.06 | 8.22 |
| Avg. B (Å2) | 19.0 | 28.2 | 36.9 |
| Avg. B macrom. (Å2) | 17.5 | 26.6 | 34.5 |
| Avg. B ligand (Å2) | − | 56.1 | 112.9 |
| Avg. B solvent (Å2) | 29.13 | 35.24 | 49.5 |
| No. of TLS grps. | 0 | 3 | 3 |
| PDB ID code | 9BF2 | 9BF0 | 9BEZ |
- a
Numbers in parentheses refer to the outermost shell.
X-ray diffraction data collection
6.Diffraction data were collected using the D8 Venture (Bruker AXS, Madison, WI) system in the Biomolecular Crystallography Facility at the Vanderbilt University Center for Structural Biology. The system includes an Excillum D2+ MetalJet X-ray source operated at 250W with Helios MX optics providing Ga Kα radiation at 1.3418 Å wavelength. The crystal was mounted on a kappa axis goniometer and maintained at 100K using an Oxford Cryosystems Cryostream 800 cryostat. The detector was a PHOTON III C14 charge-integrating pixel array detector. Data collection was performed in shutterless mode. Data were reduced using Proteum3 software (Bruker AXS, Madison, WI).
Phasing and model refinement
7.Diffraction data were phased by molecular replacement using PDB code 3LUJ. All model building and refinements were done using PHENIX (Afonine et al., 2012; Liebschner et al., 2019; Williams et al., 2018). Final checking was done using PDB-REDO (Joosten et al., 2014), which included TLS refinement. Models were placed in the unit cell using ACHESYM (Kowiel et al., 2014). Manual fitting and examination of the structures was done using COOT (Casañal et al., 2020; Emsley et al., 2010).
Basic Protocol 2: MEASUREMENT OF DISSOCIATION CONSTANT Kd BETWEEN Ago2 MID AND NUCLEOTIDES
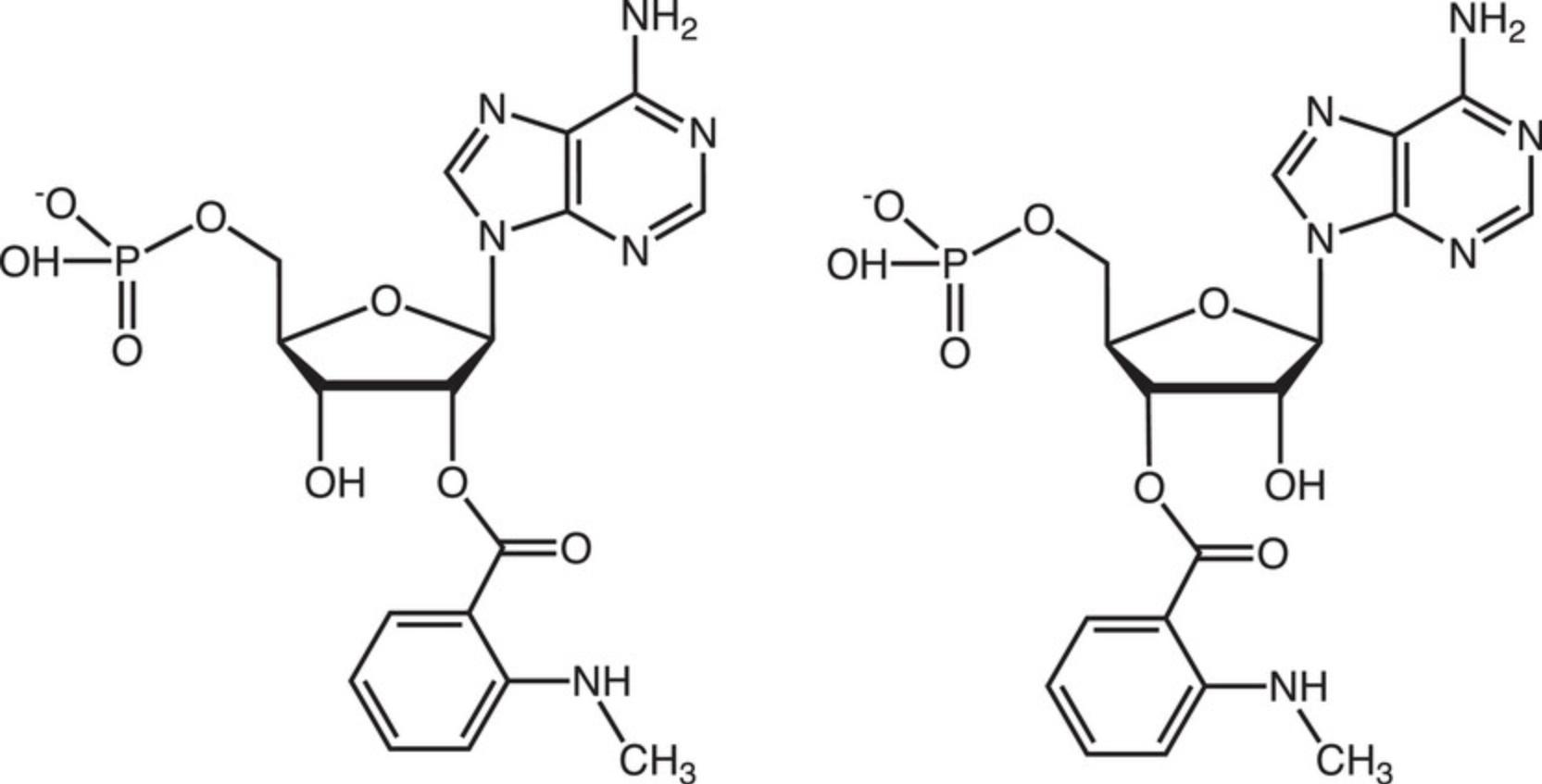
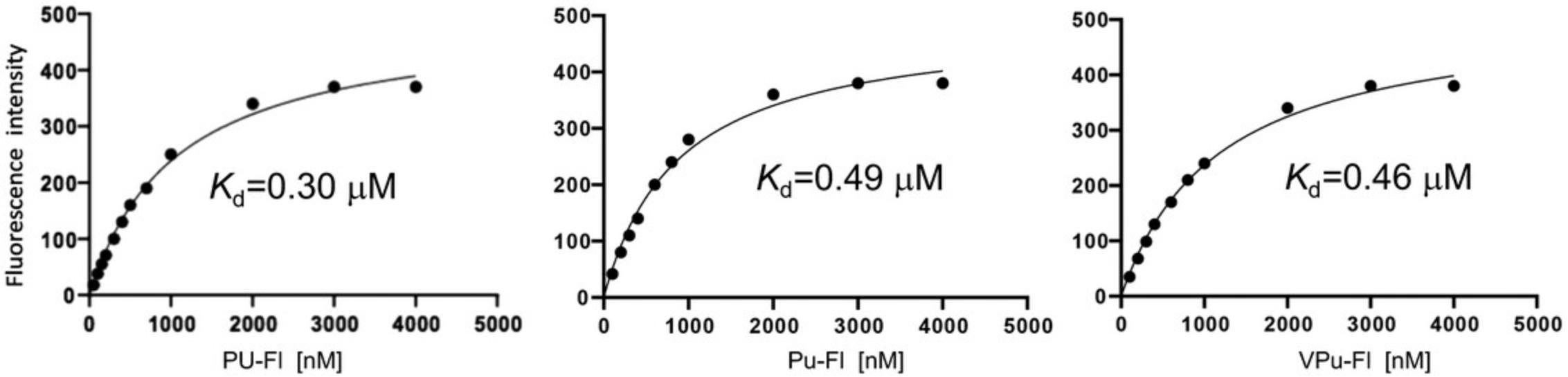
This protocol describes the determination of Kd values for complexes between hAgo2 MID and nucleoside 3′,5′-bisphosphates, i.e., uridine-3′,5′-bisphosphate, 2′-O -methyl-uridine-3′,5′-bisphosphate, and 2′-O -methyl-uridine-3′-phosphate, 5′-(E)-vinylphosphonate, that carry a 3′-terminal 1-(5-azidopentyl)-3-(fluorescein-5-yl)thiourea moiety (abbreviated here as PU-Fl, Pu-Fl and VPu-Fl, respectively, Fig. 7), and 2′/3′-(N-methyl-anthraniloyl)-adenosine-5′-monophosphate (Mant-AMP, Fig. 8). As described above, the Ago2 MID domain was expressed as a fusion protein with an N-terminal cleavable His6-SUMO tag and purified by Nickel affinity chromatography. The tag was cleaved off with Ulp1 protease and MID further purified using ion-exchange chromatography (Q and S columns), and Superdex-75 size exclusion chromatography.
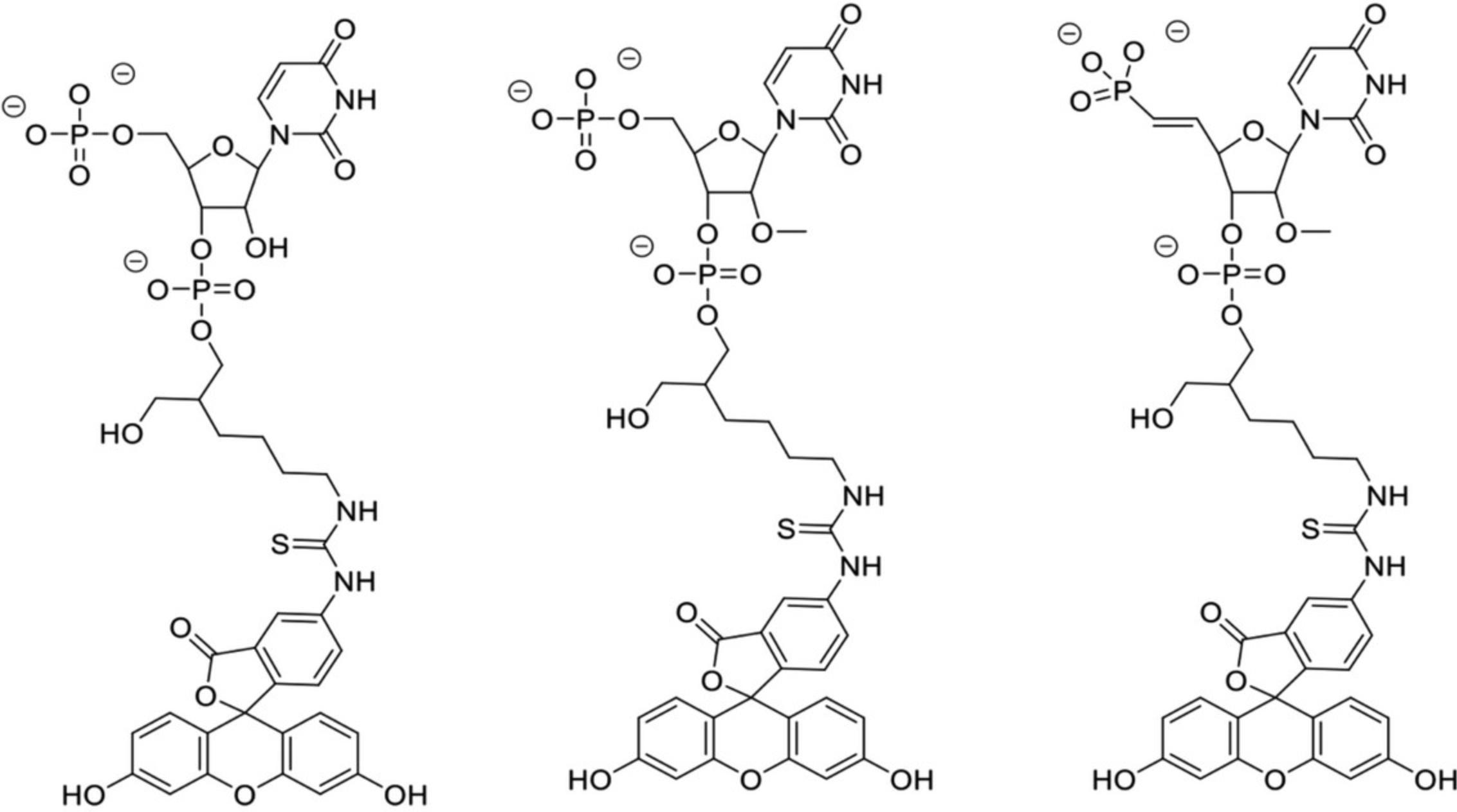
Nucleoside 3′,5′-bisphosphates were prepared on a K&A oligonucleotide synthesizer using commercially available (RNA-U, 2′-OMe-U) or in-house prepared (VPu) phosphoramidites. Coupling of each phosphoramidite was performed on a commercial fluorescein support (Glen Research) using standard protocols for 2-cyanoethyl phosphoramidites. After synthesis, cleavage from the support and deprotection of any protecting groups was accomplished with either aqueous methylamine (40% aq., PU-Fl) for 90 min at 45°C or concentrated aqueous ammonium hydroxide with 5% diethylamine (Pu-Fl and VPu-Fl) for 24 hr at 30°C. Subsequent removal of the 2′-O -silyl protecting group in PU-Fl was accomplished using triethylamine trihydrofluoride for 90 min at 40°C. Crude bisphosphates were purified using strong anion exchange over a TSKGel SuperQ-5PW column and a linear gradient of aq. triethylammonium bicarbonate buffer (pH = 8.5, buffer B) in water (buffer A). Appropriate fractions were pooled, lyophilized to dryness, and identities of the desired products confirmed via LC-MS. The purities of the three compounds as per ion exchange (IEX) chromatography and ion pair reversed-phase (IP-RP) chromatography (IEX %/IP-RP %) were 93/96 (PU-Fl), 94/95 (Pu-Fl), and 94/99 (VPu-Fl).
We employed steady-state techniques to monitor changes in the emission intensity of fluorescein (λex 493 nm, λem 517 nm) and Mant (λex 355 nm, λem 448 nm). Increasing amounts of nucleotide were added to a 1 μM solution of MID and changes in fluorescence emission intensity monitored with a Fluorolog ISA Jobin Yvon-Spex spectrometer. The temperature of the cell compartment was kept at 20°C with a refrigerated circulating water bath. Measurements were corrected for dilution and blank solutions. The fluorescence emission data served as input for GraphPad Prism (version 5.00 for Mac; Graph-Pad Software, La Jolla, CA; https://www.graphpad.com/). We selected a nonlinear regression (curve fit) in combination with the equation for “one site-specific binding” to compute Kd values for MID-nucleotide complexes.
Materials
-
1 μM Ago2 MID domain (see above)
-
PU-Fl, Pu-Fl, and VPu-Fl nucleotides (Alnylam Pharmaceuticals, synthesized)
-
H2O, nuclease free (Promega, cat. no. P119E-C)
-
Mant-AMP (Jena Bioscience, cat. no. NU-236S)
-
Binding buffer (10 mM HEPES pH 7.4, 150 mM NaCl, 3 mM EDTA, 5mM TCEP) (Current Protocols, 2006)
-
1-ml cuvette
-
Micropipette
-
Fluorometer (Fluorolog ISA Jobin Yvon-Spex spectrometer)
-
Computer with analysis software (e.g., GraphPad Prism)
1.In a cuvette that contains 1 ml of a 1 μM solution of MID protein, by adding 1 μl of a nucleotide solution (in nuclease-free water) in a stepwise fashion, increase the concentration of the latter from 0 to 100 nM, 200 nM, 300 nM, 400 nM, 600 nM, 800 nM, 1 μM, 2 μM, 3 μM, 4 μM.
2.Prior to and after adding nucleotide solution, measure the fluorescence emission intensity at 517 nm (fluorescein-labeled nucleotides PU-Fl, Pu-Fl, and VPu-Fl) or 448 nm (Mant-AMP).
3.Apply corrections for blank (buffer only) solution and dilution to the emission intensity data.
4.Use individual wavelength-emission intensity pairs as input for Graph-Pad and compute a curve fit using the nonlinear regression option in combination with the equation for “one site-specific binding” to extract the Kd value.
COMMENTARY
Background Information
Oligonucleotide therapeutics currently approved for diverse indications (Egli & Manoharan, 2023) and candidates in clinical trials at various stages (Crooke et al., 2018; Moumné et al., 2022; Shen & Corey, 2018) all feature extensive chemical modification. Modifications afford metabolic stability, facilitate cellular uptake, allow targeting of specific organs and tissues, and can mediate favorable interactions with proteins and enzymes. Five of the six siRNA therapeutics approved by the FDA since 2018, GIVLAARI, OXLUMO, LEQVIO, AMVUTTRA (Egli & Manoharan, 2023), and RIVFLOZA (Liu et al., 2022; Syed, 2023; https://www.accessdata.fda.gov/drugsatfda_docs/label/2023/215842s000lbl.pdf) are composed of fully 2′-fluoro/2′-O -methyl-modified RNA guide (antisense, AS) and passenger (sense, S) strands. The passenger strands of these siRNAs also feature GalNAc conjugates for efficient delivery to hepatocytes that is mediated by asialoglycoprotein receptor (ASGPR) (Egli & Manoharan, 2023; Kandasamy et al., 2023). In addition, phosphate groups at the 5′-ends of both guide and passenger strand and the 3′-end of the guide strand are replaced by phosphorothioates for enhanced protection against exonuclease attack (Eckstein, 1985; Egli & Manoharan, 2023; Hyjek-Skladanowska et al., 2020; Jahns et al., 2022).
siRNA strands are comprised of between 21 and 23 nucleotides. The varying nature of interactions with the Ago2 endonuclease of the RNA-induced silencing complex (RISC) along the guide and passenger siRNAs presents abundant opportunities for regiospecific chemical modification (Egli & Manoharan, 2019). Thus, modifying the 5′-terminal ends of siRNAs can enhance the discrimination between the guide and passenger strands to reduce off-target effects (Datta et al., 2023; Kumar et al., 2019). The favorable properties of the 2′-fluoro and 2′-O -methyl ribonucleotide analogs as modifiers of siRNAs were recognized early on (Manoharan et al., 2011). The 2′-fluoro modification is tolerated at all positions but does not alone afford sufficient protection against nuclease degradation. 2′-O -methyl ribonucleotides are more protective by comparison and are sterically tolerated at most locations in the two strands. The second nucleotide of the guide strand (AS2) constitutes a notable exception in this regard as modification there is limited to either 2′-deoxy or 2′-fluoro residues (Egli & Manoharan, 2019). A sharp turn in the seed region (AS2 to AS8) between nucleotides 6 and 7 seen in crystal structures of Ago2 complexes (Elkayam et al., 2012; Schirle & MacRae, 2012) led us to deploy modifications that destabilize seed pairing and preorganize the guide strand for this wrinkle, resulting in improvements in loading, mitigation of off-target effects, and better clinical safety (Egli et al., 2023; Guenther et al., 2022; Matsuda et al., 2023; Schlegel et al., 2022). Lipophilic modification in the form of a 2′-O -hexadecyl conjugation at position 16 of the sense strand was shown to result in potent and durable silencing in the central nervous system (CNS) (Brown et al., 2022).
Unlike the siRNA passenger strand, the siRNA guide strand carries a 5′-phosphate group. This phosphate group is captured by the Ago2 MID domain (Figs. 1 and 4), locks the guide strand into position during the loading process and makes a key contribution to the discrimination between the guide and passenger siRNAs. However, the 5′-phosphate group is not metabolically stable and can be removed, thereby affecting loading efficiency and the stability of the RISC complex. Replacing the phosphate with an (E)-vinylphosphonate (VP) moiety protects the 5′-end of the guide (Elkayam et al., 2017). The VP modification is an excellent electrostatic and steric mimic of the native phosphate, the latter thanks to the antiperiplanar conformation of the β torsion angle of the AS1 nucleotide in crystal structures (Fig. 4) (Egli & Manoharan, 2019). The VP modification is not yet a feature of currently approved siRNA therapeutics (Egli & Manoharan, 2023); RIVFLOZA contains a methylenephosphonate moiety at the 5′-terminus of the guide strand (Liu et al., 2022; Syed, 2023). Another noteworthy distinction of the AS1 nucleotide is its C2′-endo or C1′-exo B form–like sugar pucker (Figs. 4 and 5) (Egli & Manoharan, 2019). In addition to the negative charge, this conformational constraint is an important point to consider when designing modifications to replace the AS1 nucleotide. Besides molecular modeling, experimental structure determination is an important tool in the design of new siRNA modifications and is required to visualize the conformational properties of new analogs and their interactions with MID and other Ago2 domains (Egli & Manoharan, 2019). For example, the crystal structure of 6-mCEPh-purine bound to MID exhibits a North C3′-endo pucker (Shinohara et al., 2021) and therefore does not mimic the sugar conformation of the parent AS1 ribonucleotides (Figs. 4 and 5).
Crystals of MID complexes with NMPs are obtained by soaking crystals of the domain with phosphate trapped at the binding site in solutions of the nucleotide to displace phosphate (Frank, 2011; Frank et al., 2010). Such crystals are of space group P 1 with three molecules per asymmetric unit (Frank et al., 2010). Concentrating the MID protein following removal of the His6-SUMO tag to levels typically used for crystallization, i.e., several mg/ml, particularly under low salt conditions, is hampered by MID frequently precipitating. To circumvent this issue, we explored the use of the MID fusion protein without removal of the tag for crystallization of nucleotide complexes in the presence of Ulp1 protease in a single step (Basic Protocol 1). As an alternative to the published protocol that used solution NMR (Frank et al., 2010), we relied on fluorescence spectrometry in combination with suitably modified nucleotides to measure dissociation constants of MID-nucleotide complexes (Basic Protocol 2).
Critical Parameters and Troubleshooting
The solubility of the His6-SUMO-MID protein is much higher than that of the MID domain alone and solutions of the fusion protein can be concentrated to 15 mg/ml without risking precipitation under relatively low salt conditions and stored at −80°C for several months. Instead of removing the His6-SUMO tag in a separate step and purifying the MID domain chromatographically, our protocol directly uses the purified fusion protein for crystallization setups. In situ proteolysis to free the MID domain in the presence of nucleotide and crystallization buffer and precipitant requires optimization of the relative concentrations of MID, Ulp1 protease and nucleotide. A 1000:1 ratio of His6-SUMO-MID:Ulp1 produced the best crystals.
In a previously developed protocol to crystallize membrane-bound cytochrome P450 enzymes in the presence of protease to shorten the N-terminal tail that had been modified with a leader sequence of reduced hydrophobicity to boost expression, we tested 12 different proteases and found that trypsin and subtilisin produced viable crystals of N-terminally truncated enzymes (Lei & Egli, 2016). The ratios of P450:protease tested ranged between 100:1 to 2000:1 and optimal crystals of zebrafish P450 17A1 (with subtilisin) and P450 17A2 (with trypsin) were obtained using a P450:protease ratio of 1000:1.
Unlike the Ulp1 protease that is used in much lower concentrations than the His6-SUMO-MID fusion protein (consistent with the catalytic activity of the former), the nucleotide needs to be present in the crystallization drop at much higher concentrations than the MID fusion protein. Diffraction-quality crystals were obtained with a nucleotide:His6-SUMO-MID molar ratio of between 700:1 to 1000:1. Because the stock solution of the fusion protein is ∼1 mM, the nucleotide concentration needs to reach 1 M or higher. This can pose problems with nucleotides that are not as soluble as UMP or UTP, e.g., 5′-pUpU-3′ dimers or 5′-pUpUpU-3′ trimers. An excess of nucleotide will ensure that MID protein without the His6-SUMO tag encounters enough ligand molecules to drive crystal formation of complex rather than causing MID to precipitate. Unlike with soaks of MID-phosphate crystals in solutions of the nucleotide ligand (Frank et al., 2010), the all-in-one drop crystallization protocol described here and using His6-SUMO-MID fusion protein may not require displacement of intrinsically bound phosphate from MID to produce crystals of the MID-nucleotide complex. However, we cannot rule out that His6-SUMO-MID protein contains some bound phosphate following expression and purification.
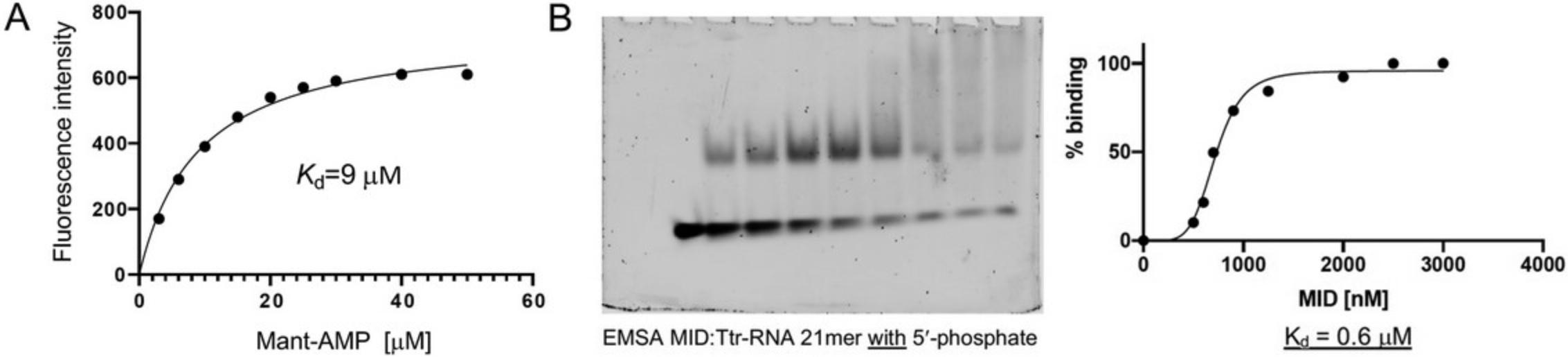
The need for very high concentrations of the nucleotide ligand may limit the chances to grow crystals of dimers and trimers bound to MID using our approach. The presence of at least one 5′-terminal phosphate group (NMP) in the ligand is another requirement for successful crystallization of MID complex. For example, we could not grow crystals of MID with uridine (U) or 5′-UpUpU. Clearly, the phosphodiester groups of 5′-UpUpU cannot serve as substitutes for a terminal phosphate, unlike the first phosphodiester (bold font) in m7Gp ppG (Fig. 3). However, in the latter compound that phosphate is accompanied by two more phosphates that glue together the complex. The crystal structure of the complex with m7GpppG may raise hopes that MID can be crystallized with di- and trinucleotides, such as 5′-pUpU and 5′-pUpUpU, respectively. In other words, because G of m7GpppG is ordered in the electron density despite the absence of interactions with MID residues or those from neighboring MID domains or dinucleotides in the crystal lattice (Fig. 3; Frank et al., 2011), it might be possible to visualize U2 or U2 and U3 in structures of complexes with 5′-pUpU or 5′-pUpUpU, respectively. However, we could not produce viable crystals of such complexes. This may in part be due to limitations with reaching high enough concentrations of dimers and trimers as alluded to above. Also, the orientation of the ppG moiety in the structure of the complex with m7GpppG (Fig. 3) extends in the opposite direction and away from the locations of AS2 and AS3 in the complex of guide siRNA with Ago2 (Fig. 5). Similarly, we could not grow crystals of MID with 5′-pUp using our protocol and attempts to obtain crystals of a complex with Mant-AMP (Fig. 8) also failed.
Interestingly, the dissociation constants between MID and ligands that feature both 5′- and 3′-phosphate groups, such as PU-Fl, Pu-Fl, and VPu-Fl (Fig. 7), indicate tighter binding compared to the complex between MID and Mant-AMP (see Understanding Results). This suggests that the 3′-phosphate may contribute to binding, perhaps by interacting with Q548, K550, and N551 as seen in the structure of Ago2 bound to miR-20a (Fig. 5). For now, it seems that crystal structures of complexes of the Ago2 MID domain alone can only be obtained with mononucleotides and select compounds like m7GpppG. MID complexes with longer pieces of RNA have all resisted crystallization, which we rationalize with the inability of the domain alone to sufficiently constrain the second and third nucleotide.
Understanding Results
We crystallized complexes of human Ago2 MID with UMP, UTP, and tTTP and determined their structures at resolutions of up to 1.59 Å (Table 1). The structures with UMP and UTP closely resemble those previously reported for the same monophosphate (Fig. 4A) (Frank et al., 2010) and ATP (Fig. 2A and B) (Frank et al., 2011), respectively. The space group in all cases is P 1 as reported for other MID complexes. Our Basic Protocol 1 using in situ proteolysis of the His6-SUMO-MID fusion protein to grow crystals of MID-nucleotide complexes does not affect crystal form and packing. Using pure fusion protein in combination with Ulp1 protease in the crystallization drop circumvents the need for extra purification steps following cleavage of the His6-SUMO tag. Further advantages of our protocol are the reduced risk of purified MID precipitating at the higher concentrations needed for crystallization and having to compete with bound phosphate when soaking MID crystals in solutions of nucleotides. However, the all-in-one-drop approach requires high concentrations of the nucleotide ligand. This requirement does not pose problems with UMP, UDP, UTP, tTTP, etc., but less soluble ligands could prove more challenging in this regard. As indicated above, dimers and trimers did not produce viable crystals of MID complexes. We obtained crystals of the complex with nicotinamide adenine dinucleotide phosphate (NADPH) that exhibited a different morphology than MID-UMP complex crystals (Fig. 6). However, these crystals only diffracted to low resolution and were not suitable for structure determination.
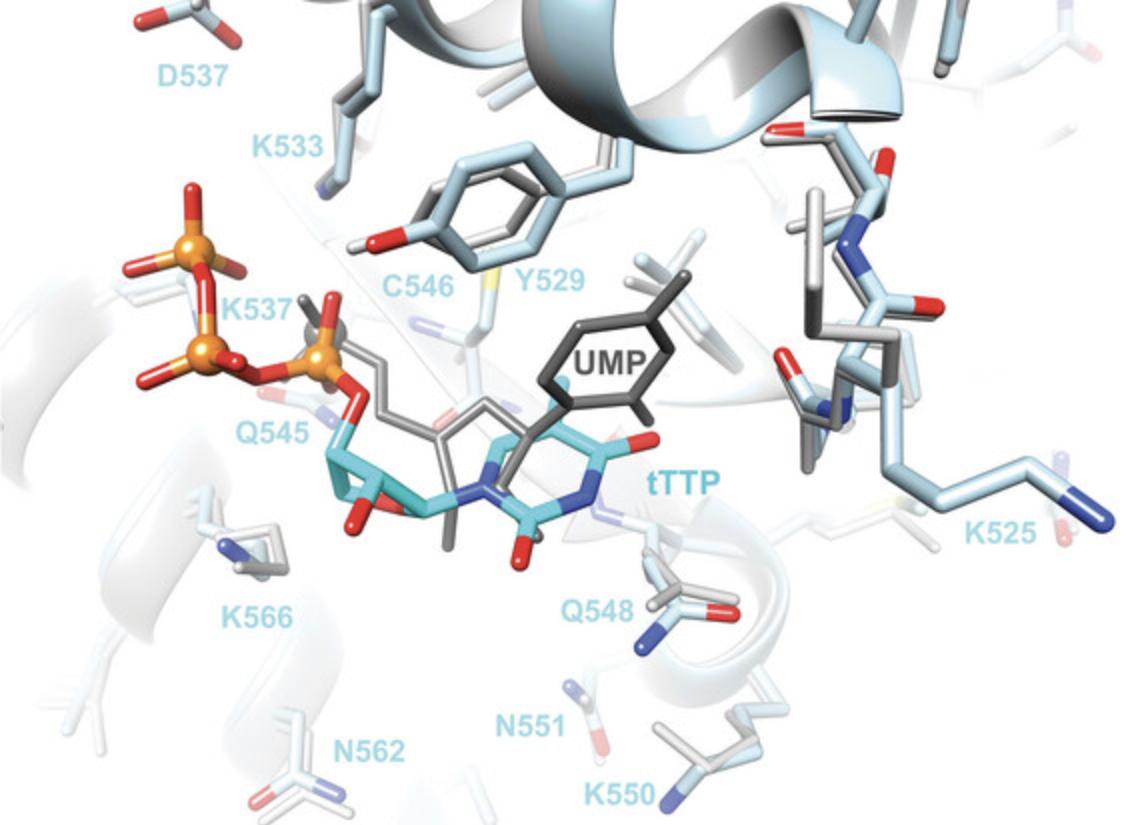
The structure of the MID complex with tTTP reveals an orientation of the TNA triphosphate that differs from that of UMP (Fig. 9). The positions of the α-phosphate groups are separated by ∼1.3Å, whereby the UMP phosphate sits deeper inside the MID binding pocket. Their sugar conformations differ in that tTTP adopts the expected C4′-exo pucker whereas UMP has a C2′-endo pucker. In addition, the tTTP base does not stack on T529; instead, the nucleoside has shifted somewhat relative to uridine and thymine engages in a partial stacking interaction with the side chain of Q548 (Fig. 9). The latter residue usually forms a H-bond with the first bridging phosphate of the guide strand along with N551 (Fig. 5). The two MID residues contribute to stabilizing a sharp turn in the RNA strand between AS1 and AS3. The absence of the second nucleotide together with PIWI residues of Ago2 that interact with the 5′-terminal region of the guide strand expose limitations of MID complexes alone with regards to studying the conformation of that region of modified guides and interactions with Ago2. Thus, a computational model of miR-20a with a 5′-tTMP that adopts a C4′-exo pucker in place of UMP and bound to Ago2 demonstrates that TNA can stack on Y529 and mimic the tight turn between AS1 and AS3 in principle (Matsuda et al., 2023).
Kd values for MID-nucleotide complexes based on concentration-dependent fluorescence emission intensity measurements as described in Basic Protocol 2 are consistent with sub-μM affinities for PU-Fl, Pu-Fl, and VPu-Fl (Figs. 7 and 10). The dissociation constants of between 0.30 and 0.49 μM and using a 1 μM solution of MID indicate a significantly tighter binding than that reported for UMP based on 1H-15N HSQC NMR and using 0.1 to 0.25 mM MID (Kd = 120 μM; Frank et al., 2010). To examine whether this difference could at least in part be related to the presence of a 3′-phosphate in the three nucleotides we had tested, we also measured the Kd for the MID complex with Mant-AMP (Fig. 8). Indeed, in the absence of the 3′-phosphate, the stability of the complex is reduced to 9 μM (Fig. 11A). However, this Kd still indicates ca. 30-fold tighter binding of AMP compared to the previously reported stability of the MID-AMP complex using HSQC NMR: Kd = 260 μM (Frank et al., 2010). The tighter binding as seen with the bisphosphates (Fig. 10) is supported by an independent electrophoretic mobility shift assay (EMSA) for a complex between Ago2 MID and an RNA 21-mer (Fig. 11B) using a previously published protocol (Seo et al., 2019).
Time Considerations
Expression and purification of the His6-SUMO-MID protein can be accomplished in 2 days. The preparation and purification of the fluorescently labeled nucleotides PU-Fl, Pu-Fl, and VPu-Fl takes ∼5 days. Collecting the fluorescence emission data for a MID-nucleotide complex including solution preparation and titration requires ∼2 hr, and data transfer and computing the Kd value using Graph-Pad Prism takes 1 hr.
Acknowledgments
This work was supported by Alnylam Pharmaceuticals, Vanderbilt University, and US National Science Foundation Award MCB-2224897. We thank Dr. Filipp Frank for helpful discussions.
Author Contributions
Li Lei : Conceptualization; formal analysis; investigation; methodology; resources; writing—original draft. Joel M. Harp : Data curation; formal analysis; writing—original draft. John C. Chaput : Resources. Kelly Wassarman : Methodology; resources; writing—original draft. Mark K. Schlegel : Methodology; resources; writing—original draft. Muthiah Manoharan : Resources; supervision. Martin Egli : Conceptualization; formal analysis; methodology; resources; supervision; validation; writing—original draft; writing—review and editing.
Conflict of Interest
The authors declare no conflict of interest.
Open Research
Data Availability Statement
PDB ID codes: 9BF2 (MID-UMP complex), 9BF0 (MID-UTP complex), and 9BEZ (MID-tTTP complex).
Literature Cited
- Afonine, P. V., Grosse-Kunstleve, R. W., Echols, N., Headd, J. J., Moriarty, N. W., Mustyakimov, M., Terwilliger, T. C., Urzhumtsev, A., Zwart, P. H., & Adams, P. D. (2012). Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallographica. Section D, Structural Biology , 68, 352–367. https://doi.org/10.1107/S0907444912001308
- Berman, H. M., Westbrook, J., Feng, Z., Gilliland, G., Bhat, T. N., Weissig, H., Shindyalov, I. N., & Bourne, P. E. (2000). The protein data bank. Nucleic Acids Research , 28, 235–242. https://doi.org/10.1093/nar/28.1.235
- Brown, K. M., Nair, J. K., Janas, M. M., Anglero-Rodriguez, Y. I., Dang, L. T. H., Peng, H., Theile, C. S., Castellanos-Rizaldos, E., Brown, C., Foster, D., Kurz, J., Allen, J., Maganti, R., Li, J., Matsuda, S., Stricos, M., Chickering, T., Jung, M., Wassarman, K., … Jadhav, V. (2022). Expanding RNAi therapeutics to extrahepatic tissues with lipophilic conjugates. Nature Biotechnology , 40, 1500–1508. https://doi.org/10.1038/s41587-022-01334-x
- Casañal, A., Lohkamp, B., & Emsley, P. (2020). Current developments in Coot for macromolecular model building of electron cryo-microscopy and crystallographic data. Protein Science , 29, 1069–1078. https://doi.org/10.1002/pro.3791
- Crooke, S. T., Witztum, J. L., Bennett, C. F., & Baker, B. F. (2018). RNA-targeted therapeutics. Cell Metabolism , 27, 714–739. https://doi.org/10.1016/j.cmet.2018.03.004
- Current Protocols (2006). Commonly Used Reagents. Current Protocols in Microbiology , 00, A.2A.1–A.2A.15. https://doi.org/10.1002/9780471729259.mca02as00
- Datta, D., Theile, C., Wassarman, K., Qin, J., Racie, T., Schmidt, K., Jiang, Y., Sigel, R., Janas, M., Egli, M., & Manoharan, M. (2023). Rational optimization of siRNA to ensure strand bias in the interaction with the RNA-induced silencing complex. Chemical Communications (Cambridge, England) , 59, 6347–6350. https://doi.org/10.1039/D3CC01143G
- Eckstein, F. (1985). Nucleoside phosphorothioates. Annual Review of Biochemistry , 54, 367–402. https://doi.org/10.1146/annurev.bi.54.070185.002055
- Egli, M., & Manoharan, M. (2019). Re-engineering RNA molecules into therapeutic agents. Accounts of Chemical Research , 52, 1036–1047. https://doi.org/10.1021/acs.accounts.8b00650
- Egli, M., & Manoharan, M. (2023). Chemistry, structure and function of approved oligonucleotide therapeutics. Nucleic Acids Research , 51, 2529–2573. https://doi.org/10.1093/nar/gkad067
- Egli, M., Schlegel, M. K., & Manoharan, M. (2023). Acyclic (S)-glycol nucleic acid (S -GNA) modification of siRNAs improves the safety of RNAi therapeutics while maintaining potency. RNA , 29, 402–414. https://doi.org/10.1261/rna.079526.122
- Elkayam, E., Kuhn, C. D., Tocilj, A., Haase, A. D., Greene, E. M., Hannon, G. J., & Joshua-Tor, L. (2012). The structure of human Argonaute-2 in complex with miR-20a. Cell , 150, 100–110. https://doi.org/10.1016/j.cell.2012.05.017
- Elkayam, E., Parmar, R., Brown, C. R., Willoughby, J. L. S., Theile, C. S., Manoharan, M., & Joshua-Tor, L. (2017). siRNA carrying an (E)-vinylphosphonate moiety at the 5′ end of the guide strand augments gene silencing by enhanced binding to human Argonaute-2. Nucleic Acids Research , 45, 3528–3536. https://doi.org/10.1093/nar/gkw1171
- Emsley, P., Lohkamp, B., Scott, W., & Cowtan, K. (2010). Features and development of coot. Acta Crystallographica. Section D, Structural Biology , 66, 486–501. https://doi.org/10.1107/S0907444910007493
- Frank, F., Sonenberg, N., & Nagar, B. (2010). Structural basis for 5'-nucleotide base-specific recognition of guide RNA by human AGO2. Nature , 465, 818–822. https://doi.org/10.1038/nature09039
- Frank, F. (2011). Structural and functional characterization of Argonaute MID domain [unpublished Ph.D. thesis]. Department of Biochemistry, McGill University, Montreal, Quebec, Canada.
- Frank, F., Fabian, M. R., Stepinski, J., Jemielity, J., Darzynkiewicz, E., Sonenberg, N., & Nagar, B. (2011). Structural analysis of 5'-mRNA-cap interactions with the human AGO2 MID domain. EMBO Reports , 12, 415–420. https://doi.org/10.1038/embor.2011.48
- Guenther, D. C., Mori, S., Matsuda, S., Gilbert, J. A., Willoughby, J. L. S., Hyde, S., Bisbe, A., Jiang, Y., Agarwal, S., Madaoui, M., Janas, M. M., Charisse, K., Maier, M. A., Egli, M., & Manoharan, M. (2022). Role of a "magic" methyl: 2'-deoxy-2'-α-F-2'-β-C -methyl pyrimidine nucleotides modulate RNA interference activity through synergy with 5'-phosphate mimics and mitigation of off-target effects. Journal of the American Chemical Society , 144, 14517–14534. https://doi.org/10.1021/jacs.2c01679
- Hyjek-Skladanowska, M., Vickers, T. A., Napiorkowska, A., Anderson, B. A., Tanowitz, M., Crooke, S. T., Liang, X.-H., Seth, P. P., & Nowotny, M. (2020). Origins of the increased affinity of phosphorothioate-modified therapeutic nucleic acids for proteins. Journal of the American Chemical Society , 142, 7456–7468. https://doi.org/10.1021/jacs.9b13524
- Jahns, H., Taneja, N., Willoughby, J. L. S., Akabane-Nakata, M., Brown, C. R., Nguyen, T., Bisbe, A., Matsuda, S., Hettinger, M., Manoharan, R. M., Rajeev, K. G., Maier, M. A., Zlatev, I., Charisse, K., Egli, M., & Manoharan, M. (2022). Chirality matters: Stereo-defined phosphorothioate linkages at the termini of small interfering RNAs improve pharmacology in vivo. Nucleic Acids Research , 50, 1221–1240. https://doi.org/10.1093/nar/gkab544
- Joosten, R. P., Long, F., Murshudov, G. N., & Perrakis, A. (2014). The PDB_REDO server for macromolecular structure model optimization. IUCrJ , 1, 213–220. https://doi.org/10.1107/S2052252514009324
- Kandasamy, P., Mori, S., Matsuda, S., Erande, N., Datta, D., Willoughby, J. L. S., Taneja, N., O'Shea, J., Bisbe, A., Manoharan, R. M., Yucius, K., Nguyen, T., Indrakanti, R., Gupta, S., Gilbert, J. A., Racie, T., Chan, A., Liu, J., Hutabarat, R., … Manoharan, M. (2023). Metabolically stable anomeric linkages containing GalNAc-siRNA conjugates: An interplay among ASGPR, glycosidase, and RISC pathways. Journal of Medicinal Chemistry , 66, 2506–2523. https://doi.org/10.1021/acs.jmedchem.2c01337
- Kowiel, M., Jaskolski, M., & Dauter, Z. (2014). ACHESYM: An algorithm and server for standardized placement of macromolecular models in the unit cell. Acta Crystallographica. Section D, Structural Biology , 70, 3290–3298. https://doi.org/10.1107/S1399004714024572
- Kumar, P., Parmar, R. G., Brown, C. R., Willoughby, J. L. S., Foster, D. J., Babu, I. R., Schofield, S., Jadhav, V., Charisse, K., Nair, J. K., Rajeev, K. G., Maier, M. A., Egli, M., & Manoharan, M. (2019). 5'-Morpholino modification of the sense strand of an siRNA makes it a more effective passenger. Chemical Communications (Cambridge, England) , 55, 5139–5142. https://doi.org/10.1039/C9CC00977A
- Lei, L., & Egli, M. (2016). In situ proteolysis for crystallization of membrane bound cytochrome P450 17A1 and 17A2 proteins from zebrafish. Current Protocols in Protein Science , 84, 29.16.1–29.16.19. https://doi.org/10.1002/0471140864.ps2916s84
- Liebschner, D., Afonine, P. V., Baker, M. L., Bunkóczi, G., Chen, V. B., Croll, T. I., Hintze, B., Hung, L. W., Jain, S., McCoy, A. J., Moriarty, N. W., Oeffner, R. D., Poon, B. K., Prisant, M. G., Read, R. J., Richardson, J. S., Richardson, D. C., Sammito, M. D., Sobolev, O. V., … Adams, P. D. (2019). Macromolecular structure determination using X-rays, neutrons and electrons: Recent developments in Phenix. Acta Crystallographica. Section D, Structural Biology , 75, 861–877. https://doi.org/10.1107/S2059798319011471
- Liu, A., Zhao, J., Shah, M., Migliorati, J. M., Tawfik, S. M., Bahal, R., Rasmussen, T. P., Manautou, J. E., & Zhong, X. B. (2022). Nedosiran, a candidate siRNA drug for the treatment of primary hyperoxaluria: Design, development, and clinical studies. ACS Pharmacology & Translational Science, 5, 1007–1016. https://doi.org/10.1021/acsptsci.2c00110
- Manoharan, M., Akinc, A., Pandey, R. K., Qin, J., Hadwiger, P., John, M., Mills, K., Charisse, K., Maier, M. A., Nechev, L., Greene, E. M., Pallan, P. S., Rozners, E., Rajeev, K. G., & Egli, M. (2011). Unique gene-silencing and structural properties of 2'-fluoro-modified siRNAs. Angewandte Chemie (International ed. in English) , 50, 2284–2288. https://doi.org/10.1002/anie.201006519
- Matsuda, S., Bala, S., Liao, J. Y., Datta, D., Mikami, A., Woods, L., Harp, J. M., Gilbert, J. A., Bisbe, A., Manoharan, R. M., Kim, M., Theile, C. S., Guenther, D. C., Jiang, Y., Agarwal, S., Maganti, R., Schlegel, M. K., Zlatev, I., Charisse, K., … Manoharan, M. (2023). Shorter Is better: The α-(L)-threofuranosyl nucleic acid modification improves stability, potency, safety, and Ago2 binding and mitigates off-target effects of small interfering RNAs. Journal of the American Chemical Society , 145, 19691–19706. https://doi.org/10.1021/jacs.3c04744
- Mossessova, E., & Lima, C. D. (2000). Ulp1-SUMO crystal structure and genetic analysis reveal conserved interactions and a regulatory element essential for cell growth in yeast. Molecular Cell , 5, 865–876. https://doi.org/10.1016/S1097-2765(00)80326-3
- Moumné, L., Marie, A. C., & Crouvezier, N. (2022). Oligonucleotide therapeutics: From discovery and development to patentability. Pharmaceutics , 14, 260. https://doi.org/10.3390/pharmaceutics14020260
- Pettersen, E. F., Goddard, T. D., Huang, C. C., Couch, G. S., Greenblatt, D. M., Meng, E. C., & Ferrin, T. E. (2004). UCSF Chimera - a visualization system for exploratory research and analysis. Journal of Computational Chemistry , 25, 1605–1612. https://doi.org/10.1002/jcc.20084
- Schirle, N. T., & MacRae, I. J. (2012). The crystal structure of human Argonaute2. Science , 336, 1037–1040. https://doi.org/10.1126/science.1221551
- Schlegel, M. K., Janas, M. M., Jiang, Y., Barry, J. D., Davis, W., Agarwal, S., Berman, D., Brown, C. R., Castoreno, A., LeBlanc, S., Liebow, A., Mayo, T., Milstein, S., Nguyen, T., Shulga-Morskaya, S., Hyde, S., Schofield, S., Szeto, J., Woods, L. B., … Maier, M. A. (2022). From bench to bedside: Improving the clinical safety of GalNAc-siRNA conjugates using seed-pairing destabilization. Nucleic Acids Research , 50, 6656–6670. https://doi.org/10.1093/nar/gkac539
- Seo, M., Lei, L., & Egli, M. (2019). Label-free electrophoretic mobility shift assay (EMSA) for measuring dissociation constants of protein-RNA complexes. Current Protocols in Nucleic Acid Chemistry , 76, e70. https://doi.org/10.1002/cpnc.70
- Shen, X., & Corey, D. R. (2018). Chemistry, mechanism and clinical status of antisense oligonucleotides and duplex RNAs. Nucleic Acids Research , 46, 1584–1600. https://doi.org/10.1093/nar/gkx1239
- Shinohara, F., Oashi, T., Harumoto, T., Nishikawa, T., Takayama, Y., Miyagi, H., Takahashi, Y., Nakajima, T., Sawada, T., Koda, Y., Makino, A., Sato, A., Hamaguchi, K., Suzuki, M., Yamamoto, J., Tamari, Y., & Saito, J. I. (2021). siRNA potency enhancement via chemical modifications of nucleotide bases at the 5'-end of the siRNA guide strand. RNA , 27, 163–173. https://doi.org/10.1261/rna.073783.119
- Syed, Y. Y. (2023). Nedosiran: First approval. Drugs , 83, 1729–1733. https://doi.org/10.1007/s40265-023-01976-4
- Williams, C. J., Hintze, B. J., Headd, J. J., Moriarty, N. W., Chen, V. B., Jain, S., Prisant, M. G., Lewis, S. M., Videau, L. L., Keedy, D. A., Deis, L. N., Arendall, W. B. III., Verma, V., Snoeyink, J. S., Adams, P. D., Lovell, S. C., Richardson, J. S., & Richardson, D. C. (2018). MolProbity: More and better reference data for improved all-atom structure validation. Protein Science , 27, 293–315. https://doi.org/10.1002/pro.3330
- Zhang, S., Yu, H., & Chaput, J. C. (2013). Synthesis of threose nucleic acid (TNA) triphosphates and oligonucleotides by polymerase-mediated primer extension. Current Protocols in Nucleic Acid Chemistry , 52, 4.54.1–4.54.17. https://doi.org/10.1002/0471142700.nc0454s52

