Simple Workflow and Comparison of Media for hPSC-Cardiomyocyte Cryopreservation and Recovery
Sebastian Diecke, Sebastian Diecke, Duncan C. Miller, Duncan C. Miller, Carolin Genehr, Carolin Genehr, Narasimha S. Telugu, Narasimha S. Telugu, Silke Kurths, Silke Kurths
Abstract
Great progress has been made with protocols for the differentiation and functional application of hPSC-cardiomyocytes (hPSC-CMs) in recent years; however, the cryopreservation and recovery of hPSC-CMs still presents challenges and few reports describe in detail the protocols and general workflow. In order to facilitate cryopreservation and recovery of hPSC-CMs for a wide range of applications, we provide detailed information and step-by-step protocols. The protocols are simple and use common reagents. They are comprised of a fast dissociation, cryopreservation using standard equipment, and gentle recovery following thawing. We discuss various features of the protocols, as well as their utilization in the context of common hPSC-CM differentiation and application workflows. Finally, we compare two proprietary and two common in-house formulations of cryopreservation media used for hPSC-CMs, and despite differences in their price and composition find broadly similar recovery rates and cellular function after thawing. © 2019 The Authors.
Basic Protocol 1 : Dissociation and cryopreservation of hPSC-CMs
Basic Protocol 2 : Thawing and recovery of cryogenically frozen hPSC-CMs
INTRODUCTION
The usability of human pluripotent stem cell-derived cardiomyocytes (hPSC-CMs) for applications within regenerative medicine, toxicology screening, and disease modelling, depends heavily on efficient reproducible bioprocesses. One of these bioprocesses is hPSC-CM cryopreservation, as it enables the generation, transportation, and application of large differentiation batches, which simplifies supply chains and should reduce variability and cost (Dunn & Palecek, 2018; Preininger, Singh, & Xu, 2016). Even on a small scale, reliable freezing and thawing of hPSC-CM batches greatly facilitates project management and experimental reproducibility. However, hPSC-CMs appear to be less robust towards freeze-thawing than many other cell types, with reported viable recovery rates varying greatly from 50% to 84% (Burridge, Holmström, & Wu, 2015; Chen et al., 2015; van den Brink et al., 2020; Xu et al., 2011). Over nearly a decade, various options in terms of protocols and reagents for cryopreservation have appeared, and although a few papers provide some detail or investigate the functional effects of cryopreservation on hPSC-CMs, there is not yet a consensus with regards to workflow or best practice. For more in-depth discussion please see Background Information and Understanding Results.
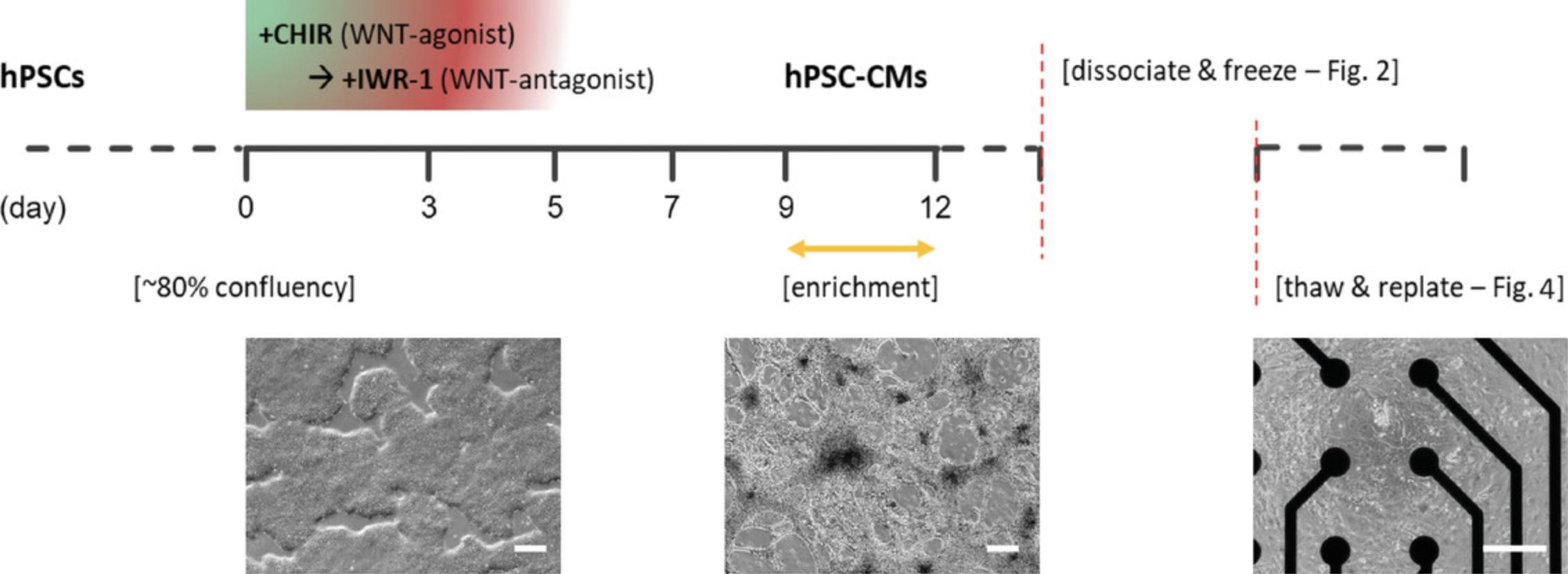
In this article, we outline a simple workflow for the step-by-step dissociation, cryopreservation, and functional recovery of hPSC-CMs that have been differentiated in adherent monolayer culture (Fig. 1). This can be performed without specialized equipment and should be applicable to many hPSC-CM differentiation protocols and downstream applications (see Strategic Planning). We provide analytical data, as well as a comparison of four common hPSC-CM freezing medium (Table 1; see Basic Protocol 2 “Sample data”). The protocols are as follows:
- Dissociation and cryopreservation of hPSC-CMs (Fig. 2)–includes sample data of effect of differentiation day on dissociation and freezing (Fig. 3)
- Thawing and recovery of cryogenically frozen hPSC-CMs (Fig. 4)–includes sample data of functional recovery and comparison of freezing media (Fig. 5)
| Freezing medium | Cost (100 ml)b | Component features | Recommended protocolc | hPSC-CM referencesd |
|---|---|---|---|---|
| CryoStor® CS10 | 360€ | Fully defined, xeno-free, protein-free, cGMP-manufactured | None available | Chen et al. (2015); Gerbin, Yang, Murry, & Coulombe (2015); Liu et al. (2018); Xu et al. (2011) |
| FBS-DMSO 10% | 121€ | NA | None available | Breckwoldt et al. (2017); Burridge et al. (2015) |
| KSR-DMSO 10% | 69€ | Defined except for lipid rich-BSA | None available | van den Brink et al. (2020) |
| STEMdiff™ cardiomyocyte freezing medium | 270€ | Fully defined, xeno-free | Freezing density of 0.5 × 106 CMs/ml. To be used in conjunction with STEMdiff CM dissociation and maintenance kits. Pipette thawed CMs directly into resuspension medium. Viability >70% (assume trypan blue exclusion). | None available |
- a All media listed contain 10% DMSO as cryoprotective agent (CPA); see Reagents and Solutions.
- b List price in Germany as of April 2020.
- c Where manufacturer instructions for hPSC-CMs are available and differ from this article.
- d Selection of primary research articles using indicated media, not including all subsequent derivative articles–see Background Information and Understanding Results for further discussion.
STRATEGIC PLANNING
The simple workflow for cryopreservation and recovery described here is ideally integrated within the wider context of monolayer differentiation, quality control (QC), and functional application of hPSC-CMs. The most commonly used differentiation protocols including the one used in experiments here are based on small molecule inhibition of WNT signaling, which may also include a CM enrichment step (Fig. 1) (Lian et al., 2012; Tohyama et al., 2013). This cryopreservation workflow should work comparably well with hPSC-CMs generated using other similar differentiation protocols or kits (see Background Information). For generating batches of hPSC-CMs for cryopreservation, we would recommend differentiation of 1-4 multi-well plates, usually 6- or 12-well, and find that this can generate up to 2.5 × 108 cells. We and others have found several key parameters affecting hPSC-CM differentiation efficiency and yield. Batch variability in certain reagents, particularly B-27 and CHIR, may have significant impacts on differentiation. For several of our hPSC lines, 6 µM CHIR for the first days of differentiation works well, but across many lines and published protocols the range is 6-12 µM, and an optimal concentration often has to be determined individually for hPSC lines and specific protocols. Cell density at the start of differentiation is also very important, with ∼80% being regarded as ideal (Allen_Institute, 2018; Burridge et al., 2014; D'Antonio-Chronowska et al., 2019; Kempf et al., 2016).
Before freezing hPSC-CMs, it is important to roughly estimate the expected cell number before dissociation and have an adequate amount of medium and number of labelled cryovials and cryostorage boxes prepared before starting. As a guideline, we generate an average of 4.01 ±1.44 × 107 cells per 6-well plate (n = 5, ±SD). Similarly before thawing and re-plating hPSC-CMs, it is important to calculate how many cells are required and in what format for their downstream application and prepare sufficient media and coated plates beforehand. See Basic Protocol 2 and Table 2 for more information on coating and plating densities.
| Plate | cm2/well | Coating/well | Cells/well (× 105) | Medium/well |
|---|---|---|---|---|
| 6-well | 9.5 | 1.5 ml | 15-30 | 3 ml |
| 12-well | 3.8 | 0.6 ml | 5-10 | 1 ml |
| 24-well | 1.9 | 0.3 ml | 2.5-5.0 | 0.5 ml |
| 48-well | 1 | 0.2 ml | 1.0-2.0 | 0.3 ml |
| 96-well | 0.32 | 0.06 ml | 0.3-0.5 | 0.1 ml |
| 384-well | 0.056 | 0.03 ml | 0.04-0.08 | 0.05 ml |
| 8-well chamber slide | 0.78 | 0.2 ml | 0.8-1.2 | 0.25 ml |
| 48-well MEAb | — | 0.008 ml droplet | 0.3-0.5 | 0.3 ml |
- a Prepare Geltrex-coated plates on the day or up to 2 weeks in advance according to manufacturer's instructions (Thermo Fisher Scientific, 2013). Suggested densities should generate a fairly complete monolayer of cells 5 days following recovery from thawing (Fig. 5B). A range is indicated to allow for optimization, the lower figure being most appropriate following optimally recovered cryogenically frozen or freshly dissociated hPSC-CMs.
- b Coating of wells and subsequent plating of hPSC-CMs in Axion Biosystems Cytoview or E-Stim+ plates must be done the same day, and may only cover the electrodes in the center of the well, hence both coating and cell plating must be done as successive 8 µl droplets, with maintenance medium topped up after 1.5-2 hr. Geltrex at half the normal dilution was found to work well. For more information refer to https://www.axionbiosystems.com/resources.
In order to determine the cell number before freezing and after thawing, cell counting including live/dead exclusion must be performed. There are various approaches and equipment available for this, which will tend to generate slightly different numbers relative to each other. It is advisable to use just one method across experiments so that numbers remain consistent. In this article we have used an automated cell counter and trypan blue exclusion; however, manual counting using a hemacytometer will work equally well, notwithstanding variability between individual users.
Basic Protocol 1: DISSOCIATION AND CRYOPRESERVATION OF hPSC-CMs
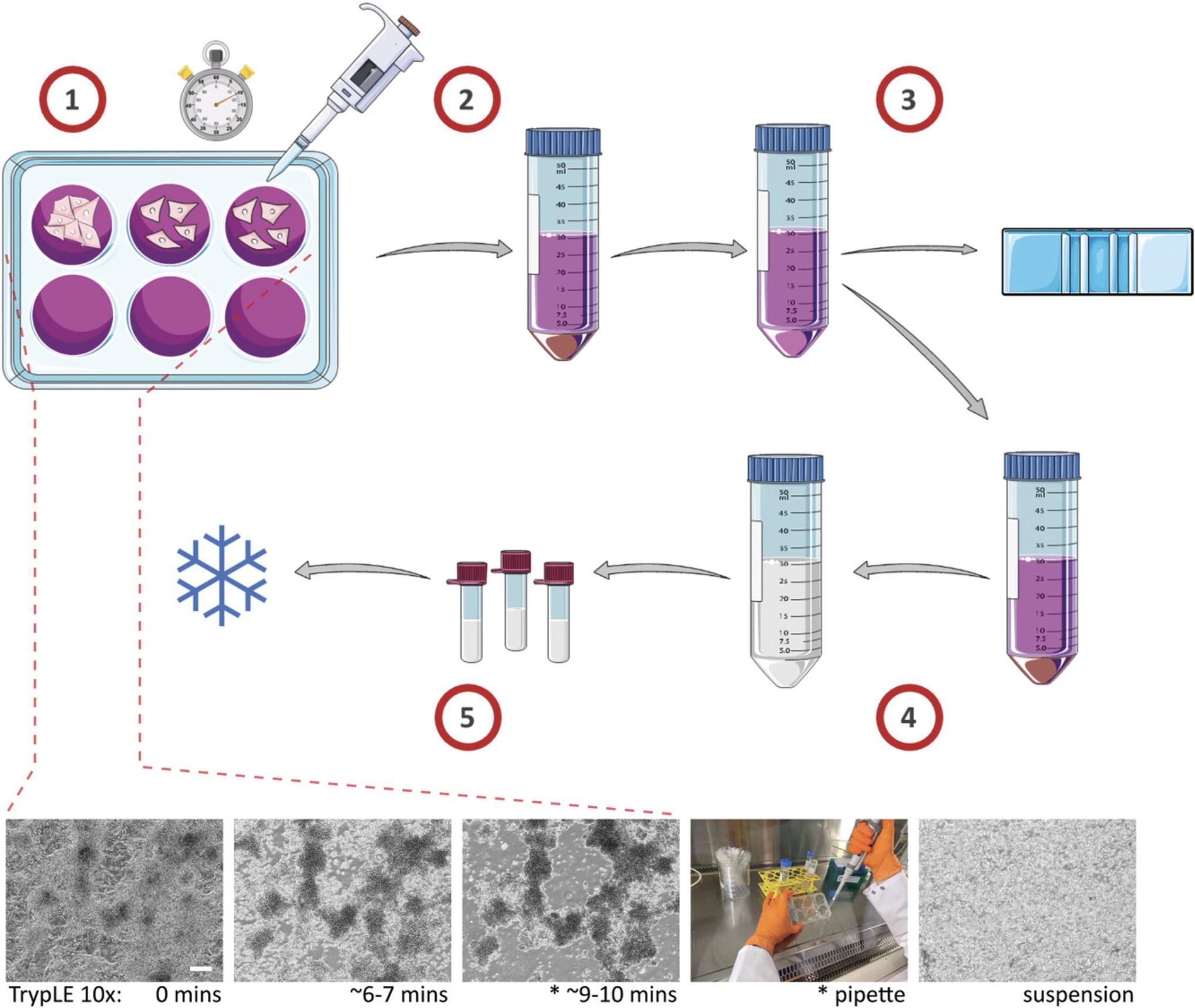
This protocol describes the careful dissociation and cryopreservation of hPSC-CMs (Fig. 2). There are several critical aspects to preserving the viability of hPSC-CMs during this process. The first is effective dissociation without degradation of the cells. This presents a challenge as often differentiating hPSC-CM monolayer cultures have a high cell density (Fig. 3A), and tend to increase the amount of extra cellular matrix proteins they secrete and cell-cell junctions they generate, which all need to be digested. The most often cited approach uses fast but powerful enzymatic dissociation, which is what is described here. An alternative is more slow but gentle dissociation, requiring different reagents and protocol (see Background Information). The next important aspect is gentle handling of the cells. Slow regular trituration generating a minimal amount of effervescence and sheer stress is strongly advised. Finally, slow but steady cooling of −1°C/min and protection against intracellular ice formation via a cryoprotective agent (CPA) is important for all conventional cell cryopreservation. Here this is handled via proprietary Mr. Frosty™ cryopreservation boxes; however, other products and even controlled rate freezing machines such as ViaFreeze™ should work well. The impact of different cryopreservation media is discussed in Basic Protocol 2 and Understanding Results.
Materials
-
hPSC-CMs in 6- or 12-well plates (see Fig. 1 and Strategic Planning)
-
CM Freezing medium (see recipe)
-
CM Suspension medium (see recipe)
-
DPBS without Ca2+/Mg2+ (PBS-) (e.g., Thermo Fisher, cat. no. 14190144)
-
TrypLE Select, 10× (Thermo Fisher, cat. no. A1217701)
-
Trypan blue solution (e.g., Sigma Aldrich, cat. no. T8154)
-
Cryogenic storage boxes (e.g., Mr Frosty™, Thermo Fisher, cat. no. 5100-0001)
-
37°C, 5% CO2 incubator
-
15- and 50-ml Falcon™ polypropylene tubes (e.g., Corning, cat. nos. 352096 and 352070)
-
Microscope (e.g. Leica DMi1)
-
1000-μl manual pipette plus 1000-µl filter tips (e.g., Eppendorf, 613-0866; Biozym, cat. no. VT0270)
-
Centrifuge (e.g., Eppendorf 5810R, Thermo Fisher, cat. no. 10012724)
-
Pipette gun and 5- to 10-ml stripettes (e.g., INTEGRA Biosciences, cat. no. 156400; Sarstedt, 86.1253.001; Sarstedt86.1254.001)
-
Countess™ II automated cell counter (Thermo Fisher, cat. no. AMQAX1000)
-
Countess™ cell counting slides (Thermo Fisher, cat. no. C10228)
-
Cryovials (e.g., CryzoTraq 2 ml, Ziath, cat. no. 590010)
NOTE : Perform all centrifugation steps at room temperature.
NOTE : The following protocol steps are based on the dissociation and cryopreservation of 2 × 6-well plates of hPSC-CMs at around day 14 of differentiation (Fig. 3A), which would generate approximately 6–10 × 107 cells. Dissociation of the two individual plates is slightly staggered to reduce disparity in 10× TrypLE incubation times.
1.Ensure a sufficient amount of the following reagents and equipment are pre-cooled by placing them for 30 to 60 min at 4°C before starting (if not already stored there): cryostorage boxes, CM Freezing medium.
2.Ensure a sufficient amount of the following reagents are brought to room temperature for 30-60 min before starting: CM Suspension medium, 10× TrypLE, PBS-.
3.Aspirate culture medium from hPSC-CMs, wash wells once with 1.5 ml PBS- each, and aspirate.
4.Add 1 ml of 10× TrypLE to each well of the first culture plate of hPSC-CMs and incubate at 37°C, 5% CO2 (Fig. 2_1).
5.After 2 min, add 1 ml of 10× TrypLE to each well of the second culture plate and incubate at 37°C, 5% CO2.
6.Prepare a 50-ml Falcon tube for collection of cells, with 1.5 ml CM Suspension medium added per well.
7.After 6-7 min incubation, remove each plate from incubator to check under the microscope for cell rounding and loosening (Fig. 2, lower panel).
8.Replace cells in incubator and check again after 1-4 min, until almost the whole culture has rounded into individual cells and clusters, but with only a few free-floating cells or small cell clusters detached from the culture plate surface (Fig. 2, lower panel).
9.Remove cells from incubator and using a 1000-μl pipette, gently triturate cells in each well, tilting the plate towards you and pipetting around the far side of the well in a semi-circular motion 3-4 times, then tilting the plate away from you and pipetting around the near side of the well in a semi-circular motion 3-4 times (Fig. 2, lower panel).
10.Carefully transfer the TrypLE-cell suspension into the 50-ml collection Falcon containing CM Suspension medium.
11.Add an additional 1 ml fresh CM suspension medium to each well and using a 1000-μl pipette gently wash and elute final remaining cells, adding them to the 50-ml collection Falcon.
Counting and Freezing
12.Centrifuge the cells for 3 min at 200 × g , (Fig. 2_2).
13.Aspirate the supernatant and very gently flick the bottom of the Falcon tube to help disperse the large cell pellet.
14.Carefully add 1 ml fresh CM Suspension medium directly to the cells at the bottom of the tube using a 1000-μl pipette, and gently pipette up and down several times.
15.Add an additional 9 ml CM Suspension medium to the cells using a stripette (Fig. 2_3).
16.Invert to mix, remove a 10-20-µl sample of cells and perform a viable cell count using Trypan Blue (Fig. 3B).
17.Centrifuge again for 3 min at 200 × g.
18.Aspirate the supernatant and resuspend hPSC-CMs in cooled CM Freezing medium to a density of 2-10 × 106 cells/ml, pipetting gently to mix (Fig. 2_4).
19.Pipette 0.5-1 ml hPSC-CM cell suspension into cryovials and place in cooled cryostorage boxes (Fig. 2_5).
20.Place cryostorage boxes at −80°C for at least 4 hr (generally overnight) before transferring cryovials to the vapor phase of liquid N2 for long-term storage.
Sample data
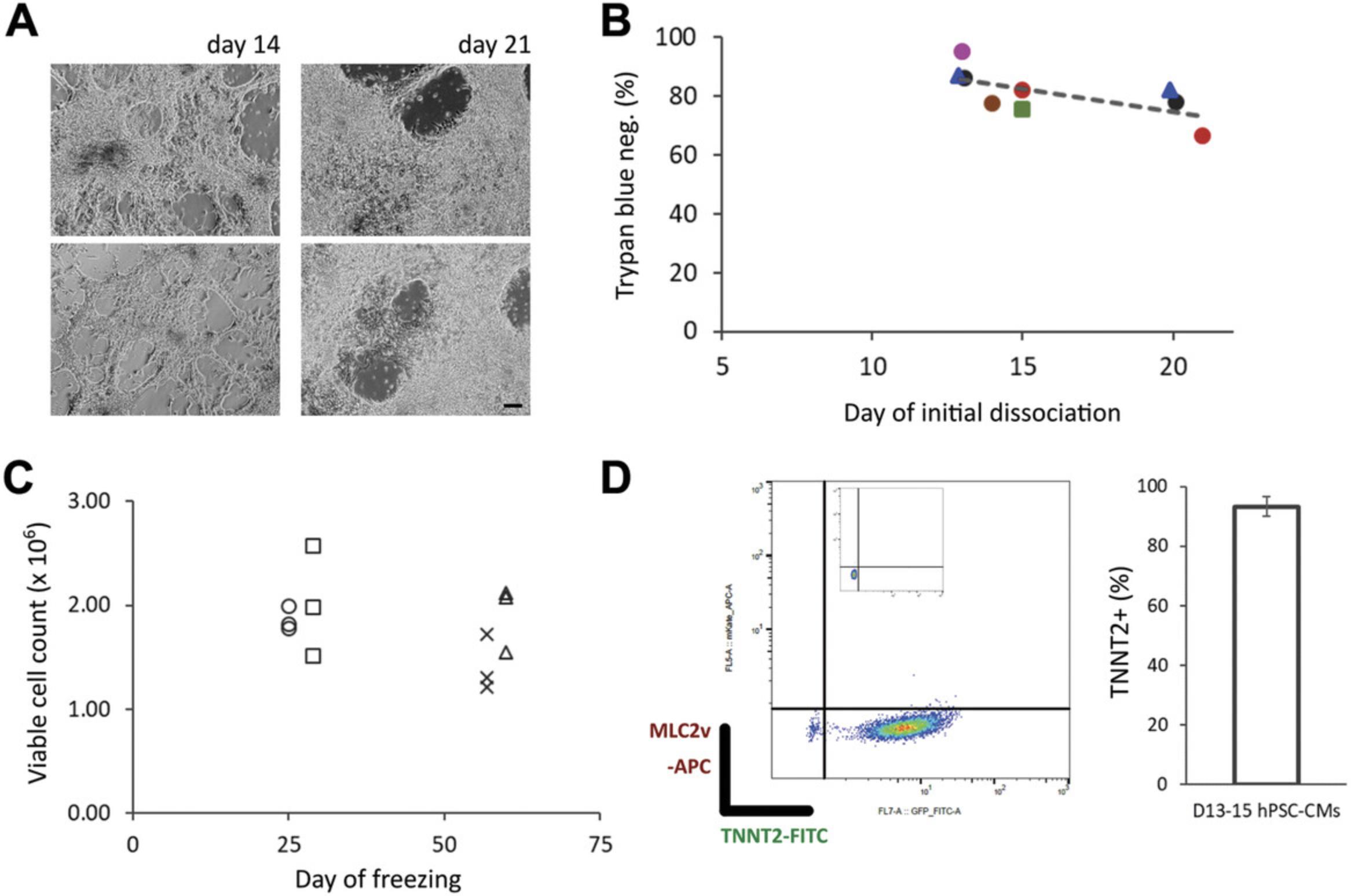
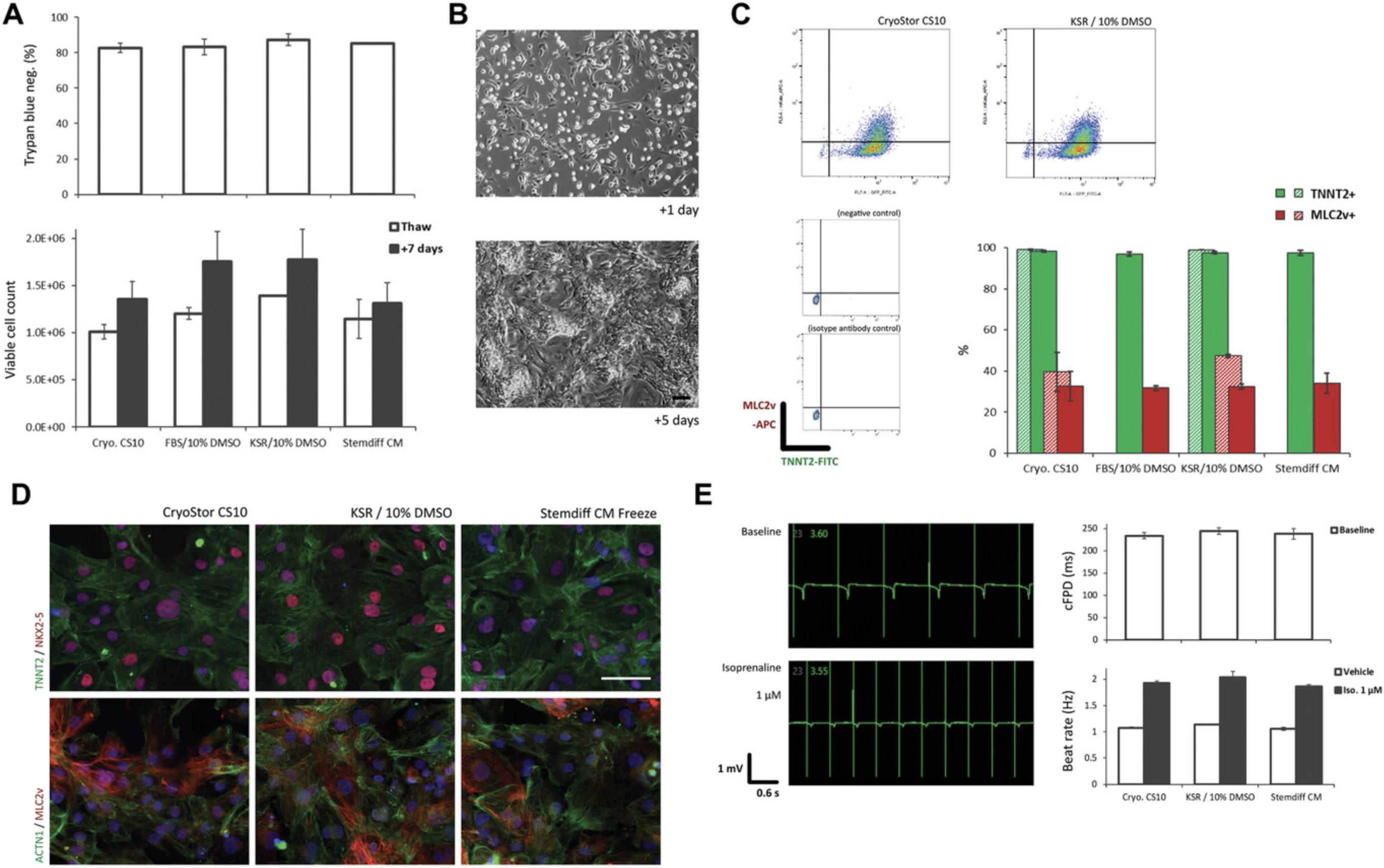
Following cardiac differentiation of hPSCs (Fig. 1), we investigated the effect of dissociation and cryopreservation on hPSC-CMs. We found that the time point (differentiation day) for initial (first) dissociation may make a difference to the viability of the hPSC-CMs. In our hands, the viability of hPSC-CMs tends to decrease with a later time point for initial dissociation, day 14 tending to be better than day 21 (Fig. 3B), with a mean viability of 84% ±2.9 (SEM). Where possible we would therefore suggest an initial dissociation at an earlier time point i.e., around day 14 of differentiation (see Background Information and Understanding Results). However, dissociation and freezing of hPSC-CMs at later time points additionally to an initial dissociation at an earlier time point does not significantly affect the recovery of cells (Fig. 3C). As a general workflow, we would also recommend taking a sample of ∼1 × 106 cells for QC by flow cytometry following dissociation (Fig. 3D) (e.g., see Berg Luecke, Waas, & Gundry, 2019). Together these data and simple protocol should facilitate the preparation and handling of hPSC-CMs for dissociation and cryopreservation at specified time points during differentiation.
Basic Protocol 2: THAWING AND RECOVERY OF CRYOGENICALLY FROZEN hPSC-CMs
Similar to many other cell types, maintaining good recovery and viability of hPSC-CMs during thawing is dependent on a controlled but fast thaw and a gentle resuspension in culture medium (Fig. 4). For hPSC-CMs, slow dropwise resuspension using a relevant culture medium is particularly important for avoiding osmotic shock and maintaining viability (see Background Information).
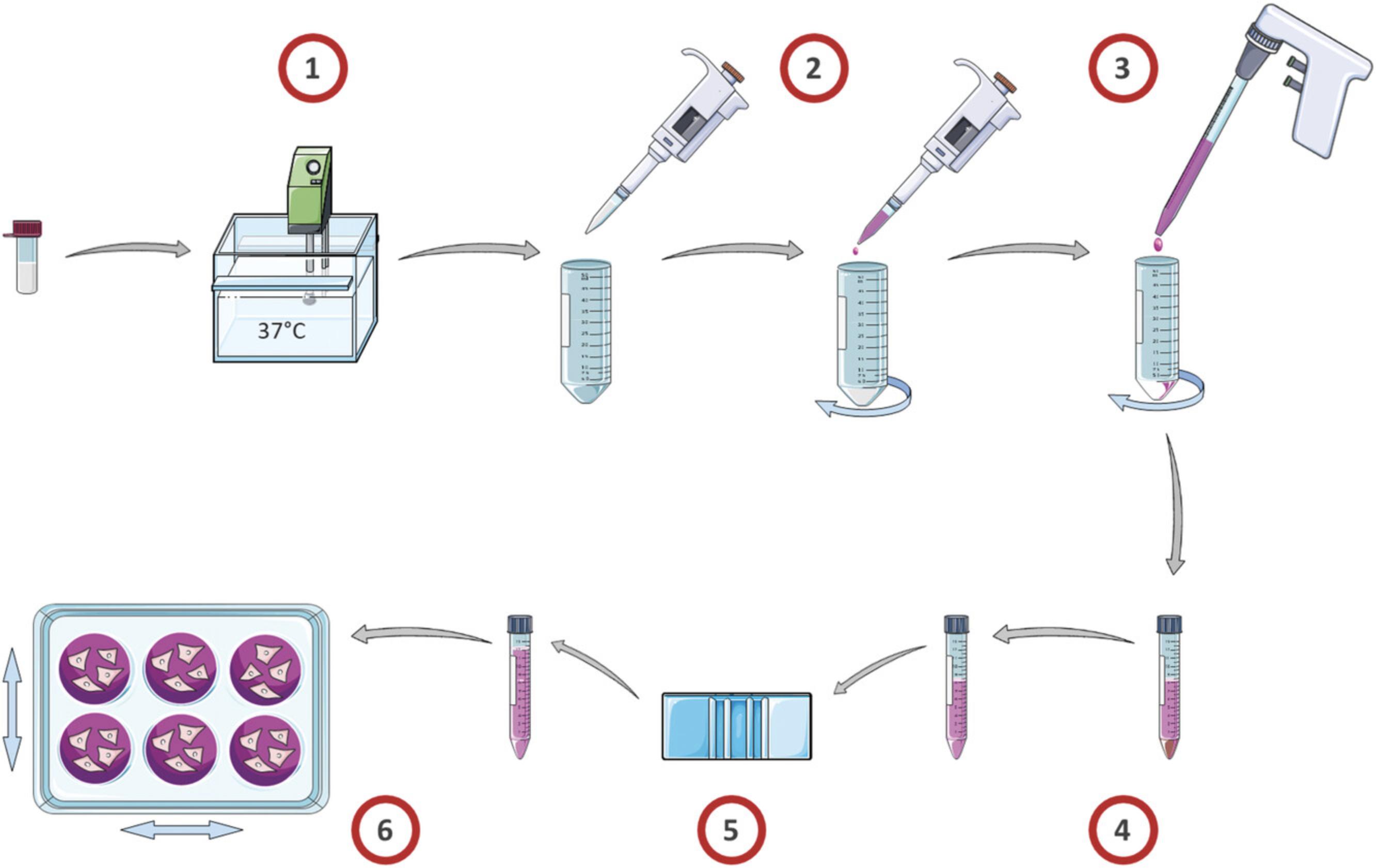
Cryopreserved cells should be kept frozen until all downstream steps for thawing and recovery have been prepared first. Most importantly this includes coated cell culture plates for the downstream application of the cells. Differentiated hPSC-CMs can be re-plated at specified densities onto different types of cell culture plate (Table 2) coated with various forms of extra cellular matrix (ECM). Re-plating can be done either immediately following dissociation or after thawing from cryopreservation. In our hands, Geltrex is largely equivalent to Matrigel and works well for applications where a complete monolayer is desired, including on glass and plastic bottom plates, and those containing electrodes. Fibronectin is also widely used, especially where a lower density of cells on glass bottom plates is required, e.g., for patch clamping. Other ECMs such as Laminin-221/211 are known to work.
| No. cryovials | Volume frozen cells | Volume initial resuspension | Top up volume |
|---|---|---|---|
| 1 | 0.5-1 ml | 1 ml | 5 ml |
| 2 | 1-2 ml | 2 ml | 8 ml |
- a
The volume of initial resuspension is to be added dropwise using a 1000 pipette-μl, with the top up volume subsequently added dropwise using a 5-10-ml stripette.
Materials
-
Geltrex (Thermo Fisher, cat. no. A1413302)
-
DMEM/F-12 (Thermo Fisher, cat. no. 11330032)
-
CM Suspension medium (see recipe)
-
Dry ice or liquid nitrogen
-
Cryovials of cryopreserved hPSC-CMs (see Basic Protocol 1)
-
RevitaCell™ Supplement 100× (Thermo Fisher, cat. no. A2644501)
-
CM Maintenance medium (see recipe)
-
Trypan blue solution (e.g., Sigma Aldrich, cat. no. T8154)
-
Multi-well culture plates (e.g., 24-well, Greiner Bio-One, cat. no. 662160)
-
Parafilm® M sealing film (Sigma, cat. no. P6543-1EA)
-
Isothermal ice bucket (e.g., Fisherbrand™ Polyurethane Ice Buckets, Fisher Scientific, cat. no. 11324085)
-
Water bath (e.g., VWR, cat. no. 462-0554)
-
1000-μl manual pipette plus 1000-µl filter tips (e.g., Eppendorf, cat. no. 613-0866; Biozym, cat. no. VT0270)
-
15- and 50-ml Falcon™ polypropylene tubes (e.g., Corning, cat. nos. 352096 and 352070)
-
Pipette gun and 5-10-ml stripettes (e.g., INTEGRA Biosciences, cat. no. 156400; Sarstedt, 86.1253.001; Sarstedt86.1254.001)
-
Centrifuge (e.g., Eppendorf 5810R, Thermo Fisher, cat. no. 10012724)
-
37°C, 5% CO2 incubator
-
Countess™ II automated cell counter (Thermo Fisher, cat. no. AMQAX1000)
-
Countess™ cell counting slides (Thermo Fisher, cat. no. C10228)
NOTE: Perform all centrifugation steps at room temperature.
NOTE: Follow safety guidance when handling cryogenically frozen material and liquid N2 storage containers.
NOTE: The following protocol steps are intended for defrosting 1× or simultaneously 2× cryovials of hPSC-CMs pooled into one Falcon tube (see Table 3 ).
Thawing and Resuspension
1.Dilute Geltrex using DMEM/F12 and prepare Geltrex-coated plates according to manufacturer's instructions (Thermo Fisher Scientific, 2013), ensuring adequate numbers of desired wells/plates are incubated at 37°C 60 min before thawing cells (see Table 2).
2.Allow an adequate volume of CM Suspension medium to reach room temperature for 30-60 min before thawing cells (Tables 2 and 3).
3.Using dry ice or liquid nitrogen, collect cryovial(s) of cryopreserved hPSC-CMs from liquid N2 and bring to cell culture.
4.Perform a quick thaw in a 37°C water bath. Occasionally gently swirl tubes in the bath, avoiding immersion above the level of the cap. Continue until about 90% defrosted (Fig. 4_1).
5.Sterilize the tube exterior before taking into laminar flow hood e.g., by wiping with 70% ethanol solution.
6.Using a 1000-μl pipette with 1000-μl pipette tip, remove cells from cryovial(s) and gently pipette into the bottom of a 50-ml conical Falcon tube.
7.Using a fresh 1-ml pipette, add 1 ml CM Suspension medium to the cryovial to recover any last microliters of leftover cells.
8.Using the same pipette tip, take up the 1 ml of CM Suspension medium now in the cryovial.
9.Dropwise and slowly, add the recovered 1 ml of CM Suspension medium into the 50-ml Falcon tube, aiming for about 1-2 drops every 3-5 s, gently swirling between (Fig. 4_2).
10.Using a stripette and pipette gun, add an additional 5 ml CM Suspension medium to the cells dropwise and slowly, again aiming for several drops every 3-5 s, swirling between (Fig. 4_3).
11.Transfer the cell suspension into a 15-ml Falcon tube.
Counting and Replating
12.Centrifuge for 3 min at 300 × g , and aspirate the supernatant (Fig. 4_4).
13.Gently resuspend the cell pellet in half the amount of CM Suspension medium + RevitaCell (1% v/v) corresponding to the cell density/number of wells desired (see Table 2).
14.Perform a cell count including trypan blue live/dead staining using Countess II and counting slides, followed by an adjustment of the volume using CM Suspension medium + RevitaCell (1% v/v) to the desired cell density (Fig. 4_5).
15.Remove Geltrex coating solution from intended culture plates and add the cell suspension to Geltrex-coated wells (Fig. 4_6).
16.Incubate the cells at 37°C, 5% CO2.
17.The next day, change medium to CM Maintenance medium (Table 2).
18.Continue culturing hPSC-CMs in CM Maintenance medium for at least 5 days before use in downstream applications (Fig. 5B-E), changing medium every 2 to 3 days.
Sample data
By following Basic Protocols 1 and 2 described here, a hPSC-CM recovery rate of between 50% and 70% can generally be expected (Fig. 3C; Fig. 5A). We investigated potential differences between certain cryopreservation media (Table 1). In our hands, different media appear to have only marginal effects on hPSC-CM viability and recovery. An exhaustive study would be required; however, in a limited study we find that viability and recovery upon thawing are comparable between four common freezing media (Fig. 5A), with a coupled spontaneously contracting monolayer exhibiting typical hPSC-CM morphology forming after 5 days (Fig. 5B), and similar levels of CM marker expression (Fig. 5C). We further investigated the three more chemically defined freezing media (Table 1) and found recovered hPSC-CMs to have a similar intracellular morphology and expression of cytoskeletal components and cardiac markers (Fig. 5D), as well as a comparable electrophysiological profile (Fig. 5E). These data show that the workflow outlined here in combination with potentially any of the four media tested can provide effective cryopreservation and functional recovery of hPSC-CMs. Please see Background Information and Understanding Results for further discussion.
REAGENTS AND SOLUTIONS
CM Freezing media
- CryoStor™ CS10 (Stem Cell Technologies, cat. no. 07930)
- FBS/10% DMSO: –Fetal bovine serum (FBS) heat-inactivated (Thermo Fisher, cat. no. 16140071); Dimethyl sulfoxide (DMSO; Sigma-Aldrich, cat. no. D2650). It is recommended to defrost stock FBS and divide into 9- or 36-ml aliquots and freeze for up to 3 months at −20°C. Defrost a 9- or 36-ml FBS aliquot until it reaches room temperature, and then add 1 or 4 ml DMSO (10% v/v) and invert to mix. Store complete medium for up to 2 weeks at 4°C.
- KSR/10% DMSO: –It is recommended to defrost stock KSR and divide into 9- or 36-ml aliquots and freeze for up to 3 months at −20°C. Defrost a 9- or 36-ml KSR aliquot until it reaches room temperature, and then add 1 or 4 ml DMSO (10% v/v) and invert to mix. Store complete medium for up to 2 weeks at 4°C.
- STEMdiff™ Cardiomyocyte Freezing Medium (Stem Cell Technologies, cat. no. 05030).
CM Maintenance medium
Add 10 ml B-27 supplement (Thermo Fisher, cat. no. 17504044) to 490 ml RPMI basal medium (Thermo Fisher, at. no. 11875-093) (2% v/v) and invert to mix. Alternatively, for less frequent use, defrost and divide 10 ml of B-27 supplement into 1-ml aliquots and freeze for up to 3 months at −20°C, and then defrost a 1-ml aliquot and add to 49 ml RPMI basal medium. Store complete medium up to 2 weeks at 4°C.
CM Suspension medium
Add 4 ml KnockOut™ serum replacement (KSR; Thermo Fisher, cat. no. 10828028) to 36 ml CM Maintenance medium (10% v/v; see recipe) and invert to mix. Store complete medium up to 2 weeks at 4°C.
COMMENTARY
Background Information
Two fundamental aspects of hPSC-CM bioprocessing for downstream applications are efficient reproducible differentiation and viable cryopreservation. In general, and particularly for diverse smaller scale research applications of hPSC-CMs, the simplicity of the workflows for these processes greatly facilitates their utilization. While simple hPSC-CM differentiation protocols have become much more precise and efficient (Denning et al., 2016), there is a slight dearth of improvement and understanding with regards to cryopreservation. The review by Preininger and colleagues is a helpful navigator (Preininger et al., 2016). Among the publications that do openly report or specifically investigate cryopreservation of hPSC-CMs, as noted in the Introduction, there are widely varying rates of recovery (Burridge et al., 2015; Chen et al., 2015; van den Brink et al., 2020; Xu et al., 2011), and anecdotally recovery rates tend to be lower. Early reports also cryopreserved hPSC-CMs that had been differentiated at a lower purity and consistency (Xu et al., 2011); hence, their methods and recovery rates may not be wholly applicable now. A high recovery rate of 84% ±5.2 is reported by Chen and colleagues; however, hPSC-CMs were differentiated as 3D clusters in stir tank bioreactors and dissociated using a two-step protocol (see below; Chen et al., 2015). Few studies address the potential effects of cryopreservation on hPSC-CMs with detailed analysis, and to our knowledge there are no in-depth comparisons of cryopreservation media. One report by Brink and colleagues investigates the effect of cryopreservation on the electrophysiology and transcripts of two hPSC lines (van den Brink et al., 2020). Interestingly, they find that following cryopreservation there is mostly no effect on key electrophysiological parameters, although one hPSC line did exhibit slight increases in ventricular, ion channel, and cytoskeletal marker expression compared with non-cryopreserved hPSC-CMs (van den Brink et al., 2020).
The reagents and techniques comprising the workflow described here derive from in-house development, as well as a variety of contemporary protocols and guidelines. The inclusion of KSR in CM Suspension and Freeze media is likely to be beneficial to hPSC-CMs by bolstering B-27 in terms of survival factors (e.g., insulin and transferrin), and increasing the amount of protein via bovine serum albumin (BSA), which among other things will help to quench protease activity following dissociation with 10× TrypLE. Slow dropwise resuspension as a way of avoiding osmotic shock to hPSC-CMs immediately following thawing is noted as critical in several protocols and papers (e.g., Breckwoldt et al., 2017; NCardia, 2018; van den Brink et al., 2020). RevitaCell is a pro-survival supplement commonly used in hPSC culture to avoid anoikis during single-cell passaging, containing a Rho-associated kinase (ROCK) inhibitor and anti-oxidant factors. The application of RevitaCell or some other pro-survival supplement for the first day following re-plating of hPSC-CMs by us and others (Breckwoldt et al., 2017; van den Brink et al., 2020) is partly based on observations by Laflamme et al., whereby inclusion of a pro-survival cocktail increased engraftment size of hESC-CMs in rat hearts (Laflamme et al., 2007). Although we have not rigorously compared hPSC-CM recovery after re-plating with or without RevitaCell, we would nonetheless recommend inclusion of some kind of pro-survival supplement in the medium on the day of thawing.
Viability and recovery of hPSC-CMs using this workflow should be comparable when using other commercial kits, published differentiation protocols, and CM maintenance medium (e.g., from NCardia, Cellular Dynamics, or Miltenyi) (Allen_Institute, 2018; Birket et al., 2015; Burridge et al., 2014). However, it is advised to follow manufacturer guidelines where available and adjust media and conditions to optimize processes. A small amount of variability can also be expected. This can derive from differences between hPSC lines, differentiation protocols, the cellular composition (CMs vs. non-CMs) of individual differentiation batches, as well as other technical details such as reagent batches and individual user technique. There are certain adjuncts to the protocols that we have also observed as potentially beneficial for cell viability and recovery although they have not been rigorously tested. In particular–addition of DNase I during dissociation with 10× TrypLE to reduce clumping and increase viability; a higher cell freezing density of >10 × 106 cells/ml and/or a freezing volume <0.5 ml/vial to provide a more consistent freezing rate, although there is slightly conflicting data regarding this (see Preininger et al., 2016). Furthermore, the use of controlled rate freezing machines such as ViaFreeze may also offer significant benefits; however, this again would require further optimization, as well as an increased financial outlay.
An alternative to the fast dissociation protocol described here exists in the form of a longer two-stage protocol. hPSC-CM monolayers or 3D clusters are partially dissociated slowly by incubating in collagenase type I (Sigma) in PBS+/20% FBS or Liberase (Roche) plus DNase I for 20-60 min, followed by a quick final digestion with trypsin-EDTA or TrypLE for 3-10 min (Berg Luecke et al., 2019; Chen et al., 2015; Mills et al., 2017). This protocol may confer one advantage in reducing the total time exposed to strong protease activity, which may be beneficial in particular for 3D clusters due to the increased density and gradient of exposure to the dissociation solution. However, a strong protease step is still required, and the workflow is more complex. Unfortunately to our knowledge there are no available direct comparisons between the two approaches.
Critical Parameters
There are several critical parameters influencing dissociation, cryopreservation, and thawing of hPSC-CMs that can each be individually addressed to maximize recovery of cells. The dissociation time with 10× TrypLE can be adjusted based on the cell density and availability of a cell shaker, and will vary based on the hPSC line, and differentiation protocol and time point. Gentle initial pipetting to dissociate the cells in TrypLE, as well as handling in subsequent steps is also important to avoid shear stress. Cell clumping as a result of excessive cell death and subsequent release of DNA and other cellular debris can be reduced by using a cell shaker during dissociation, reducing pipetting, and including DNase I. Different hPSC lines and differentiation protocols will exhibit varying amounts of cell clumping. If it does occur, use of a 70-100-µm cell strainer can partially rescue the situation. A steady rate of freezing is crucial to cryopreservation of all cells, hence all parts of the protocol especially equipment and freezing medium must be well prepared and pre-cooled, and the process conducted in a time-efficient manner. A fast thaw combined with a slow regulated resuspension avoiding osmotic shock is also crucial to maintaining viability when recovering hPSC-CMs.
Troubleshooting
See Table 4 for troubleshooting possible problems encountered during the protocols.
| Problem | Possible causes | Potential solutions |
|---|---|---|
| Severe cell clumping following dissociation of hPSC-CMs |
|
|
| Dense monolayers or clusters of hPSC-CMs not dissociating |
|
|
| Low viability upon cell count following dissociation |
|
|
| Poor functional recovery after thawing |
|
|
Understanding Results
In this article, we have described a simple workflow for the dissociation, cryopreservation, and recovery of hPSC-CMs. We have also investigated aspects of these protocols, including the effects of differentiation day and cryopreservation media. We hope this will provide a benefit for users in standardizing and simplifying hPSC-CM workflows, in order to improve reproducibility in downstream applications.
A key parameter used in this report and those discussed above is hPSC-CM viability. We use trypan blue exclusion as an indicator of viability throughout the workflow, i.e., how many cells are degraded and/or apoptotic and no longer exclude trypan blue. Viability can also be assessed by immunostaining and flow cytometry. The values derived from these two methods can slightly differ, given the different mechanism, sensitivity, and timing of the two procedures; however, in practice it is quicker and cheaper to generate an indicative number by trypan blue.
We observe a tendency to lose viability with later days for initial dissociation of hPSC-CMs (Fig. 3B). This may be due to the increase in cell density and cell-cell junctions that likely occurs with a longer period in differentiation culture (Fig. 3A), requiring longer incubation in TrypLE and/or more pipetting to dissociate. A preference for dissociating hPSC-CMs earlier to apply to more complex 3D cultures is emerging, as this may be beneficial for their maturation and functional fidelity in these contexts due to a maturation window which cells pass through soon after lineage commitment (Breckwoldt et al., 2017; Mills et al., 2017; Ronaldson-Bouchard et al., 2018). Together these observations and trends would suggest that dissociation and cryopreservation of hPSC-CMs at around day 14 of differentiation would be a good general workflow for many hPSC-CM applications. It should be noted; however, that for some applications this would then require a longer time in culture subsequent to thawing and re-plating, since older hPSC-CMs even in simple monolayer cultures exhibit a slightly more mature transcriptional and functional profile (Kumar et al., 2019), which may be important for certain assays. Interestingly, we have observed a phenomenon whereby raised expression in MLC2v (MYL2) can be seen at ∼day 21 following dissociation and re-plating of hPSC-CMs at ∼day 14, with or without cryopreservation (compare Fig. 3D and Fig. 5C), with negligible expression of MLC2v at day 21 in parallel hPSC-CMs left un-split (data not shown). This would suggest that initiation of a more mature ventricular CM identity requires dissociation and re-plating. This may be due to space restriction in differentiation cultures before dissociation and re-plating, or perhaps because dissociation and re-plating permits increased cell-cell junction and syncytia formation between hPSC-CMs.
In terms of differences between the four media we have tested for cryopreservation of hPSC-CMs, the data presented here, as well as our general observations indicate that they are largely comparable despite markedly different formulations. This may be partly due to the fact they all contain the same CPA and are applied with an optimized workflow. Our investigation does not eliminate the possibility that repeated, or more in-depth analyses may reveal some differences, e.g., at the metabolic or transcriptional level. However, any such differences may also be temporary given the brief period the cells are present in cryopreservation media. That said, we excluded FBS/10% DMSO from downstream functional analysis for several reasons. Although it has a long standing as a basis for cryopreservation medium, and is still used in some protocols and by many groups, it suffers from several drawbacks compared to the other three media investigated here–it is not the cheapest, it is the most likely to suffer from batch variability and price fluctuations, some effects on hPSC-CM function when included in maintenance medium have been observed (Dambrot et al., 2014), and it is more difficult to convert to current good manufacturing practice (cGMP).
We have studied the applicability of four common cryopreservation media in this report but would like to note several other observations and options. Despite being by far the most widely used CPA, it is acknowledged that DMSO can have some detrimental effects on cells in culture (Verheijen et al., 2019). We compared 5% rather than 10% DMSO in KSR- or FBS-based freezing media; however, found this resulted in poorer recovery of hPSC-CMs and/or a slight negative impact on TNNT2 expression (data not shown). Similarly, we found BamBanker less good for recovery of hPSC-CMs (data not shown), as noted in Burridge et al. (2015). Other media such as Synth-a-freeze, StemMACS CryoBrew, and NutriFreeze may be viable options for hPSC-CM cryopreservation but require testing.
Conclusion
Application of this simple workflow should allow cryopreservation of hPSC-CM batches with a recovery of up to 70% with no specialized or expensive equipment. The four different cryopreservation media tested here appear to give similar results in terms of critical parameters for hPSC-CM recovery and function. Cost and compatibility of media components then need to be considered for the downstream application of cells.
Time Considerations
Basic Protocol 1: ∼30 min preparation, 45-60 min handling.
Basic Protocol 2: ∼60 min preparation (including pre-coating of plates), ∼45 min handling.
Acknowledgments
This work was supported by funding from the DZHK (German Centre for Cardiovascular Research), partner site Berlin, Standortprojekt 81Z0100101. We would also like to acknowledge the use of several pieces of artwork as part of Figure 2 and 4 from Servier Medical Art (https://smart.servier.com/), obtained and used under a Creative Commons Attribution 3.0 Unported License (https://creativecommons.org/licenses/by/3.0/). Open access funding enabled and organized by Projekt DEAL.
Author Contributions
Duncan C. Miller : Conceptualization; data curation; formal analysis; investigation; methodology; project administration; resources; supervision; validation; visualization; writing-original draft; writing-review & editing. Carolin Genehr : Investigation; methodology; project administration; resources; validation; writing-review & editing. Narasimha S. Telugu : Investigation; methodology; writing-review & editing. Silke Kurths : Investigation; methodology; resources. Sebastian Diecke : Conceptualization; funding acquisition; resources; supervision; writing-review & editing.
Literature Cited
- Allen_Institute. (2018). Cardiomyocyte Differentiation Methods v1.2. Retrieved from https://www.allencell.org/methods-for-cells-in-the-lab.html.
- Berg Luecke, L., Waas, M., & Gundry, R. L. (2019). Reliable protocols for flow cytometry analysis of intracellular proteins in pluripotent stem cell derivatives: A fit-for-purpose approach. Current Protocols in Stem Cell Biology , 50(1), 1–26. doi: 10.1002/cpsc.94.
- Birket, M. J. J., Ribeiro, M. C. C., Kosmidis, G., Ward, D., Leitoguinho, A. R. R., van de Pol, V., … Mummery, C. L. L. (2015). Contractile defect caused by mutation in MYBPC3 revealed under conditions optimized for human PSC-cardiomyocyte function. Cell Reports , 13(4), 733–745. doi: 10.1016/j.celrep.2015.09.025.
- Breckwoldt, K., Letuffe-Brenière, D., Mannhardt, I., Schulze, T., Ulmer, B., Werner, T., … Hansen, A. (2017). Differentiation of cardiomyocytes and generation of human engineered heart tissue. Nature Protocols , 12(6), 1177–1197. doi: 10.1038/nprot.2017.033.
- Burridge, P. W., Holmström, A., & Wu, J. C. (2015). Chemically defined culture and cardiomyocyte differentiation of human pluripotent stem cells. Current Protocols in Human Genetics , 87, 21.3.1–21.3.15. doi: 10.1002/0471142905.hg2103s87.
- Burridge, P. W., Matsa, E., Shukla, P., Lin, Z. C., Churko, J. M., Ebert, A. D., … Wu, J. C. (2014). Chemically defned generation of human cardiomyocytes. Nature Methods , 11, 855–860. doi: 10.1038/nMeth.2999.
- Chen, V. C., Ye, J., Shukla, P., Hua, G., Chen, D., Lin, Z., … Couture, L. A. (2015). Development of a scalable suspension culture for cardiac differentiation from human pluripotent stem cells. Stem Cell Research , 15(2), 365–375. doi: 10.1016/j.scr.2015.08.002.
- D'Antonio-Chronowska, A., Donovan, M. K. R., Young Greenwald, W. W., Nguyen, J. P., Fujita, K., Hashem, S., … Frazer, K. A. (2019). Association of human iPSC gene signatures and X chromosome dosage with two distinct cardiac differentiation trajectories. Stem Cell Reports , 13(5), 924–938. doi: 10.1016/j.stemcr.2019.09.011.
- Dambrot, C., Braam, S. R., Tertoolen, L. G. J., Birket, M., Atsma, D. E., & Mummery, C. L. (2014). Serum supplemented culture medium masks hypertrophic phenotypes in human pluripotent stem cell derived cardiomyocytes. Journal of Cellular and Molecular Medicine , 18(8), 1509–1518. doi: 10.1111/jcmm.12356.
- Denning, C., Borgdorff, V., Crutchley, J., Firth, K. S. A., George, V., Kalra, S., … Young, L. E. (2016). Cardiomyocytes from human pluripotent stem cells: From laboratory curiosity to industrial biomedical platform. Biochimica et Biophysica Acta - Molecular Cell Research , 1863(7), 1728–1748. doi: 10.1016/j.bbamcr.2015.10.014.
- Dunn, K. K., & Palecek, S. P. (2018). Engineering scalable manufacturing of high-quality stem cell-derived cardiomyocytes for cardiac tissue repair. Frontiers in Medicine , 5(APR)doi: 10.3389/fmed.2018.00110.
- Gerbin, K. A., Yang, X., Murry, C. E., & Coulombe, K. L. K. (2015). Enhanced electrical integration of engineered human myocardium via intramyocardial versus epicardial delivery in infarcted rat hearts. PLoS ONE , 10(7), e0131446. doi: 10.1371/journal.pone.0131446.
- Kempf, H., Olmer, R., Haase, A., Franke, A., Bolesani, E., Schwanke, K., … Zweigerdt, R. (2016). Bulk cell density and Wnt/TGFbeta signalling regulate mesendodermal patterning of human pluripotent stem cells. Nature Communications , 7, 13602. doi: 10.1038/ncomms13602.
- Kumar, N., Dougherty, J. A., Manring, H. R., Elmadbouh, I., Mergaye, M., Czirok, A., … Khan, M. (2019). Assessment of temporal functional changes and miRNA profiling of human iPSC-derived cardiomyocytes. Scientific Reports , 9(1), 1–16. doi: 10.1038/s41598-019-49653-5.
- Laflamme, M. A., Chen, K. Y., Naumova, A. V., Muskheli, V., Fugate, J. A., Dupras, S. K., … Murry, C. E. (2007). Cardiomyocytes derived from human embryonic stem cells in pro-survival factors enhance function of infarcted rat hearts. Nature Biotechnology , 25(9), 1015–1024. doi: 10.1038/nbt1327.
- Lian, X., Hsiao, C., Wilson, G., Zhu, K., Hazeltine, L. B., Azarin, S. M., … Palecek, S. P. (2012). Robust cardiomyocyte differentiation from human pluripotent stem cells via temporal modulation of canonical Wnt signaling. Proceedings of the National Academy of Sciences , 109(27), E1848–E1857. doi: 10.1073/pnas.1200250109.
- Liu, Y. W., Chen, B., Yang, X., Fugate, J. A., Kalucki, F. A., Futakuchi-Tsuchida, A., … Murry, C. E. (2018). Human embryonic stem cell-derived cardiomyocytes restore function in infarcted hearts of non-human primates. Nature Biotechnology , 36(7), 597–605. doi: 10.1038/nbt.4162.
- Mills, R. J., Titmarsh, D. M., Koenig, X., Parker, B. L., Ryall, J. G., Quaife-Ryan, G. A., … Hudson, J. E. (2017). Functional screening in human cardiac organoids reveals a metabolic mechanism for cardiomyocyte cell cycle arrest. Proceedings of the National Academy of Sciences of the United States of America , 114(40), E8372–E8381. doi: 10.1073/pnas.1707316114.
- NCardia. (2018). Pluricyte ® Cardiomyocytes. In user manual. Retrieved from https://ncardia.com/files/documents/manuals/PluricyteCardiomyocyte_Manual_v2.pdf.
- Preininger, M. K., Singh, M., & Xu, C. (2016). Cryopreservation of human pluripotent stem cell-derived cardiomyocytes: Strategies, challenges, and future directions. In Advances in Experimental Medicine and Biology. doi: 10.1007/978-3-319-45457-3_10 .
- Ronaldson-Bouchard, K., Ma, S. P., Yeager, K., Chen, T., Song, L. J., Sirabella, D., … Vunjak-Novakovic, G. (2018). Advanced maturation of human cardiac tissue grown from pluripotent stem cells. Nature , 556(7700), 239–243. doi: 10.1038/s41586-018-0016-3.
- Thermo Fisher Scientific. (2013). Geltrex® hESC-qualified Ready-To-Use Reduced Growth Factor Basement Membrane Matrix. Retrieved from https://tools.lifetechnologies.com/content/sfs/manuals/geltrex_ready_to_use_man.pdf.
- Tohyama, S., Hattori, F., Sano, M., Hishiki, T., Nagahata, Y., Matsuura, T., … Fukuda, K. (2013). Distinct metabolic flow enables large-scale purification of mouse and human pluripotent stem cell-derived cardiomyocytes. Cell Stem Cell , 12(1), 127–137. doi: 10.1016/j.stem.2012.09.013.
- van den Brink, L., Brandão, K. O., Yiangou, L., Mol, M. P. H., Grandela, C., Mummery, C. L., … Davis, R. P. (2020). Cryopreservation of human pluripotent stem cell-derived cardiomyocytes is not detrimental to their molecular and functional properties. Stem Cell Research , 43(June 2019), 101698. doi: 10.1016/j.scr.2019.101698.
- Verheijen, M., Lienhard, M., Schrooders, Y., Clayton, O., Nudischer, R., Boerno, S., … Caiment, F. (2019). DMSO induces drastic changes in human cellular processes and epigenetic landscape in vitro. Scientific Reports , 9(1), 4641. doi: 10.1038/s41598-019-40660-0.
- Xu, C., Police, S., Hassanipour, M., Li, Y., Chen, Y., Priest, C., … Gold, J. D. (2011). Efficient generation and cryopreservation of cardiomyocytes derived from human embryonic stem cells. Regenerative Medicine , 6(1), 53–66. doi: 10.2217/rme.10.91.
Internet Resources
Repository of information and protocols for Axion Biosystems. Includes “Application Documents” and “Culture Protocols” for thawing, plating, and analyzing iCell® (Fuji Cellular Dynamics International) or Pluricyte®/Cor.4U (NCardia, formerly Pluriomics) hPSC-CMs on E-Stim+ and CytoView MEA plates.
International registry of hPSC lines including those used here (BIHi005-A, BIHi005-A-3, and BIHi050-A).
Allen Cell Explorer from the Allen Institute. Has several comprehensive SOP-style protocols including one for hPSC-CM differentiation, as well as a catalog of tagged hPSC lines and many other useful resources.
Citing Literature
Number of times cited according to CrossRef: 7
- Kavita Raniga, Aishah Nasir, Nguyen T.N. Vo, Ravi Vaidyanathan, Sarah Dickerson, Simon Hilcove, Diogo Mosqueira, Gary R. Mirams, Peter Clements, Ryan Hicks, Amy Pointon, Will Stebbeds, Jo Francis, Chris Denning, Strengthening cardiac therapy pipelines using human pluripotent stem cell-derived cardiomyocytes, Cell Stem Cell, 10.1016/j.stem.2024.01.007, (2024).
- Philipp Gmach, Marc Bathe-Peters, Narasimha Telugu, Duncan C. Miller, Paolo Annibale, Fluorescence Spectroscopy of Low-Level Endogenous β-Adrenergic Receptor Expression at the Plasma Membrane of Differentiating Human iPSC-Derived Cardiomyocytes, International Journal of Molecular Sciences, 10.3390/ijms231810405, 23 , 18, (10405), (2022).
- Maria Ercu, Michael B. Mücke, Tamara Pallien, Lajos Markó, Anastasiia Sholokh, Carolin Schächterle, Atakan Aydin, Alexa Kidd, Stephan Walter, Yasmin Esmati, Brandon J. McMurray, Daniella F. Lato, Daniele Yumi Sunaga-Franze, Philip H. Dierks, Barbara Isabel Montesinos Flores, Ryan Walker-Gray, Maolian Gong, Claudia Merticariu, Kerstin Zühlke, Michael Russwurm, Tiannan Liu, Theda U.P. Batolomaeus, Sabine Pautz, Stefanie Schelenz, Martin Taube, Hanna Napieczynska, Arnd Heuser, Jenny Eichhorst, Martin Lehmann, Duncan C. Miller, Sebastian Diecke, Fatimunnisa Qadri, Elena Popova, Reika Langanki, Matthew A. Movsesian, Friedrich W. Herberg, Sofia K. Forslund, Dominik N. Müller, Tatiana Borodina, Philipp G. Maass, Sylvia Bähring, Norbert Hübner, Michael Bader, Enno Klussmann, Mutant Phosphodiesterase 3A Protects From Hypertension-Induced Cardiac Damage, Circulation, 10.1161/CIRCULATIONAHA.122.060210, 146 , 23, (1758-1778), (2022).
- Martha E. Floy, Fathima Shabnam, Aaron D. Simmons, Vijesh J. Bhute, Gyuhyung Jin, Will A. Friedrich, Alexandra B. Steinberg, Sean P. Palecek, Advances in Manufacturing Cardiomyocytes from Human Pluripotent Stem Cells, Annual Review of Chemical and Biomolecular Engineering, 10.1146/annurev-chembioeng-092120-033922, 13 , 1, (255-278), (2022).
- Ryan Walker-Gray, Tamara Pallien, Duncan C. Miller, Andreas Oder, Martin Neuenschwander, Jens Peter von Kries, Sebastian Diecke, Enno Klussmann, Disruptors of AKAP-Dependent Protein–Protein Interactions, cAMP Signaling, 10.1007/978-1-0716-2245-2_8, (117-139), (2022).
- Vasco Sampaio‐Pinto, Jasmijn Janssen, Nino Chirico, Margarida Serra, Paula M. Alves, Pieter A. Doevendans, Ilja K. Voets, Joost P. G. Sluijter, Linda W. Laake, Alain Mil, A Roadmap to Cardiac Tissue‐Engineered Construct Preservation: Insights from Cells, Tissues, and Organs, Advanced Materials, 10.1002/adma.202008517, 33 , 27, (2021).
- Daniel Besser, Advancing Stem Cell Technologies and Applications: A Special Collection from the PluriCore Network in the German Stem Cell Network (GSCN), Current Protocols in Stem Cell Biology, 10.1002/cpsc.129, 55 , 1, (2020).

