SARS-CoV-2 incursion scenario in the city Fantastica
Benjamin Schwessinger
Abstract
This protocols is part of the ANU Biosecurity mini-research project #2 "An SARS-CoV-2 incursion scenario: Genomics, phylogenetics, and incursions." This mini-research project is modeled on the yearly Quality Assurance Program of The Royal College of Pathologists of Australia (RCPAQAP), we take part in together with ACT Pathology.
This research project is split into two major parts, identical to how the official RCPAQAP is run every year.
Part #1 is focusing on the 'wet- lab' by sequencing SARS-CoV-2 from real world RNA samples provided by ACT Pathology especially for our ANU biosecurity course (Thank YOU!). Here you will amplify and sequence five (5) RNA samples per research group. You will assess the SARS-CoV-2 genome sequences for their lineage assignments using online programs, put sequences into a global context, estimate the collection date based on genetic information, and describe mutations in the spike protein.
Part #2 is focusing on the 'dry-lab' by investigating a hypothetical incursion scenario in the so-called city Fantastica. You will combine genomic surveillance of SARS-CoV-2 with case interview data to trace the spread into of SARS-CoV-2 in the community and into high risk settings. We will provide you with real publicly available SARS-CoV-2 genome and fantasized case interviews. You will put these two together to trace the spread and suggest potential improvements in containment strategies with a focus on high risk settings.
This protocol describes the 'dry-lab' incursion scenario and its analysis for Part #2. This is a creative version of similar scenarios investigated during the official SARS-CoV-2 QAPs. The main objective of Part #2 of mini-research project #2 is to solidify concepts you learned in the lectures and tutorials around human biosecurity. We will combine fictional case interview information with a matching genomic dataset of SARS-CoV-2 genomes to investigate the incursion. Hopefully this will show you the power combining these two data types brings when compared to having only one or the other. In the larger perspective of the course, this hopefully illustrates to you that one needs to consider a multitude of perspectives and data types when operating in the biosecurity sector.
I had a lot of fun coming up with this incursion scenario and I hope you will enjoy working on it with your detective hat on. Of course this complete scenario is absolutely fictional. All the used SARS-CoV-2 sequences are publicly available on GISAID as described in this publication (Hall et. al. 2023).
The incursion scenario:
Imagine a city called Fantastica in the middle of the SARS-CoV-2 pandemic mid-2021 in a country where vaccination coverage and COVID-19 case numbers are very low. Fantastica is located on continental scale island nation and the international borders to this nation are highly regulated to prevent new COVID-19 cases from entering. The main public health measures employed to contain the spread of SARS-CoV2 are social distancing, mask wearing, mass testing, contact tracing, isolating and quarantining of confirmed cases and lock-downs.
Fantastica has two main areas of residence with A being the affluent North and B being the less well off South (Figure 1). These two areas are separated by a river. The main hospital is located right at the river.

In mid-2021 the city experiences its first COVID-19 case for a long time (Outbreak reference ID: Fantastica034), which was successfully contained in hotel quarantine for overseas travelers. The following months Fantastica experiences a larger COVID-19 outbreak that it aims to contain with lockdowns including restricting movements from 12 September 2021 till 20 November 2021. The public health unit achieves to sequence all SARS-CoV-2 genomes of all identified COVID-19 cases in this time frame.
In our simplified scenario we assume the following about SARS-CoV-2:
- Infectious period: 48 hrs before and after onset of symptoms.
- Asymptomatic cases can also cause forward transmission.
- Viral mutation rate: on average 0.5 mutations in each genome per infection cycle.
You are now provided with the following material to start your investigation and address the specific questions below. All the information is idealized and fictionalized.
Provided main material can be found here (ANU only) and listed below:
- An excel file (ContactTracingCaseInterviews) containing case interview information (not exhaustive and simplified) including the following columns:
- Outbreak Reference ID
- Area of Residence
- Age
- Date of symptom onset
- Date of specimen collection
- Symptoms
- Household contact
- Contact with known COVID case
- Case associated with known outbreak
- Locations of potential exposure
- Vaccination Status
- Overseas travel
-
A fasta file (FantasticaSARSCoV2Sequences) of SARS-CoV-2 genomes of all identified COVID-19 cases in Fatastica in the indicated study period (plus Fantastica034)
-
A PNG file (CleanedUpAlignmentAllSequencesTree) of the simple Neighbor-joining tree. You will generate the same tree in class.
What you need for the prac :
- A detective hat.
- Your computer.
- Pen and paper including different colored pens.
- A full working trial copy of Geneious https://manage.geneious.com/free-trial.
Specific questions to be addressed in the prac and your final report :
-
Describe the overall LargeClusterA1. What drove the transmission in this cluster? Was it contained successfully with public health measures such as testing, tracing, lockdowns and quarantine? Has the index case been clearly identified? Is the index case the likely first case in this cluster? Do you think most cases in this cluster have been identified? Explain your reasoning.
-
Describe the overall LargeClusterB1. What drove the transmission in this cluster? Was it contained successfully with public health measures such as testing, tracing, lockdowns and quarantine? Has the index case been clearly identified? Is the index case the likely first case in this cluster? Do you think most cases in this cluster have been identified? Include later appearing mini-clusters in your analysis: MC1: Fantastica063, Fantastica062, Fantastica058, Fantastica064 and Fantastica059.
MC2: Fantastica067, Fantastica068, Fantastica069, Fantastica072, Fantastica074, Fantastica070, Fantastica071, Fantastica073, Fantastica075In your analysis speculate how, these genetically linked subclusters could potentially physically linked (or not) to the main cluster?
-
What is a likely infection scenario for the family infection cluster containing Fantastica014, 016, 017?
-
How can you explain that case Fantastica019 is so distinct from all other cases?
-
Describe the case Fantastica033. What cluster does this case belong to? When could this case have caught COVID-19? Who could be the potential source cases? Where could this cause got infected. Explain your reasoning.
-
Describe the "HospitalCluster1 (non-COVID Ward)"? Was it a single incursion? What was the likely transmission chain? How could such an incursion scenario better managed? Explain your reasoning.
-
Describe the "ElderlyHomeClusterB"? Was it a single incursion? What was the likely transmission chain? How could such an incursion scenario be better managed? Explain your reasoning.
-
How would you have interpreted case Fantastica076 without contact tracing data? What does this case reveal for the strength and weakness of exclusive genomic surveillance?
-
There is one case that lied on the contact tracing form. Identify this case, its most likely source of infection, and who they passed it on.
For all these questions we are looking for the most parsimonious answers. The simplest and most plausible answers.
Before start
You must study the protocol carefully before you start. If anything is unclear post questions directly here on protocols.io.
Steps
Section I: Setup Genenious and import files into Geneious
Open up Geneious.
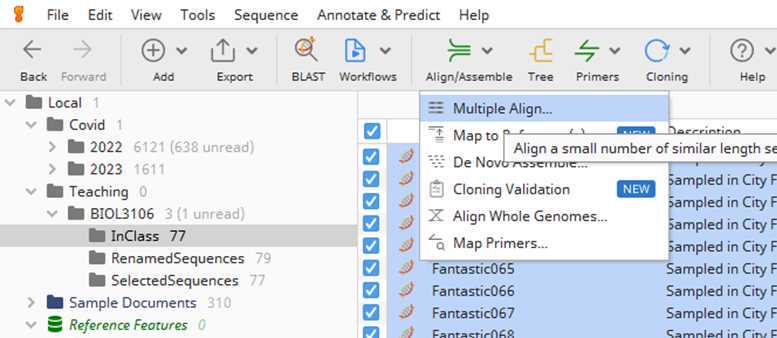
Section II: Generation of a multiple sequence alignment in Geneious
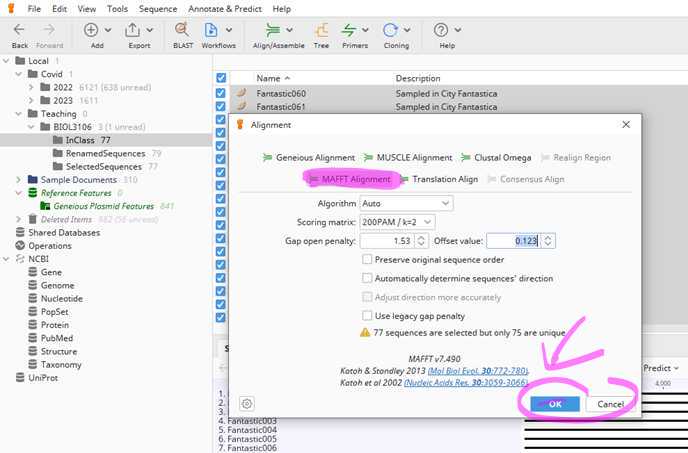
Great well done to set it all up. you are ready to generate your first whole genome alignment.
Now you have generated your first alignment. This aligns each base of all the genomes you selected to each other. I suggest you rename this alignment from "Nucleotide alignment" to something more meaningful.

With this specific display "Highlighting" setting "Disagreements" each of the black bars is a variation (mutation) compared to the consensus sequence.
Regions in the consensus sequence highlighted as red are not well covered in the aligned genomes. You can visualise this more when changing the "Highlighting" settings to something else. Play around and ask questions in class.
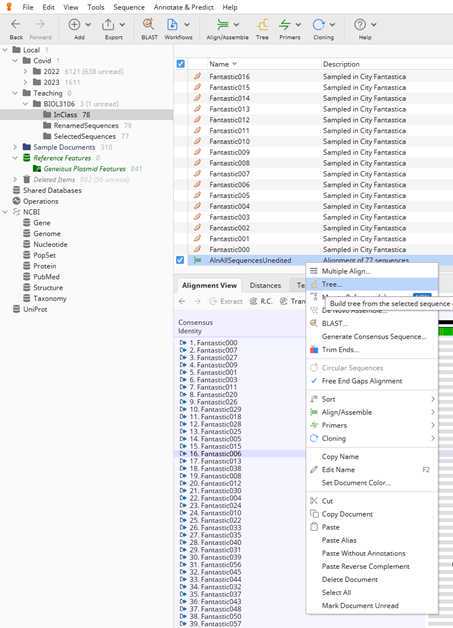
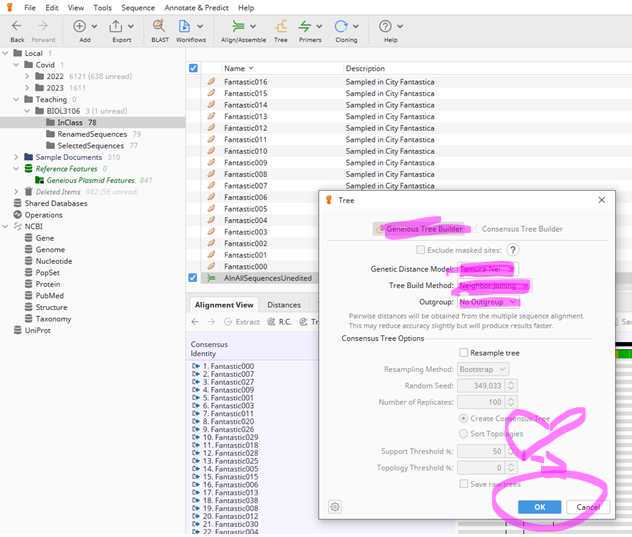
Section III: Building a very simple Neighbor-joining tree
The last thing for now that you need to do is the root your tree on the reference sequence "MN908947.3".
Select the "MN908947.3" by clicking on it and hit the "Root" bottom.
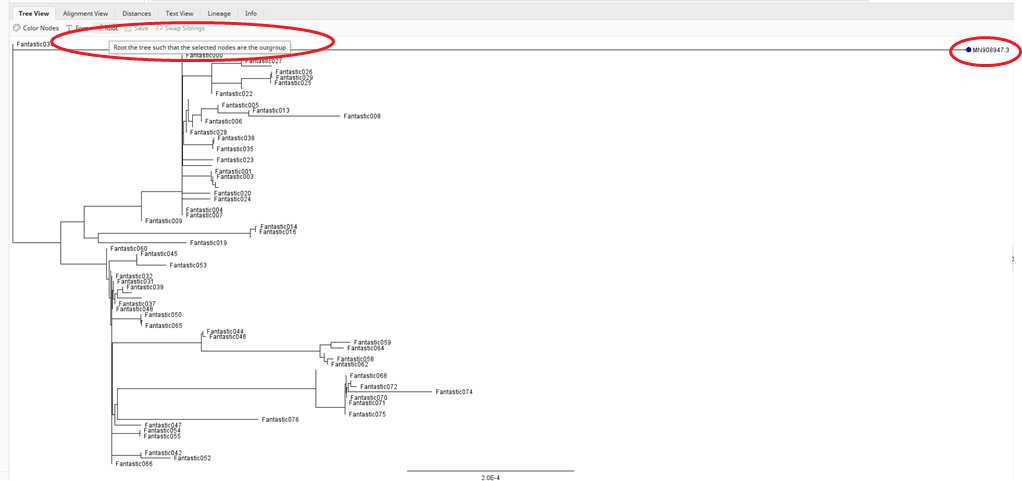
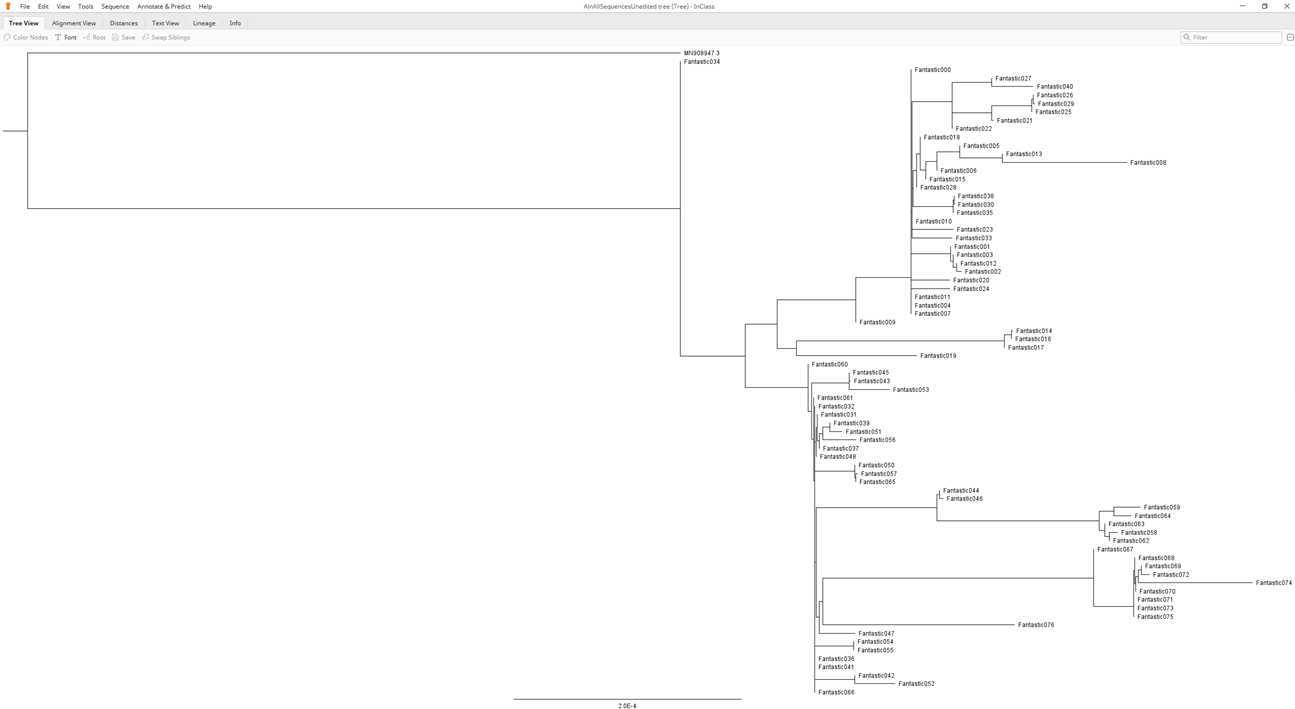
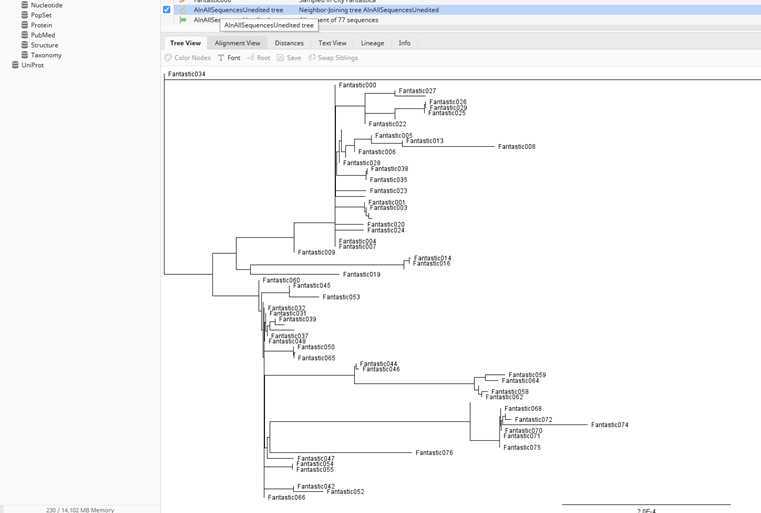
You have your first tree of all the sequences rooted with the original SARS-CoV-2 sequence. You can now overlay the case interview information to answer the questions for this part of the mini-research project #2. We will step through those in class as well. We will also explain how to interpret trees in more detail in class.
Importantly, you can generate these simple trees for subclusters as well (e.g. Hospital) if needed to address the questions better. This will be done by only selecting the sequences of interest for the alignment and tree building. Make sure to always include the reference and to root your tree on it.
Section IV: Overlay case interview information on top of the genetic data
So you have a skeleton (aka tree) of the genetic relationship of all samples and hence COVID-19 cases. We will provide you with a large print out copy as well.
Now you have to overlay the case interview data to answer the specific questions for Part #2 of the mini-research project #2. For this you can use the printed trees to draw on (with different coloured pens) or annotate it on your computer. Make sure to make good use of the sort and filter functions in Excel when going over the case interview data to ease your analysis.
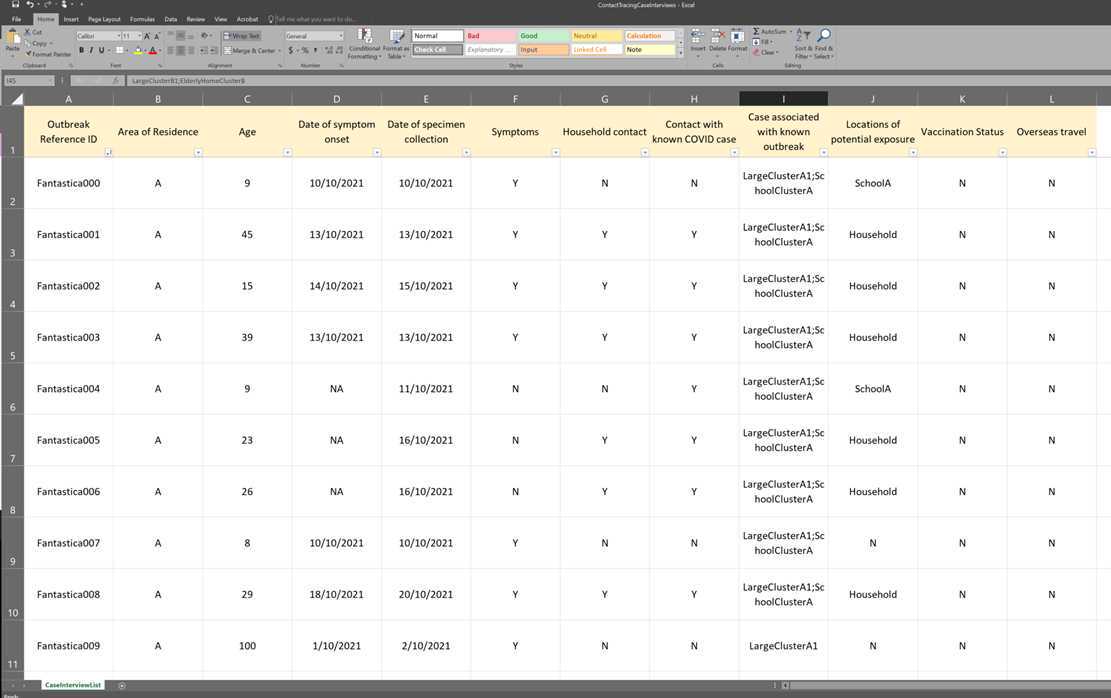
^^Screenshot of case interview data.
We will walk through some of these Part #2 questions in class guided by your questions.