SARS-CoV-2 Omicron detection RT-qPCR assay with BA.1 and BA.2/BA.3 differentitation
Nikita Yolshin, Andrey Komissarov, Kirill Varchenko
Abstract
Omicron lineage (B.1.1.529) – SARS-CoV-2 variant of concern designated on 26 November 2021 by WHO. This variant has a large number of mutations, some of which are concerning. There is already enough evidence that Omicron lineage connected with increased risk of reinfection with this variant and reduced neutralization by convalescent and vaccinated sera, as compared to other VOCs. Omicron subvariants includes several subvariants with strikingly different genetic characteristics: BA.1, BA.2, BA.3.
We developed RT-qPCR specific to omicron lineage with detemination of BA.1 and BA.2/BA.3 for epidemiological reasons. Omicron lineage B.1.1.529 and its sublineages BA.1 and BA.2/BA.3 have some indels that turned out to be a good target for its detection. Using annotations of amino acid substitutions and indels for more than 5 million samples in 1513 Pango lineages from the GISAID database available on 2021-12-03, relative frequencies of substitutions and indels by each lineage were calculated with Python v3.9.2 programming language, Pandas v1.3.5 and Numpy v1.3.5 tools. Using this data we found indels highly specific for BA.1 and BA.2/BA.3: ERS31del for Omicron (B.1.1.529) lineage, including BA.1, BA.2 and BA.3; and Ins214EPE specific only to BA.1. For these targets have been developed assays, that were then multiplexed.
Assay for detection BA.1 have been already published at protocols.io. Here we publish multiplex RT-qPCR detecting all Omicron subvariants with the opportunity to differentiate BA.1 and BA.2/BA.3.


Characteristics and analysis of specificity of Ins214EPE were already published in previous protocol. We only can add that limit of detection of the test is 1000 copies/ml
Here we describe features of newly introduced ERS31del assay.
10-fold serial dilutions of SARS-CoV-2 RNA of Omicron lineage were used to assess ERS31del assay amplification efficiency. The amplification efficiency was 105% (R2 = 0,99).

Developed RT-qPCR assay demonstrates high specificity. It was tested on 27 clinical samples (RNA extracted from oropharyngeal swabs) with previously characterized viruses belonging to 10 different SARS-CoV-2 lineages.
| A | B | C | D | E |
|---|---|---|---|---|
| P.1 | 35,35 | 15,45 | ||
| B.1 | 33,84 | 26,69 | ||
| B.1 | 24,26 | 26,98 | ||
| B.1.1.7 | 25,27 | 20,64 | ||
| B.1.1.7 | 30,54 | 28,05 | ||
| B.1.1.7 | 26,99 | 31,25 | ||
| AT.1 | 30,30 | 33,07 | ||
| AT.1 | 30,02 | 31,33 | ||
| AT.1 | 28,00 | 33,00 | ||
| B.1.1.523 | 29,12 | 27,09 | ||
| B.1.1.523 | 27,39 | 23,46 | ||
| B.1.1.523 | 30,02 | 27,09 | ||
| B.1.1.317 | 25,22 | 25,31 | ||
| B.1.1.317 | 27,58 | 22,71 | ||
| B.1.1.317 | 28,70 | 23,91 | ||
| B.1.617.2 | 26,66 | 34,03 | ||
| B.1.617.2 | 25,72 | 28,43 | ||
| B.1.617.2 | 26,78 | 25,42 | ||
| AY.122 | 23,96 | 28,95 | ||
| AY.122 | 31,16 | 35,05 | ||
| AY.122 | 24,19 | 34,93 | ||
| BA.2 | 26,50 | 19,64 | 21,07 | |
| BA.2 | 24,70 | 20,81 | 22,70 | |
| BA.1 | 27,98 | 22,20 | 23,57 | 22,08 |
| BA.1 | 27,08 | 24,93 | 27,16 | 24,65 |
| BA.1 | 25,19 | 27,48 | 29,83 | 25,57 |
Specific signal was detected only in samples with SARS-CoV-2 Omicron lineage RNA: BA.1 and BA.2 (confirmed by whole-genome sequencing). Specificity was additionally tested on clinical samples positive for other respiratory viruses from the collection of Smorodintsev Research Institute of Influenza - influenza A and B, human seasonal coronaviruses, hRSV, rhinoviruses - with no false-positive results.
Analytical sensitivity
Limit of detection of the test is 1000 copies/ml
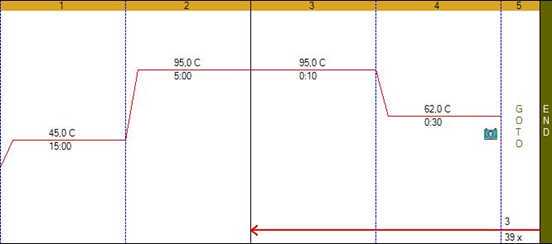
Steps
Order oligonucleotides with following sequences 5'->3':
| A | B | C |
|---|---|---|
| ORF1 | 2-SARS-CoV-2-ORF-1-F | AGAGCTATGAATTGCAGAC |
| ORF1 | 2-SARS-CoV-2-ORF-1-R | GGGAAATACAAAATTTGGACA |
| ORF1 | 2-SARS-CoV-2-ORF-1-P | FAM-AATTGGCAAAGAAATTTGACACCTTCA-BHQ1 |
| ERS31del | N31-33del F | GTTTGGTGGACCCTCAGATT |
| ERS31del | N31-33del R | CAAGACGCAGTATTATTGGGTAAAC |
| ERS31del | N31-33del P | HEX-AGTAACCAGAATGGTGGGGCGCG-BHQ1 |
| Ins214EPE | Ins214EPE F | ATATTCTAAGCACACGCCTATT |
| Ins214EPE | Ins214EPE R | GGCAAATCTACCAATGGTTCTA |
| Ins214EPE | Ins214EPE P | ROX-TGCGTGAGCCAGAAGATCTCCCT-BHQ2 |
| Human RP | RP-CB-F | AGATTTGGACCTGCGAGCG |
| Human RP | RP-CB-R | GAGCGGCTGTCTCCACAAGT |
| Human RP | RP-CB-P | Cy5-TTCTGACCTGAAGGCTCTGCGCG-BHQ2 |
SARS-CoV-2-ORF-1, ERS31del and Ins214EPE assays were developed in Smorodintsev Research Institute of Influenza (St.Petersburg, Russia). RP assay was developed by CDC and is used to check the presence of human RNA in clinical samples (internal control). Please, check compatibility of fluorescence dyes with your PCR machine and reagents
Prepare oligonucleotides mix for 100 reactions of multiplex RT-qPCR
| A | B | C |
|---|---|---|
| 2-SARS-CoV-2 F | 5 | 100 pmol/mcl |
| 2-SARS-CoV-2 R | 5 | 100 pmol/mcl |
| 2-SARS-CoV-2 P | 2,5 | 100 pmol/mcl |
| N31-33del F | 2,5 | 100 pmol/mcl |
| N31-33del R | 2,5 | 100 pmol/mcl |
| N31-33del P | 1,25 | 100 pmol/mcl |
| Ins214 F | 10 | 100 pmol/mcl |
| Ins214 R | 10 | 100 pmol/mcl |
| Ins214 P | 5 | 100 pmol/mcl |
| RP F | 5 | 100 pmol/mcl |
| RP R | 5 | 100 pmol/mcl |
| RP P | 5 | 100 pmol/mcl |
| water | 61,25 | |
| TOTAL | 120 |
Briefly vortex and centrifuge reagents before use.
Use oligonucleotides mix from previous step.
Prepare the PCR reaction mixture following the specifications below:
| A | B |
|---|---|
| 25x RT-qPCR enzyme mix | 1 |
| 2x RT-qPCR Buffer | 12,5 |
| nuclease-free water | 5,3 |
| oligonucleotides mix | 1,2 |
| RNA template | 5 |
We used Biomaster qRT-PCR mix (Biolabmix, Russia)
Interpretation of the results:
detection of fluorescence of FAM probe (2-SARS-CoV-2-ORF-1 probe) means the presence of SARS-CoV-2 RNA in the sample,
detection of fluorescence in HEX channel (ERS31del probe) means the presence of B.1.1.529/omicron (BA.1/BA.2/BA.3) RNA,
detection of fluorescence in ROX channel (Ins214EPE probe) means the presence of BA.1 RNA.
detection of fluorescence in Cy5 channel (RP probe)
| A | B | C | D | E |
|---|---|---|---|---|
| FAM | HEX | ROX | Cy5 | |
| + | - | - | + | SARS-CoV-2 positive, non-omicron lineage |
| + | + | - | + | BA.2 lineage |
| + | + | + | + | BA.1 lineage |
| - | - | - | + | SARS-COV-2 negative sample |
| - | - | - | - | Test invalid (no human template) |
swab