Resource 7: rEV immunophenotyping
Joshua A Welsh, Sean M Cook, Jennifer Jones, Joanne Lannigan, Vera A. Tang
Disclaimer
This protocol summarizes key steps for a specific type of method, which is one of a collection of methods and assays used for EV analysis in the NCI Translational Nanobiology Section at the time of submission of this protocol. Appropriate use of this protocol requires careful, cohesive integration with other methods for EV production, isolation, and characterization.
Abstract
Flow cytometry (FCM) is a common extracellular particles (EPs), including viruses and extracellular vesicles (EVs), characterization method. Frameworks such as MIFlowCyt-EV exist to provide reporting guidelines for metadata, controls, and
data reporting. However, tools to optimize FCM for EP analysis in a systematic and quantitative way are lacking. Here, we demonstrate a cohesive set of methods and software tools that optimize FCM settings and facilitate cross-platform comparisons for EP studies. We introduce an automated small particle optimization (SPOT) pipeline to optimize FCM fluorescence and light scatter detector settings for EP analysis and leverage quantitative FCM (qFCM) as a tool to further enable FCM optimization of fluorophore panel selection, laser power, pulse statistics, and window extensions. Finally, we demonstrate the value of qFCM to facilitate standardized cross-platform comparisons, irrespective of instrument configuration, settings, and sensitivity in a cross-platform standardization study utilizing a commercially available EV reference material.
Steps
Sample Preparation
Briefly centrifuge the rEVs 100x g,4°C before opening
Add 100µL of 4°C deionized water. Pipette up and down to mix.
Dilute the reconstituted rEVs 1 in 5 in PBS for staining* Take 50µL of the rEV stock and add 200µL of PBS.
Prepare the antibody dilutions for a 2x staining concentration of each of the antibodies to be used in the antibody titration. See the example below for sample calculations prepared for a staining concentration of 2 µg/ml.
| A | B | C | D | E | F |
|---|---|---|---|---|---|
| AB stock concentration (µg/ml) | 2x staining conc (µg/ml) | Stock volume for 2x staining conc (µL) | Total volume for 2x staining conc (µL) | Volume of PBS to be added (uL) | |
| anti-Human CD81-PE | 120 | 4 | 2 | 60 | 58 |
| anti-Human CD81-APC | 200 | 4 | 2 | 100 | 98 |
| anti-Human CD81-PB | 300 | 4 | 2 | 150 | 148 |
Sample calculations prepared for a staining concentration of 2 µg/ml. All Antibodies clones are 5A6.
Samples will now be prepared using the following plate map.
| A | B | C | D | E | F | G | H |
|---|---|---|---|---|---|---|---|
| CD81 PE | CD81 APC | CD81 Pacific Blue | CD81 PE | CD81 APC | CD81 Pacific Blue | ||
| rEV | rEV | rEV | PBS | PBS | PBS | ||
| 0 µg/mL | A1 | A2 | A3 | A4 | A5 | A6 | |
| 0.0625 µg/mL | B1 | B2 | B3 | B4 | B5 | B6 | |
| 0.125 µg/mL | C1 | C2 | C3 | C4 | C5 | C6 | |
| 0.25 µg/mL | D1 | D2 | D3 | D4 | D5 | D6 | |
| 0.5 µg/mL | E1 | E2 | E3 | E4 | E5 | E6 | |
| 1 µg/mL | F1 | F2 | F3 | F4 | F5 | F6 | |
| 2 µg/mL | G1 | G2 | G3 | G4 | G5 | G6 | QbSure Beads |
| PBS | PBS | PBS | PBS | PBS | PBS | 100 nm PS NIST-Traceable Beads | |
In a 96-well V-bottom plate, add 10µL 1:5 rEV solution to wells A1-A3
Add 10µL DPBS to wells A1-A3 and reverse pipet to mix. These wells will serve as the rEV controls
Add 20µL DPBS to wells A4-A6. These wells will serve as buffer only controls
In the same 96-well V-bottom plate from , add 10µL 1:5 rEV solution to wells B1-G3. Add 10µL DPBS to wells A1-G1. Add 10µL from the PE working solution tubes to wells B1-G1, add 10µL from the APC working solution tubes to wells B2-G2, add 10µL from the PB working solution tubes to wells B3-G3.
In the same 96-well V-bottom plate from , add 10µL from the corresponding PE working solutions tube to wells B4-G4. Add 10µL from the corresponding APC working solutions tube to wells B5-G5. Add 10µL from the corresponding PB working solutions tube to wells B6-G6. These wells will serve as the DBPS+AB control wells.
Cover and incubate this plate for 0h 30m 0s at RT.
Using a separate 96-well V-bottom plate, add 199µL DPBS into wells A1-G6. Pipet 1µL from the incubation plate into the new plate in the same wells and reverse pipette to mix.
Add 200µL DPBS into wells A7-G7. These wells will help reduce sample carryover after each rEV+AB combination is acquired.
Re-cover incubation plate and let incubate ON at RT to repeat measurements next day as directed in the following steps.
Label as FACS tube as 'QbSure', and add 500µL DPBS. Vortex the QbSure beads for 5 sec and add 3 drops QbSure beads into FACS tube.
Cytometer Setup
Setup cytometer to acquire at FCMPASS output gains for both light scatter and fluorescent detectors as determined by the FCMPASS detector optimization module outputs. For the Aurora, an optimal gain template is returned to import into the Spectraflo software.
On the Cytek Aurora, set window extension to 0. On the CytoFLEX platform turn on 'High Acquisition Mode'.
Set the cytometer triggering threshold to the violet SSC parameter and run a DPBS control first to ensure the cytometer is clean and thresholds/event rates have remained unchanged. The background event rate should be ~1000 events/sec. All samples should be acquired with the lowest flow rate, typically ~10-15 µL min-1. -1.
Acquire all wells for at least 60 sec.
In an open well, run the 100 nm polystyrene NIST beads at the same settings as the rEVs (diluted in DPBS to a concentration of 5E6 p/mL) until at least 10000 bead events have been acquired.
In an open well, add 200µL from the QbSure FACS tube and collect 10,000 bead events at the same settings as the rEVs.
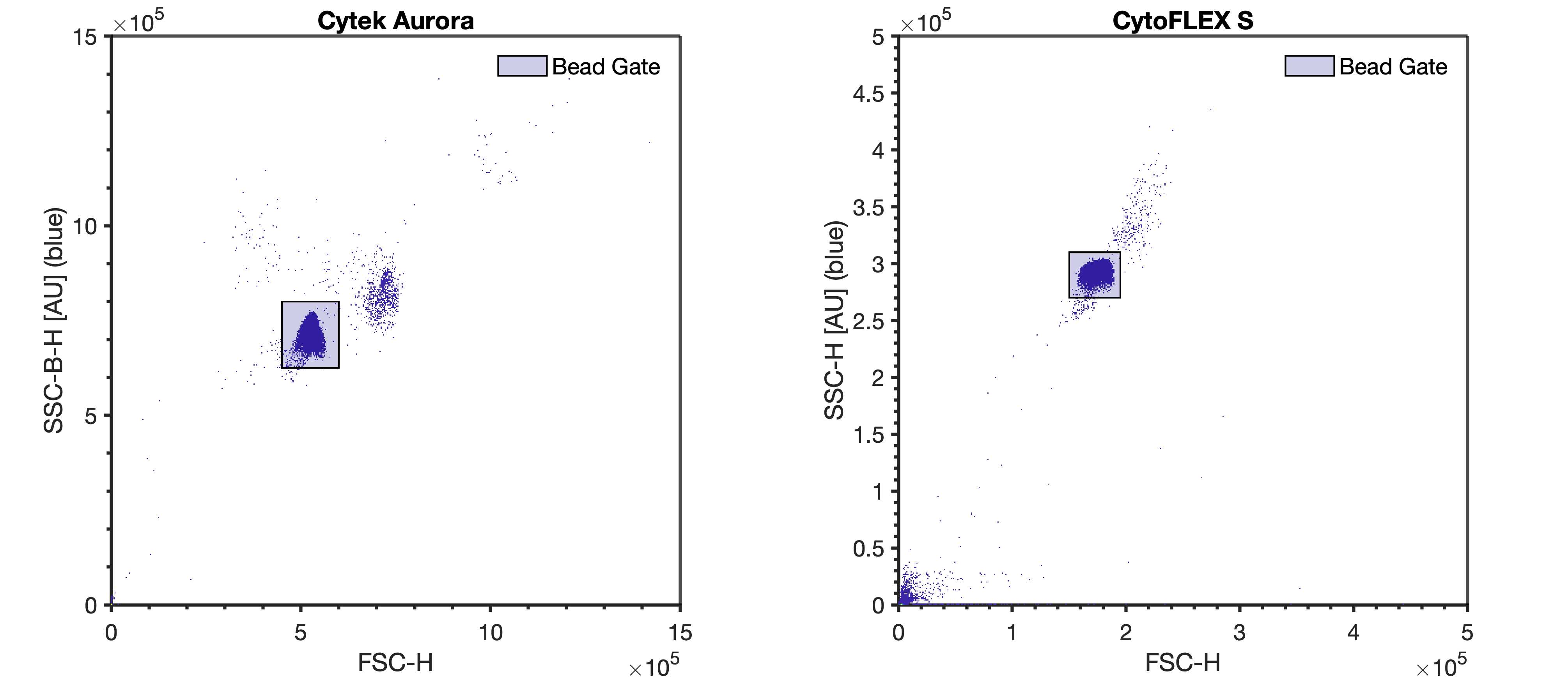
If calibration of data into standard units is desired, the 100 nm polystyrene NIST bead and QbSure beads can be used to calibrate data by following the FCMPASS experiment calibration protocols.
Don't forget to repeat the acquisition for the samples incubating ON.

