Resource 4: rEV Serial Dilution
Joshua A Welsh, Sean M Cook, Jennifer Jones, Joanne Lannigan, Vera A. Tang
Disclaimer
This protocol summarizes key steps for a specific type of method, which is one of a collection of methods and assays used for EV analysis in the NCI Translational Nanobiology Section at the time of submission of this protocol. Appropriate use of this protocol requires careful, cohesive integration with other methods for EV production, isolation, and characterization.
Abstract
Flow cytometry (FCM) is a common extracellular particles (EPs), including viruses and extracellular vesicles (EVs), characterization method. Frameworks such as MIFlowCyt-EV exist to provide reporting guidelines for metadata, controls, and
data reporting. However, tools to optimize FCM for EP analysis in a systematic and quantitative way are lacking. Here, we demonstrate a cohesive set of methods and software tools that optimize FCM settings and facilitate cross-platform comparisons for EP studies. We introduce an automated small particle optimization (SPOT) pipeline to optimize FCM fluorescence and light scatter detector settings for EP analysis and leverage quantitative FCM (qFCM) as a tool to further enable FCM optimization of fluorophore panel selection, laser power, pulse statistics, and window extensions. Finally, we demonstrate the value of qFCM to facilitate standardized cross-platform comparisons, irrespective of instrument configuration, settings, and sensitivity in a cross-platform standardization study utilizing a commercially available EV reference material.
Steps
rEV Reconstitution
Briefly centrifuge 100x g,4°C rEVs before opening.
Add 100µL of 4°C deionized water.
Pipet up and down to mix.
Aliquot 20µL into low-binding Eppendorf tubes.
Equipment
| Value | Label |
|---|---|
| Low Protein Binding Microcentrifuge Tubes | NAME |
| Microcentrifuge Tubes | TYPE |
| Thermo Scientific | BRAND |
| 90410 | SKU |
Store at 2-4°C for up to one day (up to 2 weeks at -80°C).
rEV Dilutions
Label 10 low-binding Eppendorf tubes:
| A | B |
|---|---|
| Label | Dilution |
| 1 | 50 |
| 2 | 100 |
| 3 | 200 |
| 4 | 400 |
| 5 | 800 |
| 6 | 1600 |
| 7 | 3200 |
| 8 | 6400 |
| 9 | 12800 |
| 10 | 25600 |
Add 980µL of DPBS (preferably 0.1µm filtered) to the tube 1 from .
Add 500µL of DPBS (preferably 0.1 µm filtered) to the remaining tubes.
Add 20µL of the stock reconstituted rEVs to the tube labeled 1 ( ), pipet up and down to mix.
Pipette 500µL of the 1:50 dilution in tube 1 into the 1:100 dilution in tube 2, pipet up and down to mix.
Repeat the above step for each of the remaining dilution tubes until the serial dilution has been completed for all dilutions.
Keep on ice during acquisition.
Flow Cytometer Acquisition
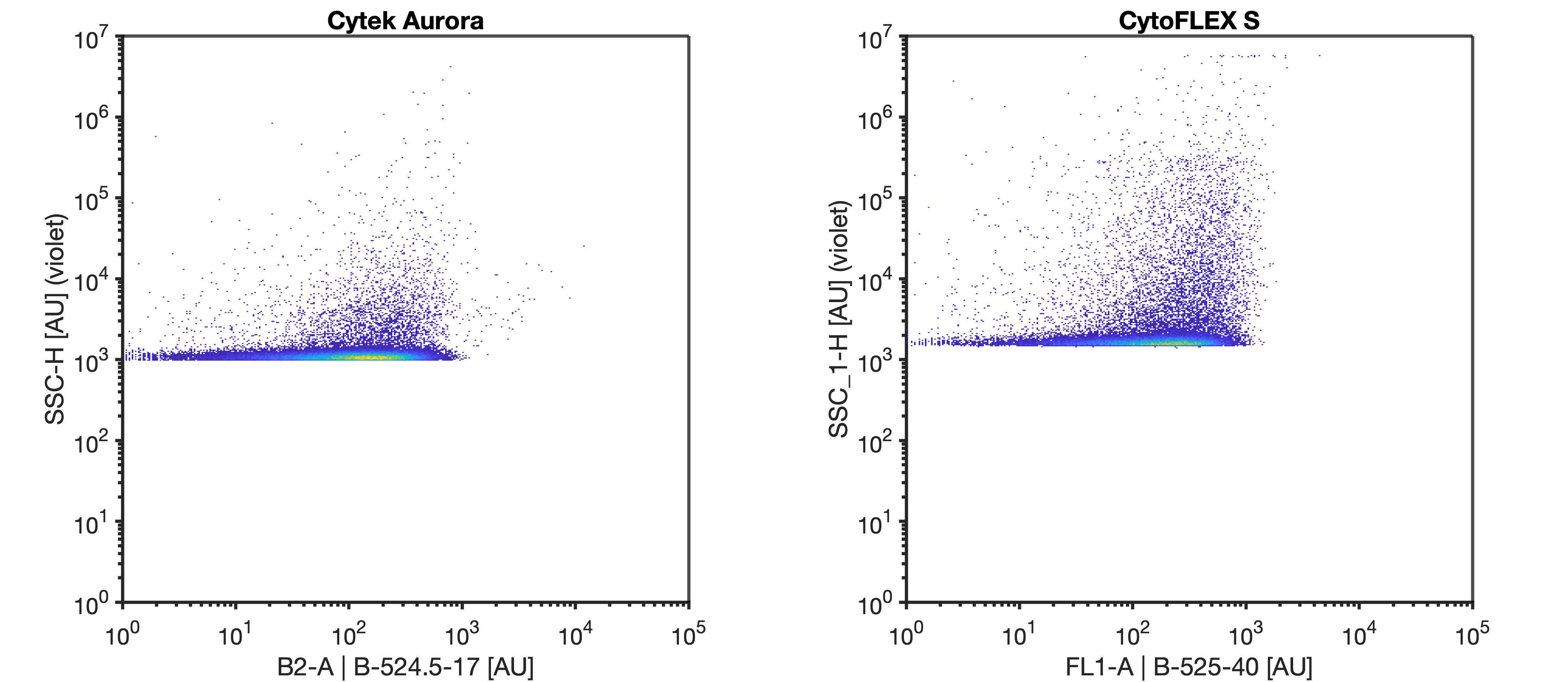
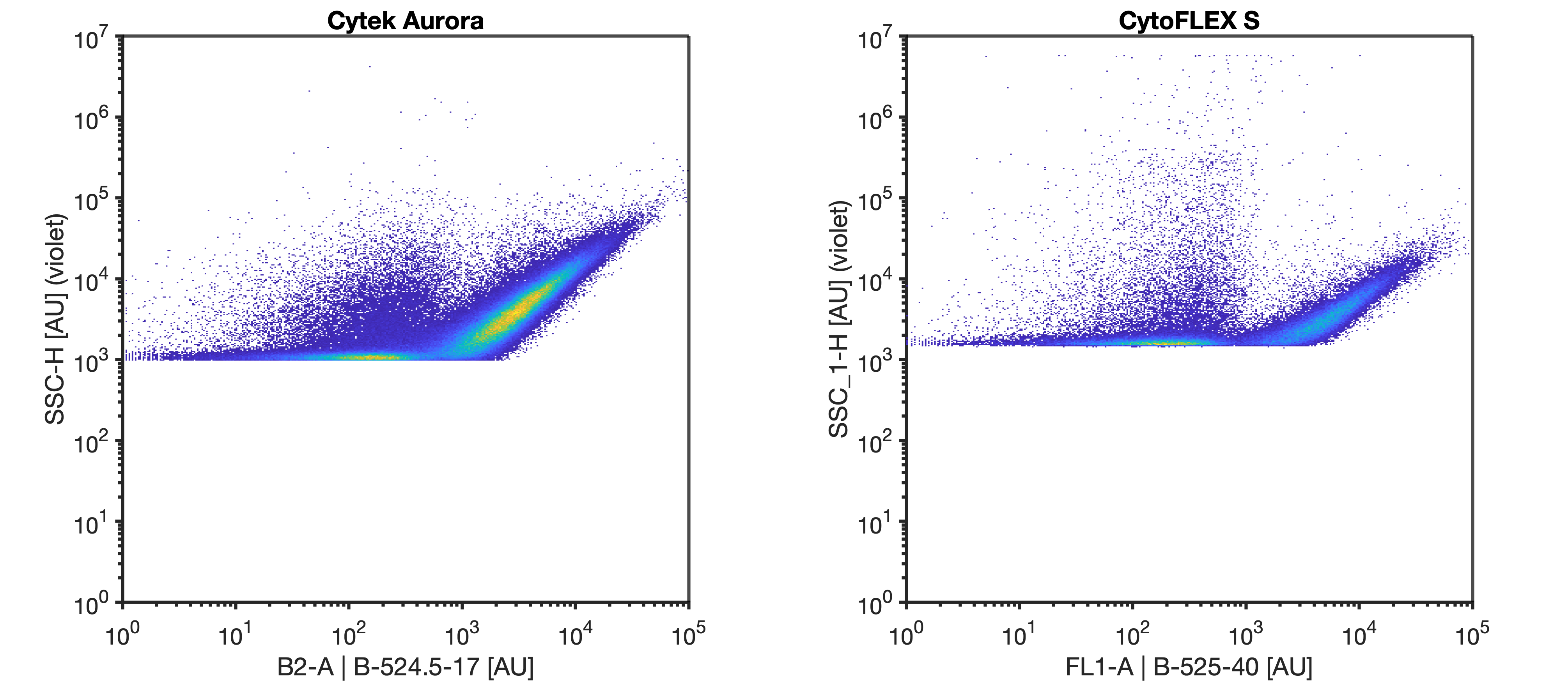
Use the instrument settings that were established from the previous gain incrementation and scatter calibration resources. On the Aurora, set the window extension to 0. On the CytoFLEX, use the high acquisition mode. Collect for at least 1 minute at a low flow rate with a 30 second recording delay if using plate mode.
Set your instrument to trigger on the most sensitive side scatter channel at the FCMPASS optimal gain output as determined by the FluoSpheres scatter voltration. Set the threshold in the noise such that the event rate is ~1000 events/sec when running DPBS.
Resource 1: Scatter Detector Setting Incrementation for FCMPASS
Set the FITC parameter to the FCMPASS optimal gain output as determined by the QbSure fluorescent voltration.
Resource 2: Fluorescence Detector Setting Incrementation for FCMPASS
Run a DPBS sample in a FACS tube to make sure there are no carryover events; there should be no events in the B1 and B2 channel.
Label as FACS tube as 'QbSure', and add 500µL DPBS. Vortex the QbSure beads for 5 sec and add 3 drops QbSure beads into FACS tube. Run QbSure beads on low and collect 10,000 bead events.

Run the 100 nm polystyrene NIST beads at the same settings as the rEVs (diluted in DPBS to a concentration of 5E6 p/mL) until at least 10000 bead events have been acquired.
If calibration of data into standard units is desired, the 100 nm polystyrene NIST bead and QbSure beads can be used to calibrate data by following the FCMPASS experiment calibration protocols.