Ratiometric Single-Molecule FRET Measurements to Probe Conformational Subpopulations of Intrinsically Disordered Proteins
Irem Nasir, Irem Nasir, Emily P. Bentley, Emily P. Bentley, Ashok A. Deniz, Ashok A. Deniz
conformational dynamics
conformational landscapes
intrinsically disordered proteins
single-molecule fluorescence
smFRET
Abstract
Over the past few decades, numerous examples have demonstrated that intrinsic disorder in proteins lies at the heart of many vital processes, including transcriptional regulation, stress response, cellular signaling, and most recently protein liquid-liquid phase separation. The so-called intrinsically disordered proteins (IDPs) involved in these processes have presented a challenge to the classic protein “structure-function paradigm,” as their functions do not necessarily involve well-defined structures. Understanding the mechanisms of IDP function is likewise challenging because traditional structure determination methods often fail with such proteins or provide little information about the diverse array of structures that can be related to different functions of a single IDP. Single-molecule fluorescence methods can overcome this ensemble-average masking, allowing the resolution of subpopulations and dynamics and thus providing invaluable insights into IDPs and their function. In this protocol, we describe a ratiometric single-molecule Förster resonance energy transfer (smFRET) routine that permits the investigation of IDP conformational subpopulations and dynamics. We note that this is a basic protocol, and we provide brief information and references for more complex analysis schemes available for in-depth characterization. This protocol covers optical setup preparation and protein handling and provides insights into experimental design and outcomes, together with background information about theory and a brief discussion of troubleshooting. © 2020 by John Wiley & Sons, Inc.
Basic Protocol : Ratiometric smFRET detection and analysis of IDPs
Support Protocol 1 : Fluorophore labeling of a protein through maleimide chemistry
Support Protocol 2 : Sample chamber preparation
Support Protocol 3 : Determination of direct excitation of acceptor by donor excitation and leakage of donor emission to acceptor emission channel
INTRODUCTION
The purpose of this article is to outline a general protocol for implementing single-molecule fluorescence (specifically, Förster resonance energy transfer, or FRET) techniques to study the conformational populations, binding interactions, and dynamics of proteins that do not possess a well-defined tertiary structure—so-called intrinsically disordered proteins (IDPs). The lack of structure allows IDPs to sample a plethora of structural states rapidly at any given time. The resulting interactions of IDPs are conceptually different from those of folded proteins: IDPs are generally involved in promiscuous interactions with moderate affinity, although there are a few exceptions (Berlow, Dyson, & Wright, 2017; Borgia et al., 2018). Due to the flexibility of IDPs, and their resulting ability to sample many different conformations, general structural characterization methods only yield information that is an average of the structural ensembles present in solution. To gain information about biological function, however, it is of utmost interest to resolve distinct populations within an ensemble, as well as the fluctuations between them. Single-molecule fluorescence methods are particularly suited to IDP research, as they allow only a handful of proteins to be detected at the same time. By reducing the number of molecules detected, single-molecule fluorescence methods allow the detection of subpopulations (if there are any) for an IDP in virtually any experimental buffer, either in isolation or in complex with their binding partners. Population-specific dynamics and the fluctuations between distinct populations can also be resolved. Overall, this method can guide us to understand the cause and the consequence of functionally important reactions at an extremely high level of detail, in terms of paths and intermediate states.
These protocols are focused on diffusion-based single-molecule Förster resonance energy transfer (smFRET). As the implementation of this method has the prerequisite that proteins must be labeled with donor and acceptor fluorescent moieties, we explain in Strategic Planning the most common labeling strategy together with details that must be attended to. In the same section, we also briefly discuss the key components of the optical detection instrument required before starting an experiment, although a detailed description of how to assemble this instrumentation is outside the scope of this protocol. For the interested reader, we provide Key References that can be consulted for more complete details about optical instrumentation, analysis, and labeling. We then continue with a detailed protocol for smFRET. The basic protocol includes subprotocols for data acquisition that will eventually yield more detailed characteristics of the system. We also provide support protocols for preparing a sample chamber and for measuring critical parameters affected by the optical setup and the sample of choice.
STRATEGIC PLANNING
Fluorophore Labeling Considerations
In single-molecule fluorescence measurements, one or more extrinsic fluorophores are generally attached to IDPs in a site-specific, covalent manner. In this section, we will briefly discuss different site-specific labeling schemes, labeling site design, controls, and choice of fluorophores. Furthermore, a method for labeling a protein is provided in Support Protocol 1.
The most common way to attach fluorophores to proteins is by maleimide chemistry. In this approach, a free thiol group (i.e., cysteine) on the protein is reacted with a fluorophore that is derivatized with a maleimide group, forming a thioether bond. It is a selective reaction around pH 7.0, though above pH 8.0, primary amines are also labeled (Nanda & Lorsch, 2014).
Cysteine (Cys) is the only natural protein amino acid with a free thiol group, which makes it suitable for this site-specific reaction. Moreover, Cys is a rare amino acid, and its underrepresentation can be used to advantage for site-specific labeling (Lodish, Berk, Zipursky, & Matsudaira, 1995). Cys can be introduced with site-directed mutagenesis methods for proteins to be studied in single-molecule fluorescence applications. If the protein has multiple accessible Cys residues, all but the ones to be labeled need to be replaced by unreactive amino acids. Disulfides (especially structurally important ones) can be problematic for this strategy. Additionally, labeling of functional Cys residues should not be performed for experiments probing function. Jensen et al. has demonstrated that it is possible to site-specifically target the free Cys residue of an ionotropic glutamate receptor that contains a disulfide bond, but the mechanism behind this reaction remains unclear (Jensen, Sukumaran, Johnson, Greger, & Neuweiler, 2011). To increase the reactivity of Cys, the residue can be appended in a peptide tag in between two basic residues (Lobocki et al., 2017).
Site-specific unnatural amino acid (uAA) incorporation in conjunction with click chemistry labeling has also become a popular approach for labeling proteins of interest for single-molecule fluorescence studies. This approach takes advantage of orthogonal tRNA and aminoacyl tRNA synthetase pairs that can recognize a specific uAA and incorporate it into the protein sequence in response to the presence of a unique codon. In bacteria, the amber stop codon is commonly used for this purpose due to its low abundance. Depending on the functionality of the uAA, the fluorophore, with a suitable click-chemistry handle, is reacted with the protein (Brustad, Lemke, Schultz, & Deniz, 2008; Lee et al., 2016; Lemke, 2011; Milles et al., 2012). Types of labeling chemistries, orthogonal tRNA/aminoacyl tRNA synthetase pairs for different organisms, and various applications have been reviewed in detail elsewhere (Nikić & Lemke, 2015).
Engineering of fluorescent probe sites for single-molecule fluorescence experiments must be accompanied by controls that ensure labeling does not overly disrupt the structure and/or function of the protein of interest. For IDPs for which no structural information is available, sequence alignment can provide information about conserved amino acids, which are likely to be vital for function. For IDPs that participate in binding reactions, or that fold as a result of binding, care should be taken to avoid modifying binding interfaces or the cores of structure-forming units. In folded regions, the surface residues should be preferred when the structure is available, because not only mutation to Cys but also fluorophore incorporation may have a destabilizing effect on the protein's structural core. The amino acids alanine, valine, serine, and threonine are preferred for replacing Cys. The stabilizing or destabilizing effect of the mutation can be quantitatively assessed by a variety of computational methods (Thiltgen & Goldstein, 2012). Finally, simple structural controls, such as assessing the difference between wild-type and mutant/labeled proteins’ circular dichroism spectrum, should be performed in conjunction with functional controls.
Inferring the distance between two fluorophores requires using two different fluorophores with overlapping donor emission and acceptor absorption spectra. This overlap is one of the deterministic factors of the Förster distance (R 0)—that is, the distance at which the FRET efficiency is 50%. Monitoring the distance between a donor and an acceptor therefore depends heavily on the specific donor-acceptor pair's R 0. When designing an smFRET experiment, a suitable donor-acceptor pair choice should be made by taking into consideration two factors: (i) the donor absorption spectrum should overlap with the excitation laser wavelength, and (ii) the sequence separation, and the changes thereof, must be within the dynamic range of FRET efficiency values that can be probed with the chosen donor-acceptor pair. It is admittedly nontrivial to design fluorophore attachment sites without a priori knowledge of approximate distances for IDPs; however, as a starting guess, one can predict the theoretical distance for an extended chain, which will be an overestimate for a native IDP in most cases.
Common fluorophores can be small-molecule dyes or chromophores embedded in proteins (e.g., visible-range-fluorescent proteins, intrinsically fluorescent amino acids, or fluorescent cofactors). Many commercially available small-molecule dyes with different reactive handles fulfill the primary requirements for suitability for single-molecule measurements: high quantum yield and photostability. These fluorophores span a wide range of excitation wavelengths, which is a crucial factor to consider in designing two- and multicolor experiments. Fluorophores vary in their relevant physical properties, such as net charge and hydrophobicity; therefore, they should be chosen to be the least disruptive possible for downstream applications. For example, if the proteins in question interact via charge-charge interactions, then the influence of the fluorescent tag's charge must be characterized. Controls that investigate the effect of fluorophore incorporation should be implemented depending on the dominant force of protein-protein interactions and physical property of the fluorophore.
Optical Setup
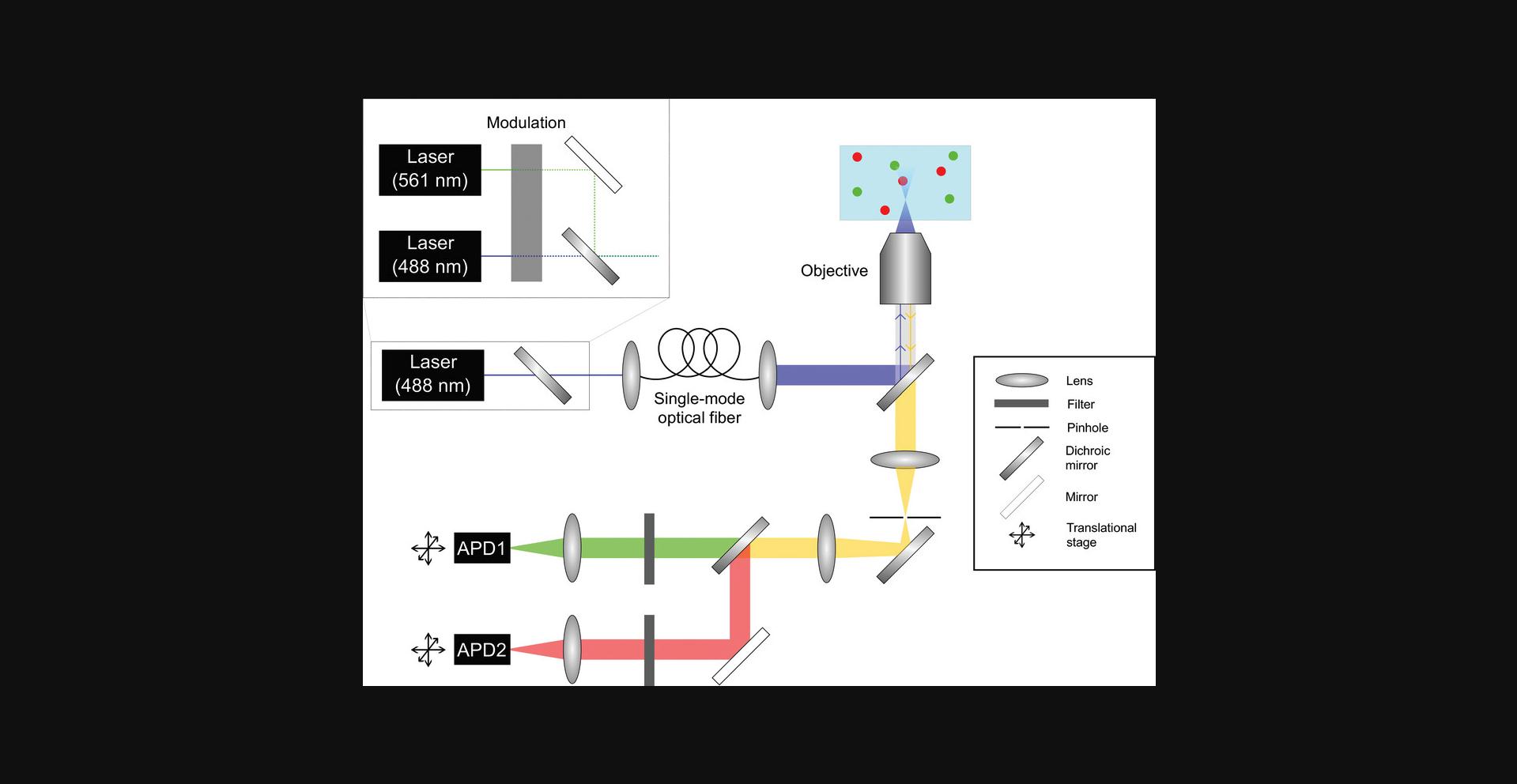
A confocal detection scheme for freely diffusing single molecules generally must satisfy the following criteria: (i) a light source for the excitation of fluorescent molecules in solution (typically a laser); (ii) a sub-femtoliter focal volume that, together with sub-nanomolar sample concentrations, permits the detection of only single molecules; (iii) highly sensitive, single-photon counting detectors that allow the number of emitted photons to be determined. Below and in Figure 1, we describe key elements of a simple layout for a confocal microscope–based single-molecule setup, though we note that variants and other layouts can be and have been used. Because the setup requires numerous components to build, a protocol for instrument building is outside the scope of this protocol. However, a short list of some key elements of the setup are provided in Table 1, and we provide references (see Key References section) with more detailed descriptions of instrumentation development that may also be helpful for the interested reader.
| Component | Product information/comments |
|---|---|
| Laser | 488-nm laser (CrystaLaser) or other stable laser emitting at the desired excitation wavelength (here, 488 nm) and providing at least a few hundred microwatts at the sample. |
| Inverted microscope | Axiovert 200 (Carl Zeiss USA). A stable inverted microscope is used to hold the sample and objective, along with defining parts of the excitation and emission light paths and optics. |
| Objective | C-Apochromat, 1.2 W, 40× (Carl Zeiss USA). |
| Photodetectors | SPCM-AQR-14 Photon Counting Modules (Perkin Elmer, now Excelitas). Photon detectors from PicoQuant (PDM) may also be considered. A MultiHarp 150 (PicoQuant) and associated software can be used to record and analyze the signals. |
| Dichroic mirrors |
Excitation: Di02-R488-25 × 36 (Semrock/IDEX) or other similar dichroic mirror for 488-nm lasers, making sure that the acceptor fluorescence is also transmitted. This dichroic mirror is placed in the microscope turret. Emission: FF580-FDi01-25 × 36 (Semrock/IDEX) or other similar dichroic mirror for splitting donor and acceptor (Alexa Fluor 488 and Alexa Fluor 594) photons. As needed, compare the spectra of the dichroic mirror and of your chosen donor and acceptor dyes, to ensure that the mirror splits the donor/acceptor photons as cleanly as possible. |
| Filters |
Alexa Fluor 488: FF01-530/55-25 (Semrock/IDEX) or other similar band-pass filter. Check that it transmits much of the donor emission, while avoiding acceptor emission and Raman scattering (∼585 nm for a 488-nm laser line). Alexa Fluor 594: FF01-593/LP-25 (Semrock/IDEX) or other similar long-pass filter. Check that it transmits much of the acceptor emission while avoiding donor emission. |
For the freely diffusing smFRET experiments used in this protocol, a laser beam is directed to an inverted microscope using mirrors and lenses for beam shaping. Neutral-density (ND) filters (variable or step-variable) can be included to vary excitation intensity on the optical table rather than at the laser source. Once in the microscope, the laser beam is focused into the sample using an objective with a high numerical aperture (NA) for excitation of the fluorophores in solution. Emitted fluorescence from one or several different fluorophores is collected through the same objective. Most of the stray excitation light that would otherwise saturate the detectors and overwhelm molecular signals is reflected by an (excitation) dichroic mirror; thus, mainly fluorescence emission light is transmitted. The light is then focused by a lens and passes through a pinhole placed at the focal plane, thereby reducing out-of-focus (background) signals and resulting in a sub-femtoliter detection volume. Outcoming light from the microscope is directed to another (emission) dichroic mirror to spatially separate donor and acceptor emission photons. These photons are filtered once more with spectral filters to minimize the background signal that might have leaked from excitation light. Finally, lenses are used to focus each photon stream onto avalanche photodiode (APD) photon-counting modules that provide excellent detection sensitivity. Detected photons are converted to electronic signals, which are directed to a counting device (the MultiHarp 150, from PicoQuant), and associated software (also from PicoQuant) can be used to conveniently record photon counts/information. Photon counting data are then processed further depending as appropriate for the application. We discuss simple data processing routines in the Data Analysis sections of respective protocols.
Basic Protocol: RATIOMETRIC smFRET DETECTION AND ANALYSIS OF IDPs
Measurement of the distance between two fluorophores (donor and acceptor) and changes thereof on an IDP can be achieved by smFRET. The strong distance dependence of smFRET can be used to distinguish distances in roughly the 30- to 70-Å range for typical single-molecule dyes. The output of a simple smFRET detection scheme is the FRET efficiency (E FRET) distribution, which is a measure of the distance between a donor and an acceptor molecule attached to an IDP of choice. Although the absolute distances are complex to obtain, it is possible to evaluate the changes in FRET efficiency in a ratiometric manner, for instance, upon folding of an IDP, binding of ligands, introduction of denaturants or crowders, and so on. Simultaneous collection of donor and acceptor bursts is essential for performing smFRET experiments. The data analysis includes the filtering of bursts originating from the same molecules and the calculation of FRET efficiency. In its simple form, FRET efficiency values can be approximated by a Gaussian distribution that will vary between 0 and 1, depending on the proximity of the donor and acceptor. After fitting the curve with a Gaussian function, an average FRET efficiency value for the population can be obtained, which can be further treated by solving an integral for an appropriate probability density function for distance distributions. Here, the protein α-synuclein and its labeling with specific dyes are used as an example, for clarity. However, general features of the protocol and variations thereof are provided at various points, so that the reader can adapt the procedure for their particular system.
Materials
-
Dropper with water or oil (for water- or oil-immersion objectives, respectively)
-
Objective lens cleaning tissue (ThorLabs, cat. no. MC-5)
-
∼10 nM stock solution of fluorescently labeled α-synuclein (or other protein; see Support Protocol 1) in dilution buffer
-
Protein dilution buffer, with appropriate photophysical protectors and reducing agents (see recipe), prepared fresh before use
-
Uniformly coated reflective surface for alignment (e.g., a small mirror; a regular coverslip can also be used, though its lower reflectivity requires higher laser power)
-
Single-molecule detection setup (see Strategic Planning for additional details)
-
Tween 20–coated borosilicate chambered glass coverslips (Nunc™ Lab-Tek™ II Chamber Slide™, ThermoFisher Scientific, cat. no. 154534)
-
Software for data analysis (e.g., OriginLab)
Beam alignment in the detection part of the setup
IMPORTANT NOTE : While APD detection is ongoing, the ambient light exposure should be minimized to avoid damage to the APDs. Also, carefully follow any instructions for the specific instrument and laser that you are using.
1.Switch the laser power on and allow it to stabilize for at least 15 min. Switch the laser power meter on, and then monitor the laser power and fluctuations until the power has stabilized.
2.Adjust the laser power to a desired value (100-150 µW should be sufficient) and remove the laser power meter from the beam path. If the light is not visible at the detection part of the setup, the intensity should be gradually increased.
3.Place a drop of water or oil (as appropriate depending on the objective type) on the objective with a dropper.
4.For alignment, place and align an appropriate surface (this can be the surface of a glass coverslip) at the sample plane to reflect excitation light back through the objective.
5.Before carrying out steps 6-10, make sure the APDs are turned off.
6.Adjust the pinhole to be as large as possible, or remove the pinhole from the detection path. This is useful for initial rough alignment of APDs.
7.Remove spectral filters and dichroic mirrors (but not lenses or other beam-shaping optics) from the beam path in the detection box.
8.With the help of a mirror, check whether the beam falls within the detector surface of the APD that detects light transmitted through the first dichroic mirror. This APD will be the least sensitive to any external alterations, as it does not require any reflection of light; hence, no mirror adjustments are needed. Change the APD position as needed until the beam is centered within the APD face.
9.Replace the first dichroic mirror and check the beam that is reflected. With a white card or piece of paper, check that the beam falls in the middle of any reflective mirrors, then toward the APD face. Adjust the dichroic mirror as needed to reflect the beam to a point that is roughly in the middle of the APD detector opening.
10.Adjust the second APD as in step 8.This operation can be extended to many APDs as needed; however, it is advisable to start from the least reflection-dependent APD and proceed accordingly. We typically use two APDs for simultaneous detection of donor and acceptor photons, though additional detectors can be used to monitor more colors or photon properties such as polarization.
11.Place all the filters back in the beam path after the objective. This must be done before any of the following steps in which APDs are turned on.
12.Insert a pinhole of the desired diameter in the light path. Replace the reflective surface with a sample chamber filled with free donor fluorophore (at low nanomolar concentration or below, and with low laser power). High fluorophore concentrations and powers could lead to APD saturation or damage. Therefore, start with as low a donor concentration as possible (5 nM is sufficient). Begin with lower laser power (20 μW), and then increase to the desired value (step 2) as needed, and decrease the frequency (i.e., increase the integration time) of data collection as necessary. A signal of ∼10-50 kHz is sufficient for alignment.
13.Turn on the APDs and data acquisition program.
14.Adjust the pinhole position until the signal from the APDs is maximized. For the two-APD detection system, at this point only one of them may respond, or both may respond but non-coincidentally (i.e., the signal may be diminished in one APD earlier than in the other one during pinhole alignment).
15.Once the signal is maximized with the adjustment of the pinhole, individually adjust APDs further to maximize the signal.
16.Repeat steps 14-15 iteratively, until no further improvements are recorded, and then turn off the ADPs and the data acquisition program.
Single-molecule FRET data acquisition
NOTE : The data acquisition is described for measurements that utilize binned photons. Be sure to carefully follow any safety precautions needed for your specific system and experiment.
17.Prepare the α-synuclein (or other protein) sample by diluting the sample with sterile-filtered protein dilution buffer and adding suitable photoprotectors (see Critical Parameters for additional information on photoprotectors). The final concentration of dual-labeled protein should be ∼100 pM. Add 200 µl of the solution to the coverslip chamber, and let it sit for 5 min before the measurement.
18.Place the coverslip in the instrument and adjust the objective to focus the beam in the sample (about 30-50 µm from the surface).
19.Turn on the APDs and data acquisition program.
20.Adjust the acquisition frequency in the data acquisition program. The integration time is the reciprocal of this frequency; therefore, it should be tested according to the molecular motions expected within the sample. However, it is also important to remember that a higher frequency will yield a lower signal-to-noise ratio.
21.If a real-time data visualization program is used, insert direct excitation, leakage, and signal threshold values. (See Support Protocol 3 on how to determine these.)
22.Run the experiment.
23.Once the experiment (or set of experiments) is complete, turn off APDs and laser, remove the sample chamber, and wipe off the objective with objective cleaning wipes.
Process binned photon counts and calculate FRET efficiency
24.For ratiometric smFRET measurements, subtract the contributions of background from the raw donor and acceptor photon streams.
25.Process background-corrected photon streams to obtain intensities corrected for leakage and direct excitation. Once these are determined, use the procedure in Support Protocol 3 to obtain corrected donor and acceptor intensities.
26.Apply a threshold (“ T ”) to the sum of donor and acceptor intensities within the same time bin (SUM = I D + I A > T) to identify qualifying bursts of donor and acceptor that originate from the same single molecule.
27.Calculate the FRET efficiency (E FRET) using Equation 1:
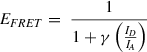
Support Protocol 1: FLUOROPHORE LABELING OF A PROTEIN THROUGH MALEIMIDE CHEMISTRY
A prerequisite for performing virtually any single-molecule fluorescence measurement is fluorophore labeling of the protein of interest. Although several labeling strategies exist that employ different chemical moieties, as explained in the Strategic Planning section, here we describe only fluorophore labeling of Cys residues via maleimide chemistry.
Materials
-
Fluorophores: maleimide-conjugated Alexa Fluor 488 (donor) and Alexa Fluor 594 (acceptor) (ThermoFisher Scientific, cat. nos. A10254 and A10256)
-
Organic solvent: dimethyl formamide (DMF) or dimethyl sulfoxide (DMSO)
-
1 M Tris•Cl, pH 7.5
-
5 N NaCl
-
Reducing agent: dithiothreitol (DTT) or tris(2-carboxyethyl)phosphine (TCEP)
-
Liquid nitrogen or dry ice and ethanol
-
Buffer-exchange device: NAP column (GE Life Sciences, cat. no. 17085201) or HiTrap desalting column (GE Life Sciences, cat. no. 29048684)
-
Centrifugal filter device (Millipore Sigma, cat. no. UFC500308) or dialysis device (ThermoFisher Scientific, cat. no. 88400)
Reagent preparation
CAUTION : Fluorophores and fluorophore-labeled proteins should be kept in the dark, avoiding direct exposure to light as far as practical to minimize photobleaching.
1.Dissolve 1 mg of each fluorophore in 200 µl of the organic solvent suggested by the manufacturer (e.g., DMSO) and measure the stock concentrations by UV-visible (UV-Vis) spectrometry. Prepare 10-µl aliquots and store up to 6 months at −80°C.
2.Prepare labeling buffer consisting of 20 mM Tris•Cl and 150 mM NaCl, pH 7.5. Sterilize it by passing it through a 0.2-μm filter.
3.Dissolve the α-synuclein (or other protein of interest) in the labeling buffer if lyophilized. Otherwise, buffer exchange it into the labeling buffer using gel filtration (NAP columns for prep scale or HiTrap desalting columns from GE) or a centrifugal filter device.
Single-fluorophore labeling
4a. Add reducing agent and incubate the protein at room temperature for 1 hr to reduce any intra- and intermolecule disulfides. We suggest adding at least 10 mM dithiothreitol (DTT) or 0.5 mM tris(2-carboxyethyl)phosphine (TCEP).
5a. If DTT is used, remove it from the solution by buffer exchange using devices outlined above. Any remaining DTT will react with maleimide fluorophores. If TCEP is used, it can remain in solution.
6a. Add 3-10 mole equivalents of the fluorophore dropwise to the protein solution and ensure homogeneous mixing by gentle pipetting. Incubate 1 hr at 37°C, 2 hr at room temperature, or overnight at 4°C.
7a. Remove the excess fluorophore by buffer exchange. If centrifugal filters are used, make sure the retentate is washed sufficiently so that no trace dye is detectable by UV absorbance of the flowthrough. Dialysis can be used for larger volumes.
Double-fluorophore labeling
Perform these steps instead of the single-fluorophore labeling detailed above to obtain dual-labeled α-synuclein (or other protein) samples. Steps 4a and 5a of the single-fluorophore labeling procedure are essentially the same for double Cys-fluorophore labeling.
4b. Add 1 mole equivalent (or substoichiometric amounts) of donor fluorophore dropwise to the protein solution. Ensure homogeneous mixing by gentle pipetting. Incubate for 1 hr at 37°C, 2 hr at room temperature, or overnight at 4°C.
5b. If only donor was added above, separate from free dye as in step 7a. It is advisable to reduce the protein again if step 4b is performed overnight, and the reducing agent (DTT) should be removed before the acceptor fluorophore is added.
6b. After donor-only single-labeled protein is obtained, measure the concentration of the protein. Add a 5× molar excess of acceptor fluorophore and incubate 1 hr at 37°C, 2 hr at room temperature, or overnight at 4°C.
7b. Perform step 7a again.
Verify purity and identity of the labeled proteins
8.Measure the labeling percentage by UV-Vis spectroscopy according to the formula
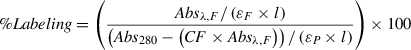
where Absλ,F is the absorbance value at the fluorophore's maximum absorbance wavelength; ε F and ε P are the fluorophore and protein extinction coefficients (in M−1 cm−1), respectively; CF is the ratio of fluorophore extinction coefficients at 280 nm and the maximum absorbance wavelength (provided by manufacturer, or can be measured using free dye); and l is the path length in centimeters. Labeling yields may be >90%, but vary depending on protein and label site.
9.If needed, further purify the labeled proteins by HPLC or high-resolution ion-exchange chromatography.
10.Verify the correct mass by LC-MS (or other MS method).
11.Aliquot the labeled protein and snap-freeze it in liquid nitrogen or a dry ice/ethanol bath.
Support Protocol 2: SAMPLE CHAMBER PREPARATION
Sample chambers for single-molecule fluorescence measurements exist in various forms. Chambered coverslip systems that are tailored for growing and imaging cells serve well for single-molecule measurements, as they come in multiwell formats. The ability to conduct measurements of different samples without changing the sample chamber on the microscope, the accessibility of well contents during the experiment, and the short preparation time make multiwell systems advantageous over sandwich-type glass-coverslip system.
Avoiding nonspecific protein adsorption is crucial to performing single-molecule fluorescence experiments as limited number of molecules are available to freely diffuse in solution. Nonspecific adsorption is reduced by treating the chamber surfaces with detergent. Many sample chambers can be prepared at once and stored in dry conditions at room temperature for 1 month in a covered box wrapped in tissue paper to prevent dust from sticking to surfaces.
Materials
-
10% (v/v) Tween 20 (see recipe)
-
Milli-Q-filtered water
-
Borosilicate chambered glass coverslip (Nunc™ Lab-Tek™ II Chamber Slide™ from ThermoFisher Scientific, cat. no. 154534) or other appropriate sample chamber(s)
-
Vacuum suction system
1.Fill each surface or well with 10% Tween 20 and incubate it 30 min at room temperature.
2.After incubation, discard the Tween 20 solution from the chamber, ideally using a vacuum suction system.
3.Wash the chamber thoroughly with Milli-Q-filtered water.
4.Dry the chambers under light vacuum and store up to 1 month at room temperature.
Support Protocol 3: DETERMINATION OF DIRECT EXCITATION OF ACCEPTOR BY DONOR EXCITATION AND LEAKAGE OF DONOR EMISSION TO ACCEPTOR EMISSION CHANNEL
The measured signals depend on contributions from emission of donor and acceptor dyes in the donor and acceptor detection channels respectively, arising from (i) donor excitation, (ii) donor emission (arising from donor excitation) leakage into the acceptor channel (L), and (iii) direct excitation of acceptor by excitation (D ex). These terms are dependent on both the inherent photophysical properties of the donor and acceptor pair and characteristics of the instrument (including filters and dichroic mirrors). To determine the leakage contribution, use a proper donor-only labeled sample.
Materials
- Donor-only and acceptor-only labeled protein samples (Support Protocol 1)
- Tween 20–coated borosilicate chambered glass coverslip (Support Protocol 2)
- Single-molecule detection setup (see Strategic Planning)
1.Place an appropriate concentration of donor-only labeled sample into the sample chamber. The concentration of the donor-only sample must be sufficiently high for a reliably measurable donor emission to leak into acceptor channel, but it should not be in the saturation or damage range for the donor channel.
2.Record the raw photon counts in both donor and acceptor channels (see Basic Protocol, steps 18-22). Perform only background correction by subtracting the average number of photons for both donor and acceptor channel, recorded for buffer-only sample. Raw photon numbers of donor (R D) and acceptor (R A) can be defined as follows (Brustad et al., 2008):
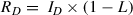
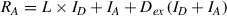
When there is no acceptor fluorophore present, the RA term in Equation 3b will reduce to R A = L × I D (because there are no acceptor fluorophores, the D ex term in Equation 3b also becomes obsolete). L can be calculated by combining Equations 3a and 3b.
In theory, D ex can even be estimated in an ensemble fluorometer by exciting acceptor-only sample with the excitation wavelength of laser. D ex can be determined in a single-molecule setup by using an equimolar mixture of donor and acceptor. The theoretical numbers found on manufacturer's website inferred from absorption spectra of donor and acceptor or determined by an ensemble fluorometer can deviate in a single-molecule setup as a function of relative detection efficiencies of detector.
3.Place an equimolar mixture of donor and acceptor fluorophore in the sample chamber. Follow the concentration guidelines suggested for determining leakage contribution.
4.Excite with a laser that is suitable for donor fluorophore excitation.
5.Assuming that there is no FRET between free dyes (which is true at low nanomolar concentrations, such as are used here), the I A term in Equation 3b, which indicates the intensity of acceptor originating from sensitized emission, should be 0, whereas R A includes contributions from leakage of donor emission and direct excitation of acceptor with donor excitation. Hence R A/R D becomes (L + D ex)/(1 – L), which can be solved for D ex with a known L.
REAGENTS AND SOLUTIONS
Protein dilution buffer
- 20 mM Tris•Cl
- 150 mM NaCl
- 2 mM DTT, pH 7.5
- Add 2-mercaptoethanol to a final concentration of 140 mM as photoprotectant. Filter sterilize with a 0.2-µm sterile filter (e.g., Sigma-Aldrich, cat. no. CLS430773).
Prepare fresh before each experiment.
10% (v/v) Tween 20 stock solution
Mix Tween 20 (Millipore Sigma, cat. no. P1379) and Milli-Q-purified water in 10:90 volume ratio—be careful to ensure proper mixing, as Tween 20 is denser than water. Sterile filter and store at room temperature for as long as the solution remains clear and homogeneous.
COMMENTARY
Background Information
A considerable proportion of the eukaryotic genome encodes proteins that contain regions without well-defined, three-dimensionally folded structures (Dyson & Wright, 2005). These so-called IDPs are biased in their amino acid composition: Polar and charged amino acids are overrepresented, whereas hydrophobic amino acids, which are normally found at the core of a folded protein, are underrepresented. Despite lacking a set three-dimensional structure, IDPs are able to perform a wide array of tasks in a cell. Their functions include transcriptional regulation, cellular signaling, and dampening of the stress response. In addition, IDPs are key players in several neurodegenerative diseases (e.g., Parkinson's and Huntington's diseases), diabetes, and heart disease (Dunker, Bondos, Huang, & Oldfield, 2015; Wright & Dyson, 2015), as well as cellular compartmentalization by liquid-liquid phase separation (Bentley, Frey, & Deniz, 2019).
IDPs are characterized by their dynamic nature. Owing to their intrinsically shallow free energy landscapes, they can rapidly sample many different conformations (Berlow, Dyson, & Wright, 2015; Csizmok, Follis, Kriwacki, & Forman-Kay, 2016). Because of this plasticity, IDPs can bind to multiple partners with tunable affinities, and often function as hubs in cellular signaling processes (Wright & Dyson, 2015). As a result of these interactions, they may either maintain their unstructured state or undergo disorder-to-order transitions (Borgia et al., 2018; Sugase, Dyson, & Wright, 2007).
A variety of structural and biophysical methods have been applied to characterize bound and unbound states of IDPs. These include nuclear magnetic resonance (NMR); small-angle x-ray scattering (SAXS) in combination with computational tools (Bernadó & Svergun, 2012; Kosol, Contreras-Martos, Cedeño, & Tompa, 2013; Sibille & Bernadó, 2012); and low-resolution techniques such as circular dichroism and a variety of ensemble fluorescence spectroscopy methods (Chemes, Alonso, Noval, & de Prat-Gay, 2012; Jain, Bhattacharya, & Mukhopadhyay, 2011; Krishnan et al., 2012). However, because of their flexibility and complexity, aspects of IDPs can be challenging to study by conventional ensemble methods. Along these lines, over the past 15 years, single-molecule tools have become indispensable in investigating IDP systems (Brucale, Schuler, & Samorì, 2014; Deniz et al., 1999; Ferreon, Gambin, Lemke, & Deniz, 2009; Greenleaf, Woodside, & Block, 2007; Hofmann et al., 2012; Mukhopadhyay, Krishnan, Lemke, Lindquist, & Deniz, 2007; Schuler, Lipman, & Eaton, 2002; Solanki, Neupane, & Woodside, 2014; Nasir, Onuchic, Labra, & Deniz, 2019). These methods offer an unprecedented ability to overcome ensemble averaging and thus resolve conformational subpopulations and complex dynamics (Deniz et al., 1999; Ferreon et al., 2009; Greenleaf et al., 2007; Hofmann et al., 2012; Schuler et al., 2002). Therefore, single-molecule tools are particularly suited for studying the conformational landscapes of IDPs either in isolation or in complex with their binding partners. The latter, in turn, can guide us to understand the causes and consequences of functionally important reactions at an extremely high level of detail in terms of paths and intermediate states.
Fluorescence-based single-molecule measurements can further be grouped into two main modes: (i) where proteins are freely diffusing in solution and fluorescence is collected via confocal detection, and (ii) where proteins are tethered to a surface and fluorescence is detected in a total internal reflection geometry.
One of the most frequently leveraged single-molecule fluorescence applications is Förster resonance energy transfer (FRET). FRET allows accurate measurements of the distance between two fluorescent molecules, namely a donor (D) and an acceptor (A), that are within a few nanometers’ proximity. Energy from the excited state of donor molecules is transferred via a dipole-dipole coupling mechanism to acceptor molecules that absorb at wavelengths overlapping those of the donor emission. The rate of energy transfer k T(r) from D to A is given by 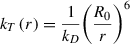
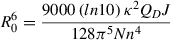
where Q D is the quantum yield of the donor in the absence of an acceptor, N is Avogadro's number, n is the refractive index of the medium, and J is a quantitative measure of spectral overlap between donor emission and acceptor absorption, known as the overlap integral. In calculating J , care should be taken to normalize the spectrum to unity with respect to its area (Lakowicz, 1983). κ 2, known as the orientation factor, is explained in the Critical Parameters section.
The fraction of photons absorbed by the donor that are transferred to the acceptor is denoted the transfer efficiency, E FRET, and can be expressed in terms of rate constants as 
Combination with Equation 4 and rearrangement yields 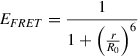
Another method of determining E FRET involves measuring the fluorescence lifetimes of donor-only and donor-acceptor molecules, 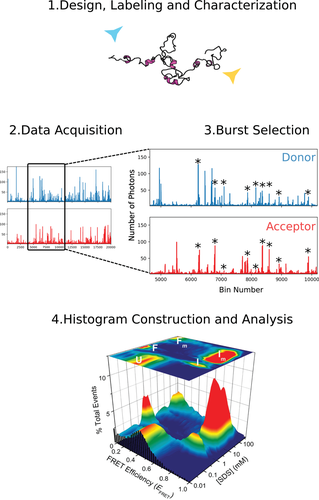
Inferring distance from FRET efficiency
Although an estimate of D-A distance can be obtained from the measured average <E FRET>, quantifying the D-A distance for IDPs is preferably done taking into consideration the dynamic nature of IDPs, because they sample many conformations rapidly. Hence, the E FRET value obtained from Gaussian fit of a histogram should be treated as an average value, which has a dependence on the D-A distance distribution as follows: 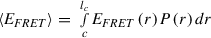
smFRET measurement modes
As discussed above, E FRET is commonly measured in two modes: (i) using the sensitized emission of A upon excitation of D (usually in a ratiometric manner with simultaneous recording of donor and acceptor signals) and (ii) using the changes in the lifetime of D in the presence of A. The principal hardware differences between the two modes are the time resolution and photon-counting capabilities of the setup, and the need for a sub-nanosecond pulsed laser for mode 2.
Mode 1, known as “ratiometric detection,” is easier to implement than mode 2. In mode 1, a counter-timer board (or other counting device) is used to count photons within time bins (integration time). Summed photons in a bin are tentative constituents of a burst, depending on the number of photon counts. Evidently, the bursts will contain more photons when the integration time is longer. However, a longer integration time also results in the inclusion of additional background and signals from multiple molecules, giving rise to decreased signal-to-noise ratio and contaminated single-molecule signals. Hence, the integration time must be chosen carefully. Historically, smFRET data have been collected with integration times ranging from 0.2 to 1 ms (Deniz et al., 2001), which work for many average-size proteins or other biomolecules. If needed, the user can determine the diffusion coefficient of the molecule through fluorescence correlation spectroscopy (FCS) and adjust the integration time for smFRET measurements to optimize the detection of single molecules. The ratiometric mode is simple and less expensive to implement, but it requires knowledge of a correction factor, γ, which can vary with solution conditions and is extremely sensitive to optical alignment of hardware components (Michalet, Weiss, & Jäger, 2006). For the purpose of this protocol, we have focused on a simple version of mode 1 for smFRET measurements.
In mode 2, a picosecond pulse laser is used for excitation, and output signals from APDs are routed to a time-correlated single-photon counting (TCSPC) device. This device registers both the arrival times of individual photons compared to the start of the experiment and the time lag between each photon and the previous exciting laser pulse. With these two parameters, the overall time trajectory of photon arrivals can be reconstructed for D and A channels separately, and correlation functions can also be computed. Furthermore, after selection of eligible florescent bursts, fluorescence decay histograms can also be computed and decay rates calculated (Eggeling et al., 2001; Sisamakis et al., 2010). Thus, mode 2 is a particularly efficient means of data collection, as the single set of basic data can be analyzed to provide several kinds of outputs, including binned or burst time traces, lifetime information, and correlation curves. Finally, with additional channels, fluorescence anisotropy for each photon (or burst) can also be recorded and used.
For readers’ information, we also briefly note a more advanced version of mode 1 (which can also be incorporated into mode 2) that involves the use of alternating laser excitation (ALEX). ALEX is a method used in combination with single-molecule FRET that introduces a second excitation source to report on the presence of the acceptor fluorophore. ALEX was developed by the Weiss laboratory to address the difficulty of distinguishing low-FRET species, with an acceptor fluorophore present but distant from the donor, from single-labeled donor-only species (Kapanidis et al., 2004; Kapanidis, Majumdar, Heilemann, Nir, & Weiss, 2015), and to provide more precise E FRET measurements.
During an ALEX experiment, molecules diffusing through the focal volume are alternately excited by two different lasers, each capable of exciting either the donor or the acceptor dye. This yields four streams of photon emission data described by the excitation (ex) wavelength used and the emitting (em) fluorophore measured: direct excitation of donor (
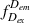
); direct excitation of acceptor (
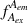
); emission from donor resulting from excitation of acceptor, which should be equal to background (
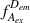
); and emission from acceptor resulting from excitation of donor, or in other words, sensitized emission by FRET (
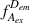
). E FRET in ALEX experiments is calculated identically to the way it is in single-excitation FRET experiments. Rewriting Equation 1 with these descriptive terms produces 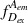
The additional information provided by directly exciting the acceptor molecule is incorporated into the stoichiometry ratio (S), where values approaching 1 represent donor-only molecules, values approaching 0 represent acceptor-only molecules, and intermediate values represent molecules dually labeled with a FRET pair; thus, 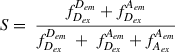
The two ALEX ratios are typically represented in a two-dimensional histogram, which allows populations of differentially labeled molecules as well as heterogeneous populations of molecules to be distinguished from one another. ALEX experiments may be conducted using most single-excitation FRET systems with some modifications. First, lasers must alternate regularly and without overlap at frequencies at least an order of magnitude faster than the transit time of a typical molecule through the focal volume. Common methods of laser alternation are electro- and acousto-optical modulation, although other strategies exist and have been reviewed elsewhere (Kapanidis et al., 2015). Second, the modulation of the laser must be synchronized with the collection method to allow the four data streams to be distinguished.
Critical Parameters
κ2
The orientation factor, κ 2, is generally taken as the value 2/3, assuming the dyes attached on a protein are rotationally unconstrained. One factor that can contribute to sufficient rotational averaging is the length of fluorophore spacers (e.g., C5, C6, etc.). However, in some cases, despite having a flexible linker, the dye can interact with amino acid side-chain moieties, constraining the rotational freedom and skewing κ 2 from 2/3. To test whether κ 2 = 2/3 is a reasonable assumption, steady-state ensemble or single-molecule anisotropy values should be measured. Ideally, anisotropy values should be <0.15 in all conditions tested in a single-molecule experiment.
γ
This parameter is a correction factor based on the detection efficiencies of detectors and quantum yields of the dyes. Although this parameter may not fluctuate very much, it is important to measure it for more precise distance measurements. γ essentially has two contributors, as follows (Ferreon et al., 2009): 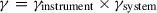
To calculate the γinstrument, Ferreon et al. reported a procedure wherein donor and acceptor intensity variation with respect to varying fluorophore concentrations were measured in both ensemble and single-molecule modes (Ferreon et al., 2009). An alternating laser excitation scheme is capable of quantitatively determining a combined γ value from parameters obtained from linear fit of 1/S versus E FRET data of singly labeled samples (Fuertes et al., 2017; Lee et al., 2005).
Additional photoprotection strategies
To maximize the signal-to-noise ratio in a single-molecule fluorescence experiment, the fluorophore attached to a protein should be kept in a stable, emitting state. Triplet states, conformational isomers, and radical ions have been found to destabilize the fluorophores, either by inducing irreversible dark states or by being reactive precursors to such states (Aitken, Marshall, & Puglisi, 2008).
To combat dark states, various strategies have been developed based on enzymatic oxygen scavenging (Aitken et al., 2008; Ha et al., 1999; Swoboda et al., 2012), triplet state quenching by 6-hydroxy-2,5,7,8-tetramethylchroman-2-carboxylic acid (commercially known as Trolox; Grunwell et al., 2001), reducing agent–induced oxygen scavenging (Kishino & Yanagida, 1988), or the combination of latter two (Campos et al., 2011; Cordes, Vogelsang, & Tinnefeld, 2009; Widengren & Schwille, 2000). Lemke and coworkers observed that simple laminar flow of the protein solution (Lemke et al., 2009) also reduces the detection of photobleached species even in the absence of additives, a method that they also combined with additives to further reduce photobleaching. All methods are relatively accessible and can be considered as needed for the experiments.
Peak broadening and shape
Although the broadening of E FRET histogram peaks may originate from IDPs’ dynamic character, E FRET histogram peaks can be broadened by other mechanisms.
One major source of broadening is shot noise, which originates from the statistics of detecting only a few photons from a single molecule. Thus, even for an ideal single distance, distributions in the detected values of I d and I a in Equation 1 would result in E FRET fluctuating around its mean value. Furthermore, the shot noise–related broadening shows a nonmonotonic dependence on E FRET, with peaks at E FRET ≈ 0.5 being the widest, and a reduction in broadening seen for peaks at low or high E FRET values (Antonik, Felekyan, Gaiduk, & Seidel, 2006; Gopich & Szabo, 2007; Nir et al., 2006). In addition, even though it is tempting to attribute excessive width of E FRET histogram peaks to the dynamic behavior of the system, it is important to keep in mind that the width can also be affected by inhomogeneities in the detection volume, variations in dye photophysical parameters or orientation factor, and labeling permutations (Schuler, 2013). Another potential contribution to broadening is that a significant fraction of detected signal arises from more than one molecule. Reduced concentration can be used to test for and minimize this source of broadening.
If the FRET peaks are broader than expected from the above sources or an appropriate control, one may conclude that this excess broadening reflects conformational heterogeneity. In this regard, the timescale of interconversion of conformational states with respect to integration time is a particularly interesting defining factor for the peak shape. For example, in the case of a two-state system with equal populations at equilibrium, states interconverting slowly with respect to the integration time (an order of magnitude lower or less) will be resolved as two separate E FRET peaks. However, an integration time comparable to the exchange rate will manifest itself as a single, wide peak with a maximum at the average of the E FRET of two states. In the case of two states with unequal distributions at the equilibrium, the single peak will be asymmetric, skewed toward the dominant population. Finally, when the exchange rate is much faster than the integration time, there will be a single peak, with width limited by shot noise (and the other factors discussed above), and centered at the average E FRET (Gopich & Szabo, 2007). To a limited extent, the integration time can be varied to test dynamical contribution to peak broadening.
Given the complexities in interpreting peak widths, inferences about conformational dynamics should preferably be validated with another method that directly quantifies fluctuations, such as FCS or FRET-FCS.
Troubleshooting
Impurities in buffer components may greatly increase the noise. In IDP research, it is quite common to vary the salt or denaturant concentrations to elicit the behavior in line with polymer physics; therefore, those components must be purchased carefully and tested. A good practice is to note the manufacturers from the publications from other laboratories conducting single-molecule FRET studies.
Occasionally, during the instrument alignment process, a local minimum in the maximum number of counts will be encountered, which will eventually skew all FRET efficiencies. Ideally, a sample with known FRET efficiency must be measured (i.e., DNA duplex with known D-A distance) to assess the viability of the alignment process.
The burst shape is an important indicator of sample condition. In an smFRET experiment, the signal will be at the noise level most of the time with occasional single, sharp bursts that ideally will appear in both donor and acceptor channels at the same time. Unusually intense or lengthy bursts can appear when the sample contains impurities that diffuse more slowly than a protein. These can be nonspecific aggregates of the IDP in question, which emit continuously while traversing the focal volume, whereas a single molecule's diffusion time is much faster. In this case further purification or optimization of the experimental conditions may be needed to avoid artifacts.
Understanding Results
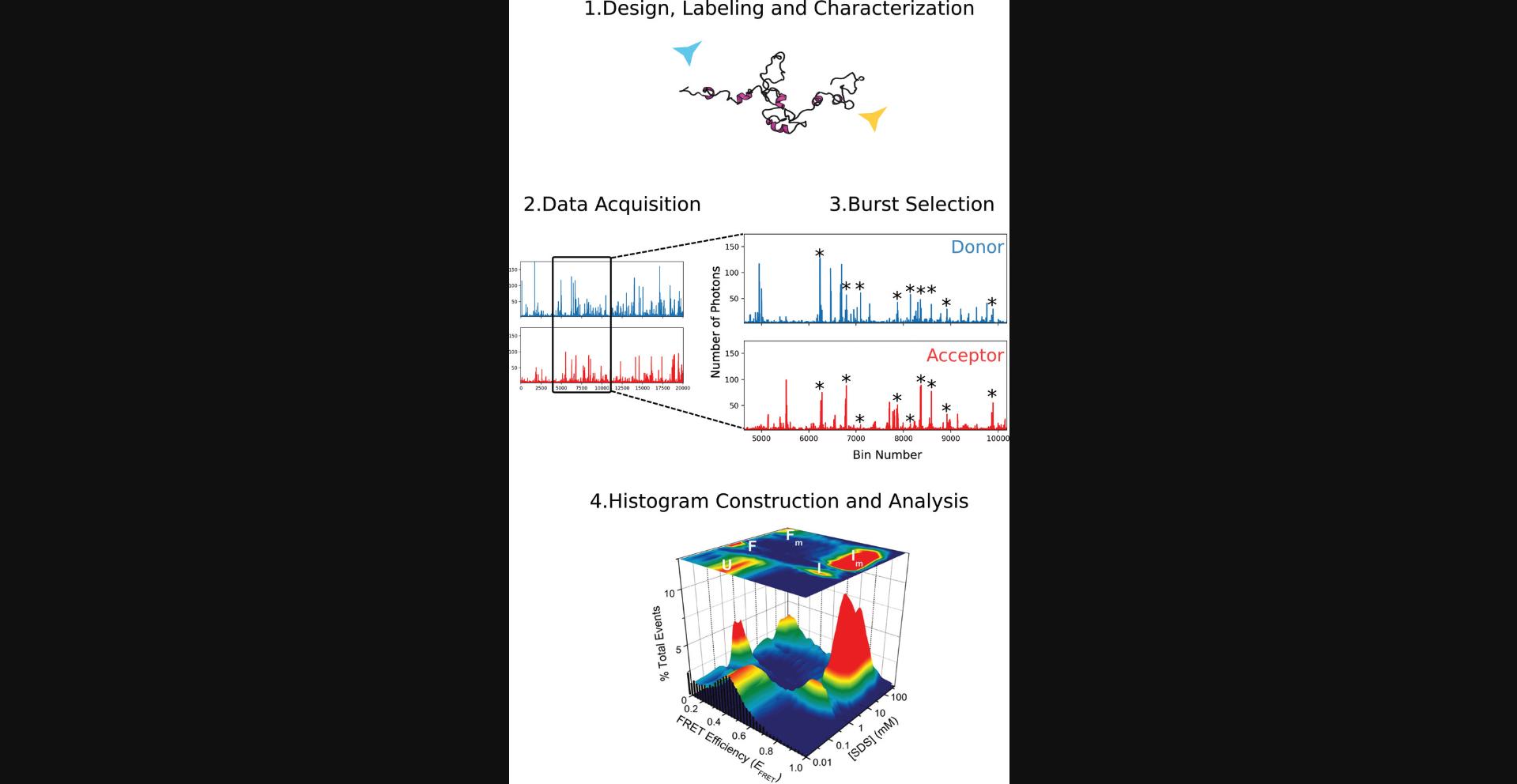
The Basic Protocol will generate a histogram of E FRET values, centered around one or multiple peaks. By sequentially titrating the binding partner, salt, denaturant, or crowder, one can observe and quantify exchange between peaks (subpopulations) or shifts in peak position and/or peak shape. See Figure 2 for a flowchart of a typical experiment. As an example, part 4 of Figure 2 contains histogram data (from Ferreon et al., 2009) as a function of the binding partner SDS, showing that the protein populates a variety of interconverting states as a function of this titration. Note that these samples also contained 10 µM unlabeled protein, which was specific to this system (so not used generally). Fitting the data will yield the median E FRET value, as well as the peak area and width, which can then be plotted against the effector concentration for further analysis and interpretation. Several additional types of analyses are possible, some of which are discussed in papers mentioned in the Critical Parameters subsection on Peak broadening and shape and listed in the Key References.
Time Considerations
Once the required protein constructs are expressed and purified, the Basic Protocol and labeling can be completed within a few weeks, including some degree of troubleshooting and an initial set of data collection and analysis. Initial beam alignment (e.g., after changing filters/dichroic mirrors to suit the particular experiment) usually takes <1 hr. Subsequent minor alignment optimization typically takes only a few minutes. Once the data acquisition process is streamlined, data acquisition takes <1 hr per sample. Multiwell chambers save a few minutes per sample preparation; however, a strength of the open multiwell chambers is the ability to carry out titrations or switch back and forth between multiple samples in the chamber. The experimental timeline can extend substantially if the protein reagents require more elaborate labeling or purification schemes, and to optimize experimental conditions for more complex IDP systems. For simple and easy-to-handle IDP systems, following optimization, the Basic Protocol may be completed in 2-3 days.
Acknowledgments
We thank the past and present members of the Deniz Lab for their contributions to developing and applying single-molecule FRET to IDPs and other systems. We gratefully acknowledge the US NIH (grants R35 GM130375, R01 CA214054, and R01 GM066833) for support of our lab's related work. Irem Nasir is a Swedish Research Council (Vetenskapsrådet) International Postdoctoral Fellow under grant number 2016-06672.
Literature Cited
- Aitken, C. E., Marshall, R. A., & Puglisi, J. D. (2008). An oxygen scavenging system for improvement of dye stability in single-molecule fluorescence experiments. Biophysical Journal , 94(5), 1826–1835. doi: 10.1529/biophysj.107.117689.
- Antonik, M., Felekyan, S., Gaiduk, A., & Seidel, C. A. M. (2006). Separating structural heterogeneities from stochastic variations in fluorescence resonance energy transfer distributions via photon distribution analysis. The Journal of Physical Chemistry. B , 110(13), 6970–6978. doi: 10.1021/jp057257.
- Bentley, E. P., Frey, B. B., & Deniz, A. A. (2019). Physical chemistry of cellular liquid-phase separation. Chemistry , 25, 5600–5610. doi: 10.1002/chem.201805093.
- Berlow, R. B., Dyson, H. J., & Wright, P. E. (2015). Functional advantages of dynamic protein disorder. FEBS Letters , 589(19 Pt A), 2433–2440. doi: 10.1016/j.febslet.2015.06.003.
- Berlow, R. B., Dyson, H. J., & Wright, P. E. (2017). Hypersensitive termination of the hypoxic response by a disordered protein switch. Nature , 543(7645), 447–451. doi: 10.1038/nature21705.
- Bernadó, P., & Svergun, D. I. (2012). Structural analysis of intrinsically disordered proteins by small-angle X-ray scattering. Molecular BioSystems , 8(1), 151–167. doi: 10.1039/C1MB05275F.
- Borgia, A., Borgia, M. B., Bugge, K., Kissling, V. M., Heidarsson, P. O., Fernandes, C. B., … Schuler, B. (2018). Extreme disorder in an ultrahigh-affinity protein complex. Nature , 555(7694), 61–66. doi: 10.1038/nature25762.
- Brucale, M., Schuler, B., & Samorì, B. (2014). Single-molecule studies of intrinsically disordered proteins. Chemical Reviews , 114(6), 3281–3317. doi: 10.1021/cr400297g.
- Brustad, E. M., Lemke, E. A., Schultz, P. G., & Deniz, A. A. (2008). A general and efficient method for the site-specific dual-labeling of proteins for single molecule fluorescence resonance energy transfer. Journal of the American Chemical Society , 130(52), 17664–17665. doi: 10.1021/ja807430h.
- Campos, L. A., Liu, J., Wang, X., Ramanathan, R., English, D. S., & Muñoz, V. (2011). A photoprotection strategy for microsecond-resolution single-molecule fluorescence spectroscopy. Nature Methods , 8(2), 143–146. doi: 10.1038/nmeth.1553.
- Chemes, L. B., Alonso, L. G., Noval, M. G., & de Prat-Gay, G. (2012). Circular dichroism techniques for the analysis of intrinsically disordered proteins and domains. Methods in Molecular Biology (Clifton, N.J.) , 895(Chapter 22), 387–404. doi: 10.1007/978-1-61779-927-3_22.
- Cordes, T., Vogelsang, J., & Tinnefeld, P. (2009). On the mechanism of Trolox as antiblinking and antibleaching reagent. Journal of the American Chemical Society , 131(14), 5018–5019. doi: 10.1021/ja809117z.
- Csizmok, V., Follis, A. V., Kriwacki, R. W., & Forman-Kay, J. D. (2016). Dynamic protein interaction networks and new structural paradigms in signaling. Chemical Reviews , 116(11), 6424–6462. doi: 10.1021/acs.chemrev.5b00548.
- Deniz, A. A., Dahan, M., Grunwell, J. R., Ha, T., Faulhaber, A. E., Chemla, D. S., … Schultz, P. G. (1999). Single-pair fluorescence resonance energy transfer on freely diffusing molecules: Observation of Förster distance dependence and subpopulations. Proceedings of the National Academy of Sciences of the United States of America , 96(7), 3670–3675. doi: 10.1073/pnas.96.7.3670.
- Deniz, A. A., Laurence, T. A., Dahan, M., Chemla, D. S., Schultz, P. G., & Weiss, S. (2001). Ratiometric single-molecule studies of freely diffusing biomolecules. Annual Review of Physical Chemistry , 52(1), 233–253. doi: 10.1146/annurev.physchem.52.1.233.
- Dunker, A. K., Bondos, S. E., Huang, F., & Oldfield, C. J. (2015). Intrinsically disordered proteins and multicellular organisms. Seminars in Cell & Developmental Biology, 37, 44–55. doi: 10.1016/j.semcdb.2014.09.025.
- Dyson, H. J., & Wright, P. E. (2005). Intrinsically unstructured proteins and their functions. Nature Reviews Molecular Cell Biology , 6(3), 197–208. doi: 10.1038/nrm1589.
- Eggeling, C., Berger, S., Brand, L., Fries, J. R., Schaffer, J., Volkmer, A., & Seidel, C. A. M. (2001). Data registration and selective single-molecule analysis using multi-parameter fluorescence detection. Journal of Biotechnology , 86(3), 163–180. doi: 10.1016/S0168-1656(00)00412-0.
- Ferreon, A. C. M., Gambin, Y., Lemke, E. A., & Deniz, A. A. (2009). Interplay of alpha-synuclein binding and conformational switching probed by single-molecule fluorescence. Proceedings of the National Academy of Sciences of the United States of America , 106(14), 5645–5650. doi: 10.1073/pnas.0809232106.
- Fuertes, G., Banterle, N., Ruff, K. M., Chowdhury, A., Mercadante, D., Koehler, C., … Lemke, E. A. (2017). Decoupling of size and shape fluctuations in heteropolymeric sequences reconciles discrepancies in SAXS vs. FRET measurements. Proceedings of the National Academy of Sciences of the United States of America , 114(31), E6342–E6351. doi: 10.1073/pnas.1704692114.
- Gopich, I. V., & Szabo, A. (2007). Single-molecule FRET with diffusion and conformational dynamics. The Journal of Physical Chemistry. B , 111(44), 12925–12932. doi: 10.1021/jp075255e.
- Greenleaf, W. J., Woodside, M. T., & Block, S. M. (2007). High-resolution, single-molecule measurements of biomolecular motion. Annual Review of Biophysics and Biomolecular Structure , 36(1), 171–190. doi: 10.1146/annurev.biophys.36.101106.101451.
- Grunwell, J. R., Glass, J. L., Lacoste, T. D., Deniz, A. A., Chemla, D. S., & Schultz, P. G. (2001). Monitoring the conformational fluctuations of DNA hairpins using single-pair fluorescence resonance energy transfer. Journal of the American Chemical Society , 123(18), 4295–4303. doi: 10.1021/ja0027620.
- Ha, T., Ting, A. Y., Liang, J., Caldwell, W. B., Deniz, A. A., Chemla, D. S., … Weiss, S. (1999). Single-molecule fluorescence spectroscopy of enzyme conformational dynamics and cleavage mechanism. Proceedings of the National Academy of Sciences , 96(3), 893–898. doi: 10.1073/pnas.96.3.893.
- Hofmann, H., Soranno, A., Borgia, A., Gast, K., Nettels, D., & Schuler, B. (2012). Polymer scaling laws of unfolded and intrinsically disordered proteins quantified with single-molecule spectroscopy. Proceedings of the National Academy of Sciences of the United States of America , 109(40), 16155–16160. doi: 10.1073/pnas.1207719109.
- Jain, N., Bhattacharya, M., & Mukhopadhyay, S. (2011). Chain collapse of an amyloidogenic intrinsically disordered protein. Biophysical Journal , 101(7), 1720–1729. doi: 10.1016/j.bpj.2011.08.024.
- Jensen, M. H., Sukumaran, M., Johnson, C. M., Greger, I. H., & Neuweiler, H. (2011). Intrinsic motions in the N-terminal domain of an ionotropic glutamate receptor detected by fluorescence correlation spectroscopy. Journal of Molecular Biology , 414(1), 96–105. doi: 10.1016/j.jmb.2011.09.037.
- Joo, C., & Ha, T. (2012). Preparing sample chambers for single-molecule FRET. Cold Spring Harbor Protocols , 2012(10), 1104–1108. doi: 10.1101/pdb.prot071530.
- Kapanidis, A. N., Lee, N. K., Laurence, T. A., Doose, S., Margeat, E., & Weiss, S. (2004). Fluorescence-aided molecule sorting: Analysis of structure and interactions by alternating-laser excitation of single molecules. Proceedings of the National Academy of Sciences , 101(24), 8936–8941. doi: 10.1073/pnas.0401690101.
- Kapanidis, A., Majumdar, D., Heilemann, M., Nir, E., & Weiss, S. (2015). Alternating laser excitation for solution-based single-molecule FRET. Cold Spring Harbor Protocols , 2015(11), 979–987. doi: 10.1101/pdb.top086405.
- Kishino, A., & Yanagida, T. (1988). Force measurements by micromanipulation of a single actin filament by glass needles. Nature , 334(6177), 74–76. doi: 10.1038/334074a0.
- Kosol, S., Contreras-Martos, S., Cedeño, C., & Tompa, P. (2013). Structural characterization of intrinsically disordered proteins by NMR spectroscopy. Molecules , 18(9), 10802–10828. doi: 10.3390/molecules180910802.
- Krishnan, R., Goodman, J. L., Mukhopadhyay, S., Pacheco, C. D., Lemke, E. A., Deniz, A. A., & Lindquist, S. (2012). Conserved features of intermediates in amyloid assembly determine their benign or toxic states. Proceedings of the National Academy of Sciences of the United States of America , 109(28), 11172–11177. doi: 10.1073/pnas.1209527109.
- Lakowicz, J. R. (1983). Principles of fluorescence spectroscopy. Boston, MA: Springer US. doi: 10.1007/978-1-4615-7658-7.
- Lee, N. K., Kapanidis, A. N., Wang, Y., Michalet, X., Mukhopadhyay, J., Ebright, R. H., & Weiss, S. (2005). Accurate FRET measurements within single diffusing biomolecules using alternating-laser excitation. Biophysical Journal , 88(4), 2939–2953. doi: 10.1529/biophysj.104.054114.
- Lee, T. C., Kang, M., Kim, C. H., Schultz, P. G., Chapman, E., & Deniz, A. A. (2016). Dual unnatural amino acid incorporation and click-chemistry labeling to enable single-molecule FRET studies of p97 folding. Chembiochem , 17(11), 981–984. doi: 10.1002/cbic.201500695.
- Lemke, E. A. (2011). Site-specific labeling of proteins for single-molecule FRET measurements using genetically encoded ketone functionalities. Methods in Molecular Biology , 751(Chapter 1), 3–15. doi: 10.1007/978-1-61779-151-2_1.
- Lemke, E. A., Gambin, Y., Vandelinder, V., Brustad, E. M., Liu, H.-W., Schultz, P. G., … Deniz, A. A. (2009). Microfluidic device for single-molecule experiments with enhanced photostability. Journal of the American Chemical Society , 131(38), 13610–13612. doi: 10.1021/ja9027023.
- Lobocki, M., Zakrzewska, M., Szlachcic, A., Krzyscik, M. A., Sokolowska-Wedzina, A., & Otlewski, J. (2017). High-yield site-specific conjugation of fibroblast growth factor 1 with monomethylauristatin E via cysteine flanked by basic residues. Bioconjugate Chemistry , 28(7), 1850–1858. doi: 10.1021/acs.bioconjchem.7b00158.
- Lodish, H., Berk, A., Zipursky, S. L., & Matsudaira, P. (1995). Molecular Cell Biology ( 3rd ed.). New York, NY: W. H. Freeman & Co.
- Michalet, X., Weiss, S., & Jäger, M. (2006). Single-molecule fluorescence studies of protein folding and conformational dynamics. Chemical Reviews , 106(5), 1785–1813. doi: 10.1021/cr0404343.
- Milles, S., Tyagi, S., Banterle, N., Koehler, C., Vandelinder, V., Plass, T., … Lemke, E. A. (2012). Click strategies for single-molecule protein fluorescence. Journal of the American Chemical Society , 134(11), 5187–5195. doi: 10.1021/ja210587q.
- Mukhopadhyay, S., Krishnan, R., Lemke, E. A., Lindquist, S., & Deniz, A. A. (2007). A natively unfolded yeast prion monomer adopts an ensemble of collapsed and rapidly fluctuating structures. Proceedings of the National Academy of Sciences , 104(8), 2649–2654. doi: 10.1073/pnas.0611503104.
- Nanda, J. S., & Lorsch, J. R. (2014). Labeling of a protein with fluorophores using maleimide derivitization. Methods in Enzymology 536 , 79–86. doi: 10.1016/B978-0-12-420070-8.00007-6.
- Nasir, I., Onuchic, P. L., Labra, S. R., & Deniz, A. A. (2019). Single-molecule fluorescence studies of intrinsically disordered proteins and liquid phase separation. Biochimica et Biophysica Acta - Proteins Proteomics , 186, 980–987. doi: 10.1016/j.bbapap.2019.04.007.
- Nir, E., Michalet, X., Hamadani, K. M., Laurence, T. A., Neuhauser, D., Kovchegov, Y., & Weiss, S. (2006). Shot-noise limited single-molecule FRET histograms: Comparison between theory and experiments. The Journal of Physical Chemistry. B , 110(44), 22103–22124. doi: 10.1021/jp063483n.
- Schuler, B. (2013). Single-molecule FRET of protein structure and dynamics—a primer. Journal of Nanobiotechnology , 11(Suppl 1), S2. doi: 10.1186/1477-3155-11-S1-S2.
- Schuler, B., Lipman, E. A., & Eaton, W. A. (2002). Probing the free-energy surface for protein folding with single-molecule fluorescence spectroscopy. Nature , 419(6908), 743–747. doi: 10.1038/nature01060.
- Sibille, N., & Bernadó, P. (2012). Structural characterization of intrinsically disordered proteins by the combined use of NMR and SAXS. Biochemical Society Transactions , 40(5), 955–962. doi: 10.1042/BST20120149.
- Sisamakis, E., Valeri, A., Kalinin, S., Rothwell, P. J., & Seidel, C. A. M. (2010). Accurate single-molecule FRET studies using multiparameter fluorescence detection. Methods in Enzymology , 475, 455–514. doi: 10.1016/S0076-6879(10)75018-7.
- Solanki, A., Neupane, K., & Woodside, M. T. (2014). Single-molecule force spectroscopy of rapidly fluctuating, marginally stable structures in the intrinsically disordered protein α-synuclein. Physical Review Letters , 112(15), 158103. doi: 10.1103/PhysRevLett.112.158103.
- Sugase, K., Dyson, H. J., & Wright, P. E. (2007). Mechanism of coupled folding and binding of an intrinsically disordered protein. Nature , 447(7147), 1021–1025. doi: 10.1038/nature05858.
- Swoboda, M., Henig, J., Cheng, H.-M., Brugger, D., Haltrich, D., Plumeré, N., & Schlierf, M. (2012). Enzymatic oxygen scavenging for photostability without pH drop in single-molecule experiments. ACS Nano , 6(7), 6364–6369. doi: 10.1021/nn301895c.
- Thiltgen, G., & Goldstein, R. A. (2012). Assessing predictors of changes in protein stability upon mutation using self-consistency. PLoS One , 7(10), e46084. doi: 10.1371/journal.pone.0046084.
- Widengren, J., & Schwille, P. (2000). Characterization of photoinduced isomerization and back-isomerization of the Cyanine dye Cy5 by fluorescence correlation spectroscopy. The Journal of Physical Chemistry A , 104(27), 6416–6428. doi: 10.1021/jp000059s.
- Wright, P. E., & Dyson, H. J. (2015). Intrinsically disordered proteins in cellular signalling and regulation. Nature Reviews Molecular Cell Biology , 16(1), 18–29. doi: 10.1038/nrm3920.
Key References
- Nikić, I., & Lemke, E. A. (2015). Genetic code expansion enabled site-specific dual-color protein labeling: Superresolution microscopy and beyond. Current Opinion in Chemical Biology , 28, 164–173. doi: 10.1016/j.cbpa.2015.07.021.
A well-crafted compilation of methods involving unnatural amino acids for fluorophore labeling.
- Holmstrom, E. D., Holla, A., Zheng, W., Nettels, D., Best, R. B., & Schuler, B. (2018). Accurate transfer efficiencies, distance distributions, and ensembles of unfolded and intrinsically disordered proteins from single-molecule FRET. Methods in Enzymology , 611, 287–325. doi: 10.1016/bs.mie.2018.09.030.
A comprehensive book chapter detailing how to perform a more complicated single-molecule fluorescence experiment, with details on controls.
- Schuler, B., Soranno, A., Hofmann, H., & Nettels, D. (2016). Single-molecule FRET spectroscopy and the polymer physics of unfolded and intrinsically disordered proteins. Dx.Doi.org , 45(1), 207–231. doi: 10.1146/annurev-biophys-062215-010915.
A review discussing the polymer models that can be leveraged to describe IDPs.
- Sisamakis et al. (2010). See above.
A reference discussing advanced multiparameter single-molecule fluorescence detection (including FRET), as well as instrumentation used for this purpose.
- Hellencamp, B., Schmid, S., Doroshenko, O., Opanasyuk, O., Kühnemuth, R., Rezaei Adariani, S., … Hugel, T. (2018). Precision and accuracy of single-molecule FRET measurements—a multi-laboratory benchmark study. Nature Methods , 15, 669–676.
A reference discussing advanced smFRET data collection and analysis methods, and including descriptions of alternative instrumentation setups from several laboratories.
Internet Resources
Website providing R0 values of commonly used FRET pairs.
A manual for calculating fluorophore labeling ratios.
A general handbook that includes various labeling strategies, product information, and technical notes.
Citing Literature
Number of times cited according to CrossRef: 5
- Sudip Nepal, Erik D. Holmstrom, Single-molecule-binding studies of antivirals targeting the hepatitis C virus core protein, Journal of Virology, 10.1128/jvi.00892-23, 97 , 10, (2023).
- Ashish Joshi, Anuja Walimbe, Anamika Avni, Sandeep K. Rai, Lisha Arora, Snehasis Sarkar, Samrat Mukhopadhyay, Single-molecule FRET unmasks structural subpopulations and crucial molecular events during FUS low-complexity domain phase separation, Nature Communications, 10.1038/s41467-023-43225-y, 14 , 1, (2023).
- Unmesh D. Chowdhury, Arnav Paul, B.L. Bhargava, Interaction of the tau fibrils with the neuronal membrane, Biophysical Chemistry, 10.1016/j.bpc.2023.107024, 298 , (107024), (2023).
- Qiong-Qiong Yao, Jitao Wen, Sarah Perrett, Si Wu, Distinct lipid membrane-mediated pathways of Tau assembly revealed by single-molecule analysis, Nanoscale, 10.1039/D1NR05960B, 14 , 12, (4604-4613), (2022).
- Eitan Lerner, Anders Barth, Jelle Hendrix, Benjamin Ambrose, Victoria Birkedal, Scott C Blanchard, Richard Börner, Hoi Sung Chung, Thorben Cordes, Timothy D Craggs, Ashok A Deniz, Jiajie Diao, Jingyi Fei, Ruben L Gonzalez, Irina V Gopich, Taekjip Ha, Christian A Hanke, Gilad Haran, Nikos S Hatzakis, Sungchul Hohng, Seok-Cheol Hong, Thorsten Hugel, Antonino Ingargiola, Chirlmin Joo, Achillefs N Kapanidis, Harold D Kim, Ted Laurence, Nam Ki Lee, Tae-Hee Lee, Edward A Lemke, Emmanuel Margeat, Jens Michaelis, Xavier Michalet, Sua Myong, Daniel Nettels, Thomas-Otavio Peulen, Evelyn Ploetz, Yair Razvag, Nicole C Robb, Benjamin Schuler, Hamid Soleimaninejad, Chun Tang, Reza Vafabakhsh, Don C Lamb, Claus AM Seidel, Shimon Weiss, FRET-based dynamic structural biology: Challenges, perspectives and an appeal for open-science practices, eLife, 10.7554/eLife.60416, 10 , (2021).

