Purification of Adeno-Associated Viral Vector Serotype 9 Using Ceramic Hydroxyapatite Chromatography and its Analysis
Yae Kurosawa, Yae Kurosawa, Yuji Tsunekawa, Yuji Tsunekawa, Mikako Wada, Mikako Wada, Yuko Aizen, Yuko Aizen, Yuko Nitahara-Kasahara, Yuko Nitahara-Kasahara, Takashi Okada, Takashi Okada
Abstract
Adeno-associated virus (AAV) vectors can efficiently transduce exogenous genes into various tissues in vivo. Owing to their convenience, high efficiency, long-term stable gene expression, and minimal side effects, AAV vectors have become one of the gold standards for investigating gene functions in vivo, especially in non-clinical studies. However, challenges persist in efficiently preparing a substantial quantity of high-quality AAV vectors. Commercial AAV vectors are typically associated with high costs. Further, in-laboratory production is hindered by the lack of specific laboratory equipment, such as ultracentrifuges. Therefore, a simple, quick, and scalable preparation method for AAV vectors is needed for proof-of-concept experiments. Herein, we present an optimized method for producing and purifying high-quality AAV serotype 9 (AAV9) vectors using standard laboratory equipment and chromatography. Using ceramic hydroxyapatite as a mixed-mode chromatography medium can markedly increase the quality of purified AAV vectors. Basic Protocols and optional methods for evaluating purified AAV vectors are also described. © 2024 The Author(s). Current Protocols published by Wiley Periodicals LLC.
Basic Protocol 1 : Production of AAV9 vectors in 293EB cells
Basic Protocol 2 : Concentration and buffer exchange of AAV9 vectors from 293EB cell culture supernatants using tangential flow filtration
Basic Protocol 3 : Purification of AAV9 vectors from TFF samples using ceramic hydroxyapatite chromatography
Basic Protocol 4 : Analysis of the purified AAV9 vectors
INTRODUCTION
Adeno-associated virus (AAV), a member of the Parvoviridae family, is one of the smallest non-envelope viruses with a single-stranded DNA genome (Samulski & Muzyczka, 2014). AAV has been extensively explored as an in vivo gene delivery vector due to its weak immune response, high infective efficiency, and stable long-term gene expression. Consequently, AAV vectors have emerged as promising gene delivery systems for gene therapy modalities (Van Vliet et al., 2008). Traditionally, laboratories rely on ultracentrifugation to purify the AAV vectors (Belova et al., 2022). Although ultracentrifugation is a sophisticated technique, this process is time consuming, requires expertise, and has low scalability. Although we developed an improved, scalable, and short-term ultracentrifugation method that is compliant with Good Manufacturing Practice (Wada et al., 2023), proof-of-concept experiments in the laboratory usually do not require the removal of empty capsids and instead focus on eliminating contaminated proteins and nucleotides. Wada et al. (2023) developed a buffer-exchange method using a ceramic hydroxyapatite column as an alternative to dialysis after ultracentrifugation. Thus, we introduced a simple and scalable purification method using ceramic hydroxyapatite chromatography (Kurosawa et al., 2012, Kurosawa et al., 2019). This method enables the purification of AAV vectors at neutral pH, reducing the denaturation and deactivation of AAV vectors that often occur with affinity chromatography (Lins-Austin et al., 2020; Miyaoka et al., 2023). Here, we describe the detailed protocols from production to purification and quality quantification of AAV serotype 9 (AAV9) vectors. Basic Protocol 1 covers 293EB cell culture, transfection, and harvesting of AAV9 vector particles. Basic Protocol 2 covers concentration, diafiltration, and buffer exchange of AAV vectors from cell culture supernatant using tangential flow filtration (TFF). Basic Protocol 3 covers purification of AAV9 vectors using ceramic hydroxyapatite chromatography. Basic Protocol 4 covers quantification and evaluation of the purified AAV vectors.
Basic Protocol 1: PRODUCTION OF AAV9 VECTORS IN 293EB CELLS
Previously, we presented our adeno-associated virus serotype 9 (AAV9) production protocol, which outlined the steps for cell culture, triple transfection, and harvesting of the culture supernatant (Wada et al., 2023). Using Basic Protocol 1, the culture supernatant can be prepared for the purification process outlined in Basic Protocol 2.
Materials
-
HEK293EB cells (Tomono et al., 2018)
-
Culture medium (see recipe)
-
pHelper (pAdDeltaF6; Addgene, cat. no. 112867)
-
pAAV2/9n (Addgene, cat. no. 112865)
-
pAAV-ZsGreen1 vector (Takara Bio, cat. no. 6231)
-
Transfection medium (see recipe)
-
2 mg/ml PEI MAX (see recipe)
-
CELLdisc, 4 layers, 1000-cm2 (Greiner Bio-One International, cat. no. 678104)
-
CO2 incubator, 37°C
-
Vortex
-
Easy grip polystyrene storage bottle with 45-mm caps, 150-ml (Corning, cat. no. 431175)
-
Centrifuge bottle, 1000PP bottle (WM) (Eppendorf Himac Technologies, cat. no. 5721411025)
-
Centrifuge (Eppendorf Himac Technologies, cat. no. himac CR30NX)
-
500-ml rapid flow bottle top filter, 0.45-μm PES membrane (Thermo Fisher Scientific, cat. no. 295-4545) and vacuum
Preparation of 293EB cells (Day 1)
1.Seed 293EB cells in culture medium in a CELLdisc (4 × 107 cells per 150 ml per CELLdisc) and incubate at 37°C in a CO2 incubator (Day 1).
Transfection of the AAV9 vectors into cells (Day 4)
2.On Day 4, transfect three plasmids (pHelper, pAAV2/9n, and pAAV-ZsGreen1) into 293EB cells as described in steps 3 to 8.
3.Prepare DNA mixture comprising 38.66 μg pHelper, 19.36 μg pAAV2/9n, and 19.36 μg pAAV-ZsGreen1 in 4.4 ml transfection medium for one CELLdisc.
4.Dilute 116 μl of 2 mg/ml PEI MAX (232 μg total) with 4.4 ml transfection medium for one CELLdisc.
5.Combine the DNA mixture and diluted PEI MAX solution, vortex, and allow the mixture to settle for at least 15 min at room temperature.
6.Add the DNA and PEI MAX mixture to 150 ml transfection medium in a storage bottle like an Easy grip polystyrene storage bottle.
7.Remove the supernatant from the CELLdisc and add 150 ml transfection medium containing the DNA and PEI MAX mixture.
8.Culture cells at 37°C in a CO2 incubator.
Harvesting of the AAV9 vector particles (Day 9)
9.On Day 9, collect and decant the supernatant in a centrifuge bottle.
10.Centrifuge the supernatant 15 min at 2000 × g , 4°C, and filter the solution using a 0.45-µm bottle-top filter and a vacuum to remove cells and cell debris.
11.Store the clarified supernatant at 4°C until use.
Basic Protocol 2: CONCENTRATION AND BUFFER EXCHANGE OF AAV9 VECTORS FROM 293EB CELL CULTURE SUPERNATANTS USING TANGENTIAL FLOW FILTRATION
A description of the protocol for performing the initial step of AAV vector purification is provided below. A tangential flow filtration (TFF) system is used to concentrate and diafiltrate the AAV9 crude sample. This process yields the AAV9 vector solution required for use in Basic Protocol 3.
Materials
-
Clarified cell culture supernatant from Basic Protocol 1
-
TFF buffer (see recipe)
-
H2O, ultrapure
-
Ethanol (Nacalai Tesque, cat. no. 14813-95)
-
Water bath (Thermax Water Bath TM-2A, AS ONE, cat. no. 1-4594-32)
-
ReadyFilter hollow fiber cartridge, 500 kDa, membrane surface area of 650 cm2 (Cytiva, cat. no. RTPUFP-500-C-4MS)
-
KrosFlo KR2i TFF system (Repligen Corporation)
-
500-ml rapid-flow bottle top filter with 0.45-μm PES membrane (Thermo Fisher Scientific, cat. no. 295-4545)
1.Pre-warm 500 ml clarified cell culture supernatant from Basic Protocol 1 and TFF buffer in a water bath at 37°C.
2.Connect the hollow fiber cartridge to the TFF system.
3.Rinse the hollow fiber cartridge with ultrapure water at a flow rate of 200 ml/min to remove 30% ethanol.
4.Concentrate ∼500 ml of the cell culture supernatant to 50 to 60 ml at a flow rate of 800 ml/min.
5.Diafiltrate the concentrated culture supernatant with 2 L of pre-warmed TFF buffer at a flow rate of 800 ml/min.
6.Collect the diafiltrated sample and add 50 ml of the fresh TFF buffer to the sampling bottle.
7.Circulate the buffer at a flow rate of 200 ml/min for 10 min by closing the permeate vent to collect the remaining AAV vector in the system.
8.Repeat steps 6 and 7.
9.Collect the remaining AAV vector in the system.
10.Filter the diafiltrated culture supernatant (200 ml total) through a 0.45-μm bottle top filter.
11.After TFF, store the sample at 4°C until use.
Basic Protocol 3: PURIFICATION OF THE AAV9 VECTORS FROM TFF SAMPLES USING CERAMIC HYDROXYAPATITE CHROMATOGRAPHY
The protocol used to perform the second step of AAV vector purification is outlined below. Ceramic hydroxyapatite chromatography is employed to purify AAV9 vectors from the concentrated and diafiltrated crude supernatants using TFF. This step yields AAV9 vectors with high purity. The evaluation protocol is described in Basic Protocol 4.
Materials
-
CHT ceramic hydroxyapatite, Type I, 40-μm (Bio-Rad Laboratories)
-
Equilibration buffer (see recipe)
-
6 N HCl
-
CIP solution (see recipe)
-
Wash buffer (see recipe)
-
Elution buffer (see recipe)
-
H2O, ultrapure
-
Ethanol (Nacalai Tesque, cat. no. 14813-95)
-
Empty stainless-steel column, 4.6-mm i.d. × 35-mm, 10-μm mesh filter [Sugiyama Shoji, cat. no. LCIT-WF-4.6×35-10 (mesh filter)]
-
Balance
-
Poly funnel, polypropylene (AS ONE, cat. no. 6-316-01)
-
2- to 3-cm tube that can fit to a column and a funnel
-
Cutter blade
-
Vantage L laboratory column, VL16 × 250 (Merck, cat. no. 96160250)
-
ÄKTA avant 25 (Cytiva)
Packing the empty column with ceramic hydroxyapatite medium
1.To create a small ceramic hydroxyapatite column, pack CHT medium into an empty stainless-steel column using a dry method as described in steps 2 to 7.
2.Attach an end fitting to only one side of the empty column and tare it with a balance.
3.Connect a small funnel to the other side of the column with 2 to 3 cm of tube and add powder of CHT medium.
4.Tap the column on the table.
5.Scrape off any CHT powder protruding from the column with a cutter blade.
6.Measure its weight on a balance. The recommended weight is ∼0.35 to 0.37 g.
7.Set the end fitting.
Setting up the chromatography system: ÄKTA avant 25
8.Place each pump and buffer line in the chromatography system in the appropriate buffer. Equilibrate the sample loop, pH probe, and fraction collector line with equilibration buffer.
Purification of the AAV9 vectors using ceramic hydroxyapatite chromatography
If TFF buffer is not used for the TFF step, the pH of the sample should be adjusted to ∼6.5 with diluted HCl, e.g., 6 N HCl after TFF.
9.Place a small ceramic hydroxyapatite column in the ÄKTA avant 25, wash the column with 5 ml CIP solution and 10 ml wash buffer, and equilibrate with the equilibration buffer (>15 ml) at a flow rate of 1.0 ml/min.
10.Load 10 ml of the sample onto the small ceramic hydroxyapatite column.
11.Wash the column with 10 ml equilibration buffer.
12.Elute the AAV9 vector using 10 ml buffer (the B pump set at 15% elution buffer).
13.Wash the column with 5 ml wash buffer.
Basic Protocol 4: ANALYSIS OF THE PURIFIED AAV9 VECTORS
The protocol for quantifying and evaluating the purified AAV vectors is outlined below. The copy number of the viral genome (vg) for the purified AAV vector can be quantified using TaqMan quantitative polymerase chain reaction (qPCR). The amount of protein contaminants in the AAV vector solution can be determined using the HEK293 HCP ELISA kit and sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), according to the manufacturer's instructions. The amount of double-stranded DNA (dsDNA) contaminants derived from the host cell dsDNA and plasmid DNA can be quantified using a Quant-iT PicoGreen dsDNA assay kit. Finally, the full/empty ratio of purified AAV9 vectors can be determined using transmission electron microscopy (TEM) (Wada et al., 2023).
Materials
-
Sample purified in Basic Protocols 2 and 3
-
Recombinant DNase I, RNase-free, with buffer (Takara Bio, cat. no. 2270A)
-
DEPC-treated H2O (Nacalai Tesque, cat. no. 36415-54)
-
Buffer AL, lysis buffer (QIAGEN, cat. no. 19075)
-
THUNDERBIRD probe qPCR mix (TOYOBO, cat. no. QPS-101)
-
Detection probe for the ZsGreen1 transgene (Bryda et al., 2019), synthesized by Integrated DNA Technologies:
- 5’-/56-FAM/TT CAT CCA G/ZEN/C ACA AGC TGA C/3IABkFQ/-3’
-
Primers for the ZsGreen1 transgene (Bryda et al., 2019), synthesized by Eurofin Genomics:
- 5’-GTG TAC AAG GCC AAG TCC GT-3’
- 5’-CCA CTT CTG GTT CTT GGC GT-3’
-
NuPAGE LDS sample buffer, 4× (Thermo Fisher Scientific, cat. no. NP0007)
-
NuPAGE reducing agent, 10× (Thermo Fisher Scientific, cat. no. NP0009)
-
NuPAGE MOPS SDS running buffer, 20× (Novex, cat. no. NP0001)
-
NuPAGE 4% to 12% Bis-Tris gel, 1.5-mm, mini protein gel, 10-well (Thermo Fisher Scientific, cat. no. NP0335BOX)
-
PageRuler unstained protein ladder (Thermo Fisher Scientific, cat. no. 26614)
-
Oriole fluorescent gel stain (Bio-Rad Laboratories, cat. no. 1610496)
-
H2O, ultrapure
-
HEK293 HCP ELISA kit 3G (Cygnus Technologies, cat. no. F650S)
-
Sample diluent buffer (Cygnus Technologies, cat. no. I700)
-
Quant-iT PicoGreen dsDNA assay kit (Thermo Fisher Scientific, cat. no. P7589)
-
Phosphotungstic acid EM (TAAB Laboratories Equipment, cat. no. P013)
-
Thermal cycler (C1000 Touch; Bio-Rad Laboratories)
-
MicroAmp fast optical 96-well reaction plate, 0.1-ml (Thermo Fisher Scientific, cat. no. 4346907)
-
MicroAmp optical adhesive film (Thermo Fisher Scientific, cat. no. 4311971)
-
QuantStudio 3 (Applied Biosystems by Thermo Fisher Scientific)
-
Heat block (block incubator) (Astec, cat. no. BI-516S)
-
1.5-ml microcentrifuge tube (Thermo Fisher Scientific, cat. no. 3448)
-
Electrophoresis system (XCell SuperLock Mini-Cell; Thermo Fisher Scientific, cat. no. EI0001)
-
PowerStation Ghibli I (ATTO, cat. no. WSE-3100)
-
ChemiDoc Touch imaging system (Bio-Rad Laboratories, cat. no.1708370)
-
Plate reader (Varioskan Flash; Thermo Fisher Scientific)
-
Black-type fluorescent plate S (Sumitomo Bakelite, cat. no. MS-8496K)
-
Collodion membranes (Nissin EM, cat. no. 6512)
-
Ion bombarder (Nissin EM, type PIB-10)
-
Filter paper (Advantec Toyo Kaisha, cat. no. 00011090)
-
TEM (Hitachi High-Tech, cat. no. HT7800)
qPCR
1.Add recombinant DNase I (RNase-free; 1 μl), 10× DNase I buffer (2 μl) and DEPC-treated water (15 μl) to the samples (2 μl each) and incubate the mixture at 37°C for 15 min and 95°C for 10 min in a thermal cycler.
2.Lyse the vector particles (DNase-treated sample, 2 μl) with AL lysis buffer (40 μl) and DEPC-treated water (38 μl) at 56°C for 10 min in a thermal cycler.
3.Dilute the lysed sample 100-fold using DEPC-treated water.
4.Prepare the pre-mixed reagents in Table 1.
| Reagents | Vol (μl) |
|---|---|
| TOYOBO Thunderbird probe qPCR mix (2×) | 10 |
| ZsGreen1-probe (10 μΜ) | 0.4 |
| ZsGreen1-F primer (100 μΜ) | 0.06 |
| ZsGreen1-R primer (100 μΜ) | 0.06 |
| ROX reference dye (50×) | 0.04 |
| DEPC-treated water | 7.44 |
| Total | 18 |
5.Dispense 18 μl of the pre-mixed reagents and 2 μl of the diluted samples into the wells of a MicroAmp fast optical 96-well microplate.
6.Place the sealed microplate in the QuantStudio 3 instrument, perform PCR according to the conditions outlined in Table 2 (recommended by TOYOBO), and determine the titers.
| Step | Temperature (°C) | Time (s) |
|---|---|---|
| Initial denaturation | 94 | 20 |
| PCR (40 cycles): | ||
| Denaturation | 95 | 3 |
| Extension | 60 | 30 |
SDS-PAGE
Perform the following steps using 0.5–5.0 × 1010 vg/lane of samples.
7.Preheat heat block to 70°C.
8.Prepare the sample buffer mixture by mixing 8.75 μl NuPAGE LDS sample buffer (4×) and 3.5 μl NuPAGE reducing agent (10×) per sample.
9.Dispense 12.25 μl of the sample buffer mixture into 1.5-ml microcentrifuge tubes.
10.Dispense 22.75 μl of the sample into microcentrifuge tubes.
11.Heat the mixture at 70°C for 10 min on the heat block.
12.Dilute the NuPAGE MOPS SDS running buffer 20-fold to prepare the running buffer.
13.Assemble the electrophoresis system by adding the gel to the system and pouring the running buffer into the tank.
14.Dispense 2 μl/lane of marker (ladder) and 35 μl/lane of samples into the appropriate lanes.
15.Place the lid on the system, connect the cords to the power station, and run electrophoresis at 200 V (constant voltage) for 50 min.
16.Remove the gel and gel plates from the system.
17.Add enough Oriole fluorescent gel stain to soak the gel and stain by shaking for 90 min at room temperature.
18.Lightly rinse the gel with ultrapure water.
19.Place the gel in the ChemiDoc Touch imaging system and capture an image.
Quantifying the level of the host cell protein contaminant
Use the HEK293 HCP ELISA kit according to the manufacturer's instructions, with minor modifications.
20.Allow all reagents to equilibrate to room temperature.
21.Prepare the wash buffer (1 L) by diluting 50 ml of the wash concentrate (20×) with ultrapure water.
22.Dispense 100 μl of anti-HEK:HRP into each of the required number of wells for the kit standards and samples.
23.Dispense 50 μl of the kit standards and samples diluted with sample diluent buffer to be within the standard curve into the appropriate wells and incubate the microplate at room temperature in the dark for 1.5 hr.
24.Wash the plate four times with 200 μl/well of wash buffer.
25.Dispense 100 μl of the TMB substrate into each well and incubate the microplate at room temperature for 30 min.
26.Stop the reaction to by adding 100 μl stop solution to each well.
27.Measure the absorbance at 450/650 nm using a microplate reader.
28.Calculate the concentration of host cell protein (HCP) in each sample using the standard curve.
Quantifying the level of the dsDNA contaminant
Use the Quant-iT PicoGreen dsDNA assay kit according to the manufacturer's instructions.
29.Dilute the TE supplied in the kit by 20-fold with ultrapure water (1 ml TE + 19 ml ultrapure water, per plate).
30.Dilute the DNA standard curve for calibration with TE to a final volume of 100 μl in wells of a black-type fluorescent plate.
31.Dilute the samples with TE to a final volume of 100 μl in microplate wells.
32.Add 100 μl of PicoGreen diluted 200-fold to each sample.
33.Incubate the samples at room temperature in the dark for 5 min.
34.Measure the fluorescence of the samples using a microplate reader (excitation ∼480 nm, emission ∼520 nm).
35.Determine the concentration of dsDNA in the samples using a standard curve.
| dsDNA | HCP | |||||
|---|---|---|---|---|---|---|
| Sample | Concentration (ng/ml) | Removal (%) | Total removal (%) | Concentration (ng/ml) | Removal (%) | Total removal (%) |
| Supernatant | 7309 | 36,755 | ||||
| TFF | 1446 | 92.085 | 21.6 | 99.976 | ||
| Chromatography | 3.0 | 99.959 | 99.997 | 8.0 | 92.613 | 99.998 |
Full/empty ratio analysis of the AAV9 vectors using transmission electron microscopy (optional)
Perform transmission electron microscopy (TEM), according to previously described method (Wada et al., 2023). If TEM facilities are not available, it may be convenient to outsource TEM imaging.
36.Hydrophilize the collodion membranes using an ion bombarder as described in steps 37 to 40.
37.Place the collodion membranes with tweezers on the metal or plastic plate.
38.Transfer the plate with the membranes to the ion bombarder.
39.Set the parameters: Hard and 1.5 min.
40.Press the green bottom to start hydrophilizing the membranes.
41.Place the samples (3 μl) on a hydrophilized grid (collodion membranes) for 1 min.
42.Dispense ∼3 μl of ultrapure water and adsorb using a filter paper; repeat three times.
43.Stain the samples with 3 μl of 2% phosphotungstic acid solution pH 7.0 for 10 s and then adsorb with a filter paper.
44.Place the membrane on its face and place the sample on a filter paper for 2 min.
45.Analyze the samples loaded onto the collodion membranes using TEM (HC-1 mode; high resolution; acceleration voltage, 100 kV; magnification, 25.0 k).
46.Analyze the full/empty ratio of AAV vectors by counting the full and empty capsid numbers.
REAGENTS AND SOLUTIONS
CIP solution: 0.5 M NaOH
- 20 g NaOH (Nacalai Tesque, cat. no. 31511-05)
- Bring up volume to 1 L with ultrapure water
- Filter the solution using a 0.22-μm filter
- Degas for chromatography
- Store up to 3 months at room temperature
Culture medium
- 500 ml Dulbecco's modified Eagle medium (DMEM), high-glucose, supplemented with L-glutamine and phenol red (Fujifilm Wako Pure Chemical Corporation, cat. no. 044-29765)
- 50 ml fetal bovine serum (FBS) (Sigma-Aldrich, cat. no. F0926-500ML)
- 5 ml penicillin-streptomycin solution (Nacalai Tesque, cat. no. 26253-84)
- Prepare under sterile conditions
- Store up to 3 weeks at 4°C
Elution buffer
Prepare a solution of 150 mM NaCl/100 mM sodium phosphate buffer using Na2HPO4·12H2O (Nacalai Tesque, cat. no. 31722-45), NaH2PO4·2H2O (Nacalai Tesque, cat. no. 31718-15), and NaCl (Nacalai Tesque, cat. no. 31320-05) following instructions in Current Protocols (2006). Adjust the pH to 7.2. Filter the solution using a 0.22-μm filter. Degas for chromatography. Store up to 3 weeks at room temperature.
Equilibration buffer
- 4.77 g HEPES (Dojindo Laboratories, cat. no. 346-08235) (10 mM)
- 17.53 g NaCl (Nacalai Tesque, cat. no. 31320-05) (150 mM)
- 1.9 L H2O, ultrapure
- Adjust to pH 7.2 using NaOH
- Bring up volume to 2 L with ultrapure water
- Filter the solution using a 0.22-μm filter
- Degas for chromatography
- Store up to 3 weeks at room temperature
PEI MAX, 2 mg/ml
- 1 g PEI MAX transfection grade linear polyethyleneimine hydrochloride, MW 40,000 (Polysciences, cat. no. 24765-1)
- 500 ml H2O, ultrapure
- Adjust pH to ∼7.0 using NaOH or HCl
- Filter the solution using a 0.22-μm filter
- Store aliquots up to 3 years at −30°C
TFF buffer
Dilute the 10× TFF stock solution (see recipe) 10-fold with ultrapure water and add a 1/1000 volume of 1 M MgCl2 (Nacalai Tesque, cat. no. 20942-34) (1 mM) to the TFF buffer. Store up to 3 months at room temperature.
Of note, 10 mM HEPES, 150 mM NaCl pH 7.2, and 1 mM MgCl 2 can also be used as the TFF buffer. However, the pH of the sample must be adjusted to 6.5 before loading onto a ceramic hydroxyapatite column.
TFF stock solution, 10×
- 21.33 g MES (Nacalai Tesque, cat. no. 21623-26) (100 mM)
- 87.66 g NaCl (Nacalai Tesque, cat. no. 31320-05) (1.5 M)
- 800 ml H2O, ultrapure
- Adjust to pH 6.5 using NaOH
- Bring volume up to 1 L with ultrapure water
- Store up to 3 months at room temperature
Transfection medium
- 500 ml Dulbecco's modified Eagle medium (DMEM), high-glucose, supplemented with L-glutamine and phenol red (Fujifilm Wako Pure Chemical Corporation, cat. no. 044-29765)
- 5 ml penicillin-streptomycin solution (Nacalai Tesque, cat. no. 26253-84)
- 5 ml of 200 mmol/L L-alanyl-L-glutamine solution, 100× (Nacalai Tesque, cat. no. 04260-64)
- 8.4 ml of 7.5% NaHCO3 (Nacalai Tesque, cat. no. 31213-15), filtered using a 0.22-μm filter
- 6.7 ml of 10% D-(+)-glucose (Nacalai Tesque, cat. no. 16806-25), filtered using a 0.22-μm filter
- Prepare under sterile conditions
- Store up to 3 weeks at 4°C
Wash buffer
Prepare a solution of 400 mM sodium phosphate buffer using Na2HPO4·12H2O (Nacalai Tesque, cat. no. 31722-45) and NaH2PO4·2H2O (Nacalai Tesque, cat. no. 31718-15) following instructions in Current Protocols (2006). Adjust the pH to 7.2. Filter the solution using a 0.22-μm filter. Degas for chromatography. Store up to 3 weeks at room temperature.
COMMENTARY
Critical Parameters
Using the correct quantity of plasmids and PEI is critical in the production of AAV9 vectors (Basic Protocol 1) as different ratios or amounts can affect the cytotoxicity and efficiency of production.
TFF is an effective tool for removing contaminants, such as host cell proteins (HCPs), DNA, and plasmids, and for concentrating the AAV vector in the culture supernatant (Basic Protocol 2). Crucial considerations include pre-warming the buffer and sample at 37°C. This step increases the fluidity of the sample and buffer and efficiently removes small molecules. A TFF with a 500-kDa cutoff hollow fiber is recommended in this protocol. Performing TFF with a smaller cutoff will increase recovery under the same conditions but will decrease the efficiency of small molecule removal. As this protocol is optimized for a 500 ml sample and a hollow fiber with a membrane surface area of 650 cm2, washing with 2 L buffer should be performed to effectively remove small molecules.
The critical elements in Basic Protocol 3 include the sodium phosphate buffer and sample pH for ceramic hydroxyapatite chromatography. Anhydrous sodium dihydrogen phosphate or anhydrous disodium hydrogen phosphate should not be used to prepare sodium phosphate buffer due to the potential presence of polyphosphoric acid. This acid can chelate Ca in the apatite and damage the column. The pH of the samples should be adjusted to ∼6.5. However, replacing the buffer (pH 6.5) with TFF buffer eliminates the need for pH control. A high sample pH decreases the adsorption efficiency while a low sample pH (below pH 6.5) damages the column. To avoid damaging the column, the addition of EDTA to the buffer is discouraged.
TEM analysis enables the differentiation between full and empty capsids with small sample volumes (Basic Protocol 4). Tweezers can be used to grasp the membrane at the edge of the grid. When the sample is placed on the membrane, subsequent steps proceed smoothly. An AAV vector concentration of >1.0 × 1012 vg/ml is ideal for TEM with negative staining. The prepared samples can be stored in a desiccator for an extended duration.
Troubleshooting
The troubleshooting procedure and corresponding solutions are presented in Table 4.
| Problem | Possible cause | Solution |
|---|---|---|
| Basic Protocol Basic Protocol 2 | ||
| Increased pressure in TFF | Membrane is clogged | Wash hollow fiber well with pre-warmed NaOH at 55°C and rinse with ultrapure water; follow the manufacturer's manual |
| Basic Protocol Basic Protocol 3 | ||
| Low adsorption efficiency on the column | Sample pH is not suitable | Check the sample pH and re-adjust to 6.5 |
| Excessive loading volume | Decrease and optimize the volume of sample | |
| Basic Protocol Basic Protocol 4 | ||
| AAV capsids not found | Low concentration of AAV | Concentrate samples via ultrafiltration (e.g., ultrafiltration column) |
Understanding Results
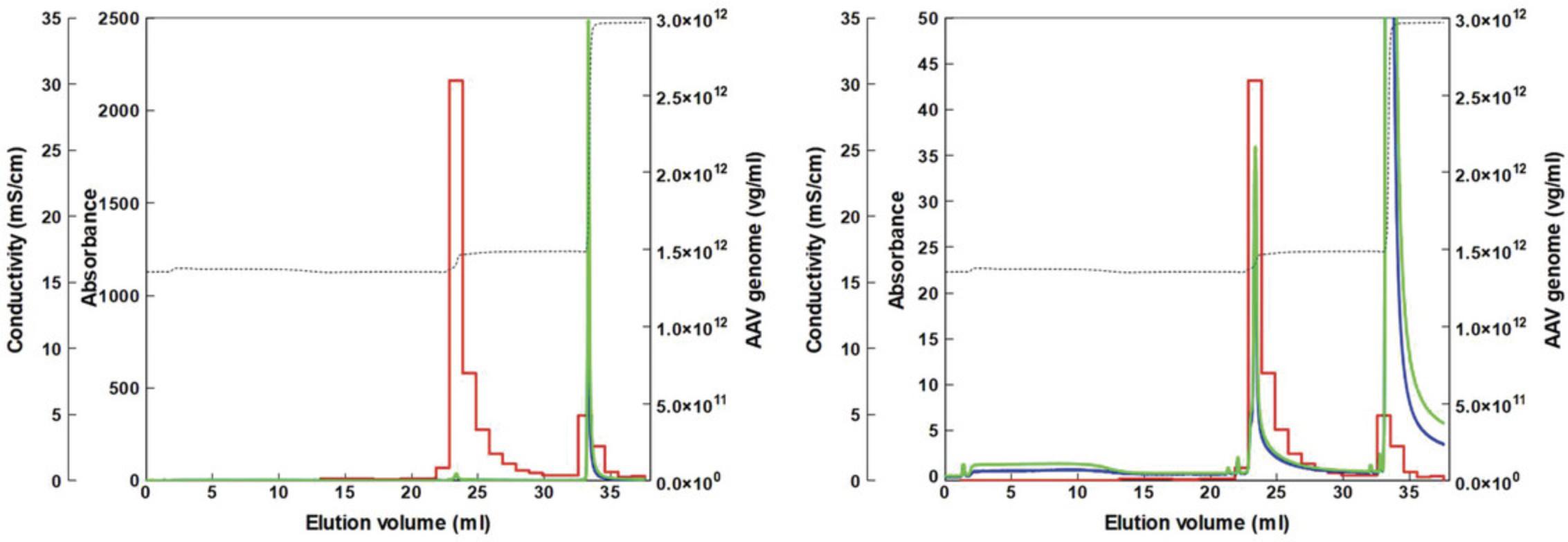
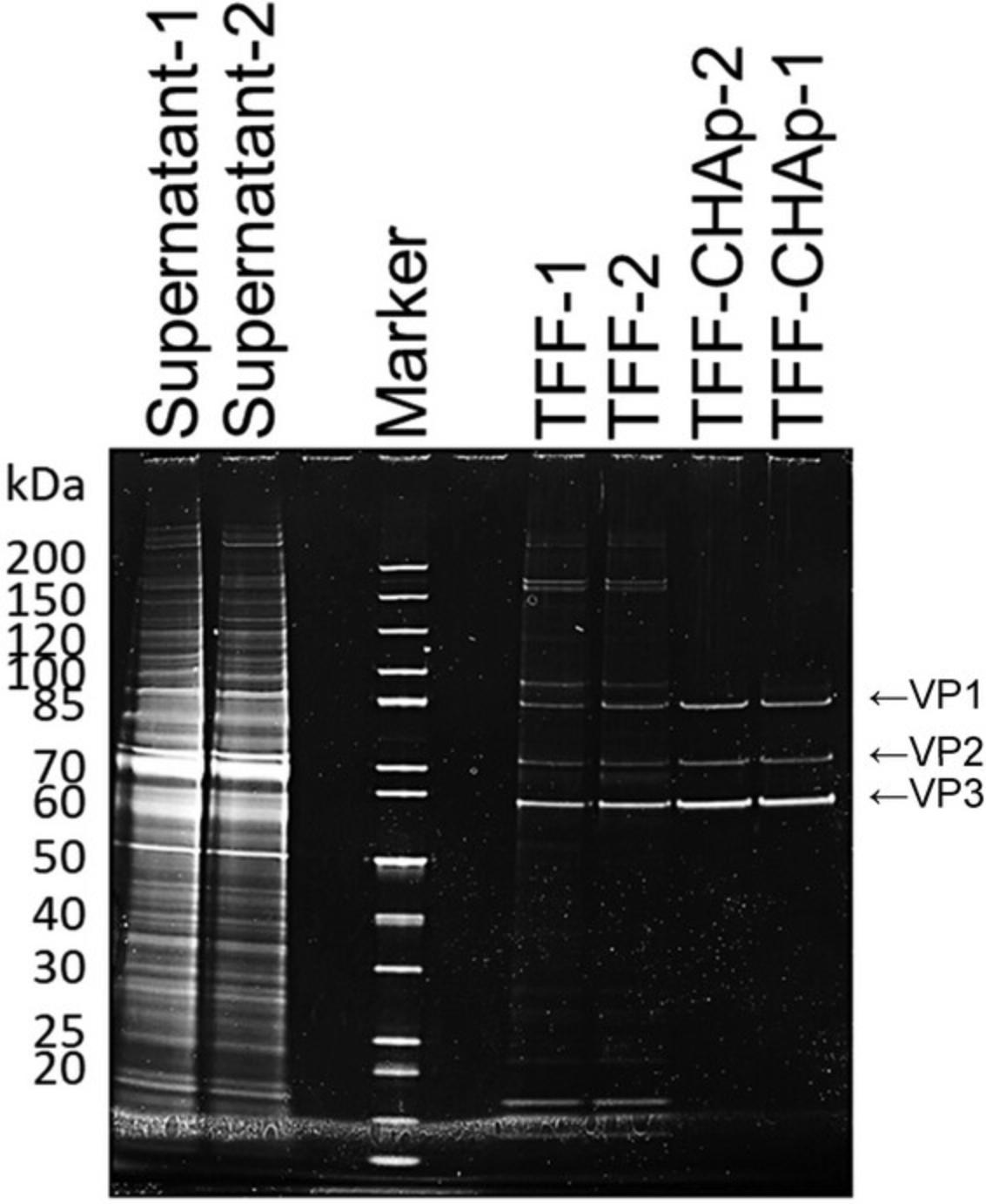
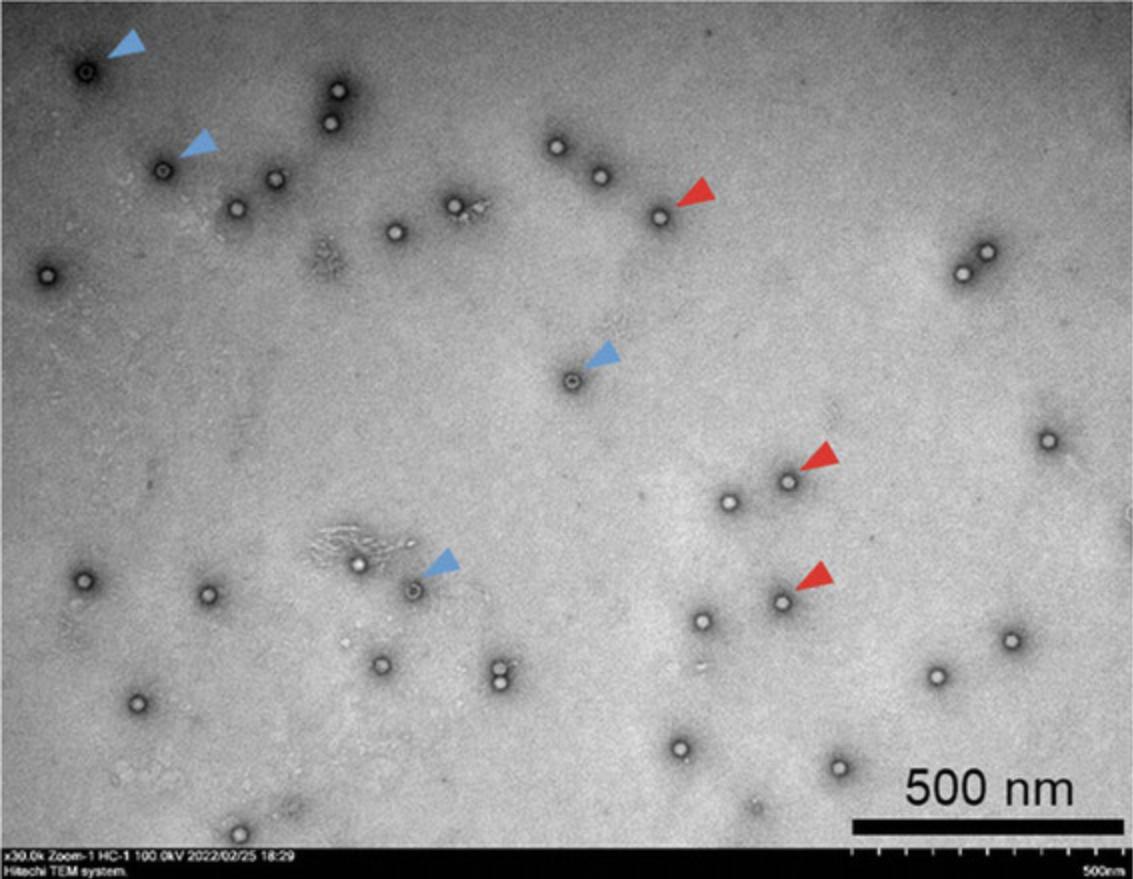
AAV9 vectors with a high titer were produced using Basic Protocol 1 and then purified using Basic Protocols 2 and 3. Figure 1 shows a representative chromatogram of the AAV9 vector after TFF using a ceramic hydroxyapatite column. The eluates were pooled and analyzed via SDS-PAGE (Fig. 2). The contaminant bands in the sample decreased after TFF but remained detectable. Following two-step purification with TFF and ceramic hydroxyapatite chromatography, good purity was obtained and only three AAV structural proteins: VP1, VP2, and VP3, were detected. The purity was further evaluated (Table 3). In terms of contaminants, HCP was efficiently removed by TFF while dsDNA was efficiently removed by ceramic hydroxyapatite chromatography, with removal efficiencies of 99.998% and 99.997%, respectively (Table 3). Morphological analysis of the purified AAV9 vector particles was performed using TEM (Fig. 3). Although full and empty capsids were not separated using this protocol, good particles with good shape and high purity were obtained.
As TFF and ceramic hydroxyapatite chromatography have already been used in biopharmaceutical manufacturing, both are scalable methods. The scalability can be adjusted according to the equipment, facility capabilities, and sample requirements.
DNase pre-treatment is usually performed during purification to remove dsDNA derived from host cells. This pre-treatment step is not required, thereby promoting time and cost-effectiveness. However, when DNA removal is insufficient during scale-up, a pre-treatment step should be considered.
Basic Protocols 1, 2, and 4 can be applied to other serotypes using other serotype plasmids on the transfection. Basic Protocol 3 is suitable for AAV9 and AAV8. For other serotypes, it requires to optimize the purification conditions of ceramic hydroxyapatite chromatography.
Time Considerations
For Basic Protocol 1, the seeding of cells takes 30 min on Day 1, transfection takes 1 hr on Day 4, and harvesting is completed in 40 min on Day 9; thus, a total of 9 days is required to carry out this protocol.
For Basic Protocol 2, the preparation (pre-warming buffer and sample, and setting and washing TFF) requires ∼1 hr, the TFF run takes ∼2 to 3 hr, and the typical cleaning process lasts 1 to 1.5 hr.
For Basic Protocol 3, packing ceramic hydroxyapatite powder into the column takes ∼5 min and setting up the chromatography system takes 20 min. An additional 30 to 40 min is required to load the column onto the chromatography system and then wash and equilibrate the column. Purification requires ∼40 min, and sterilization and cleaning take ∼20 to 30 min.
For Basic Protocol 4, qPCR lasts ∼2.5 hr; SDS-PAGE takes ∼3 to 4 hr; dsDNA detection takes 30 min; and HCP ELISA takes 2.5 to 3 hr. For TEM analysis, ∼5 min is required to prepare the sample, ∼1 hr is required to set up the TEM system, and ∼1.5 hr is required for observation.
Acknowledgments
We thank Dr. Rumi Miyaoka (Asahi Kasei Medical Co., Ltd.) for providing technical support for TFF, and Dr. Yukihiko Hirai (The Institute of Medical Science, The University of Tokyo) and Dr. Tomohiko Yoshitake (HOYA Technosurgical Corporation) for the helpful discussions. This research was funded by the Japan Agency for Medical Research and Development (AMED) with the grant numbers JP21ae0201001, JP21ae0201005, and JSPS KAKENHI with the grant number 20K06464 and 20H03788.
Author Contributions
Yae Kurosawa : Conceptualization; data curation; formal analysis; investigation; methodology; project administration; resources; validation; visualization; writing original draft; writing review and editing. Yuji Tsunekawa : Conceptualization; data curation; formal analysis; funding acquisition; investigation; methodology; project administration; resources; supervision; validation; writing original draft; writing review and editing. Mikako Wada : Investigation; methodology; visualization; writing original draft; writing review and editing. Yuko Aizen : Investigation. Yuko Nitahara-Kasahara : Investigation; methodology; writing original draft; writing review and editing. Takashi Okada : Conceptualization; funding acquisition; project administration; supervision; writing review and editing.
Conflict of Interest
This work was supported by research grant from HOYA Technosurgical Corporation. Kurosawa receives a salary from HOYA Technosurgical Corporation.
Open Research
Data Availability Statement
The data, tools, and materials that support the protocol are available from the corresponding author upon reasonable request.
Literature Cited
- Belova, L., Kochergin-Nikitsky, K., Erofeeva, A., Lavrov, A., & Smirnikhina, S. (2022). Approaches to purification and concentration of rAAV vectors for gene therapy. BioEssays , 44(6), e2200019. https://doi.org/10.1002/bies.202200019
- Bryda, E. C., Men, H., Davis, D. J., Bock, A. S., Shaw, M. L., Chesney, K. L., & Hankins, M. A. (2019). A novel conditional ZsGreen-expressing transgenic reporter rat strain for validating Cre recombinase expression. Scientific Reports , 9, 13330. https://doi.org/10.1038/s41598-019-49783-w
- Current Protocols. (2006). Commonly used reagents. Current Protocols in Microbiology , 00, A.2A.1–A.2A.15. https://doi.org/10.1002/9780471729259.mca02as00
- Kurosawa, Y., Saito, M., Kobayashi, S., & Okuyama, T. (2012). Purification of dengue virus particles by one-step ceramic hydroxyapatite chromatography. World Journal of Vaccines , 2(3), 155–160. https://doi.org/10.4236/wjv.2012.23020
- Kurosawa, Y., Sato, S., Okuyama, T., & Taoka, M. (2019). Sequential two-step chromatographic purification of infectious poliovirus using ceramic fluoroapatite and ceramic hydroxyapatite columns. PLoS One , 14(9), e0222199. https://doi.org/10.1371/journal.pone.0222199
- Lins-Austin, B., Patel, S., Mietzsch, M., Brooke, D., Bennett, A., Venkatakrishnan, B., van Vliet, K., Smith, A. N., Long, J. R., McKenna, R., Potter, M., Byrne, B., Boye, S. L., Bothner, B., Heilbronn, R., & Agbandje-McKenna, M. (2020). Adeno-associated virus (AAV) capsid stability and liposome remodeling during endo/lysosomal pH trafficking. Viruses , 12(6), 668. https://doi.org/10.3390/v12060668
- Miyaoka, R., Tsunekawa, Y., Kurosawa, Y., Sasaki, T., Onodera, A., Sakamoto, K., Kakiuchi, Y., Wada, M., Nitahara-Kasahara, Y., Hayashita-Kinoh, H., & Okada, T. (2023). Development of a novel purification method for AAV vectors using tangential flow filtration. Biotechnology and Bioengineering , 120(11), 3311–3321. https://doi.org/10.1002/bit.28524
- Samulski, R. J., & Muzyczka, N. (2014). AAV-mediated gene therapy for research and therapeutic purposes. Annual Revier of Virology , 1(1), 427–451. https://doi.org/10.1146/annurev-virology-031413-085355
- Tomono, T., Hirai, Y., Okada, H., Miyagawa, Y., Adachi, K., Sakamoto, S., Kawano, Y., Chono, H., Mineno, J., Ishii, A., Shimada, T., Onodera, M., Tamaoka, A., & Okada, T. (2018). Highly efficient ultracentrifugation-free chromatographic purification of recombinant AAV serotype 9. Molecular Therapy. Methods and Clinical Development , 11, 180–190. https://doi.org/10.1016/j.omtm.2018.10.015
- van Vliet, K. M., Blouin, V., Brument, N., Agbandje-McKenna, M., & Snyder, R. O. (2008). The role of the adeno-associated virus capsid in gene transfer. In K. K. Jain (Ed.), Drug Delivery Systems. Methods in Molecular Biology (Vol. 437, pp. 51–91). Humana Press. https://link.springer.com/protocol/10.1007/978-1-59745-210-6_2
- Wada, M., Uchida, N., Posadas-Herrera, G., Hayashita-Kinoh, H., Tsunekawa, Y., Hirai, Y., & Okada, T. (2023). Large-scale purification of functional AAV particles packaging the full genome using short-term ultracentrifugation with a zonal rotor. Gene Therapy , 30(7–8), 641–648. https://doi.org/10.1038/s41434-023-00398-x
Internet Resources
Adeno-associated virus gene therapy landscape (Alcrita Consulting).

