Protocol for harvesting and dissociating mouse brain neurons for single cell RNA Sequencing on the 10X Genomics platform
Viktor Feketa, Elena O. Gracheva
Abstract
Single-cell RNA sequencing has emerged as a powerful method to characterize gene expression on a single cell level. Producing useful data with this method critically relies on obtaining a suspension of dissociated cells with high concentration and viability from the tissue of interest. This protocol allows to isolate and dissociate mouse brain cellsinto a concentrated cell suspension that is compatible with the 10X Genomics library preparation and sequencing pipeline and enables capturing up to 10,000 single cells.
Attachments
Steps
Part 1: Advance preparation of solutions
Prepare stock solutions of media supplements
Lactic acid : prepare 1Molarity (M) (90mg/mL) solution of lactic acid in nuclease-free water and aliquot in50µL aliquots. Store at -20°C.
GlutaMAX : aliquot original GlutaMAX (200millimolar (mM)) solution into 100µL aliquots. Store at-20°C.
B27 (minus insulin) supplement : thaw original solution and aliquot into 700µL aliquots. Store at -20°C.
Prepare 1L of Brain Perfusion Solution (composition: 2.5millimolar (mM) KCl, 7millimolar (mM) MgCl2, 1.25millimolar (mM) NaH2PO4, 28millimolar (mM) NaHCO3, 0.5millimolar (mM) CaCl2, 7millimolar (mM)glucose, 1millimolar (mM)ascorbate, and 3millimolar (mM) pyruvate in nuclease-free water). Adjust the osmolarity to ~300mOsm using approximately 67g/L sucrose. Oxygenate by bubbling with 95% O2 /5% CO2 for 0h 15m 0sand adjust the7.4. Filter with a filter unit. Aliquot in 200mL aliquots (each experiment will require approximately 200mL of perfusion solution). Aliquots can be stored at-20°C for up to 2 months. The day before experiment: thaw an aliquot of the Brain Perfusion Solution at4°C``0h 15m 0s.
We prepare the solution from the following specific components (for 1L solution):
| A | B | C | D | E |
|---|---|---|---|---|
| # | Component | Final Conc. (mM) | MW (g/mol) | Solid weight (mg/1000 mL) |
| 1 | Sucrose | 96 | 342.3 | 67000 |
| 2 | KCl | 2.5 | 74.55 | 186.4 |
| 3 | NaHCO3 | 28 | 84.01 | 2352 |
| 4 | NaH2PO4 * H20 | 1.25 | 137.99 | 172.5 |
| 5 | Glucose | 7 | 180.16 | 1261 |
| 6 | Sodium Ascorbate | 1 | 198.11 | 198.1 |
| 7 | CaCl2 * 2H20 | 0.5 | 147.01 | 73.5 (or 0.5ml of 1 M stock) |
| 8 | MgCl2 | 7 | 95.21 | 666.5 (or 7ml of 1 M stock) |
| 9 | Sodium Pyruvate | 3 | 110.04 | 30 ml of 100 mM stock |
The day before experiment, prepare a fresh 30mL aliquot of Hibernate A media with 50millimolar (mM) glucose, osmolarity of280mOsm, and 100U/ml penicillin-streptomycin.
In the sterile culture hood, transfer 30mL of stock Hibernate A (pre-made by the supplier with 2.5millimolar (mM) glucose and nominal osmolarity of275mOsm) to the 50mL culture flask.
Add 300µL of 100X Pen-strep stock solution to 30mL Hibernate A aliquot.
Add NaCl (cell culture grade) to adjust osmolarity to 280mOsm (~0mg to 5mg).
Store media aliquot at 4°C in dark until the day of experiment.
Fire-polish the tips of eight 9-inch glass Pasteur pipettes: 4 pipettes barely polished until the tips are no longer sharp (for cell transfers) and 4 pipettes for trituration with the following approximate tip opening diameters: 0.9 mm; 0.7 mm; 0.5 mm; 0.3 mm. The largest should be barely polished and the smallest should be about a third of that diameter.
Part 2. Experimental procedure on the day of experiment.
Maintain RNAse-free conditions throughout the procedure. Wipe down all surfaces and tools with the "RNAse-away" reagent.
Thaw frozen aliquots of lactic acid, GlutaMax and B27 (minus insulin) supplements, vortex.
Prepare working solution of the Hibernate A media by adding the supplements for the following final working concentrations:
Lactic acid : 1millimolar (mM) (add 30µL of 1Molarity (M) stock solution to the30mL media aliquot).
GlutaMax : 0.5millimolar (mM) (add 75µLof 200millimolar (mM) stck solution to the 30mL media aliquot) .
B27 : 2% (add 600µL of the stock solution to the 30mL media aliquot).
Mix media aliquot by inversion. Adjust pH to 7.4using 1Normality (N) NaOH.
Oxygenate the thawed 200mLaliquot of the Brain Perfusion Solution with 95% O2/5% CO24On icefor at least 0h 15m 0s.
Prepare a 50mL culture flask labeled "digestion" and 9 polystyrene 50mL conical tubes labeled "harvest", "dissection", "papain", "DNAse", "BSA", "BSA filtered", "BSA centrifugation", "trituration", "cell suspension".
Distribute Hibernate-A media : transfer2mL to the "harvest" tube,9mLto "dissection" tube, 4mL to "papain" tube, 5.5mL to "DNAse" tube; 5mL to "trituration" tube. Preserve the rest of Hibernate A media for next steps.
Place "harvest" and "dissection" tubes with media 34On ice near the animal dissection area.
Prepare RNA-se free area for animal dissection: wipe all surfaces and tools with "RNAse Away" reagent.
Prepare vibratome : get a new blade, wash with 70% ethanol followed by ultrapure water, load the blade in the vibratome blade holder.
Prepare area and tools for cardiac perfusion and animal dissection: bath for cardiac perfusion with absorbent pad, surgical tools, cardiac perfusion system with an oxygenation line inserted into perfusion reservoir.
Fill cardiac perfusion system with ultrapure water and let it run through to clean.
Add isoflurane to induction chamber for animal anesthesia.
Fill outer vibratome bath with ice and install on the vibratome.
Place 2 glass Petri dishes 4On ice.
Prepare oxygenation line (with 95% O2/5% CO2) to be later inserted into inner vibratome bath.
Place a mouse into induction chamber with isoflurane and wait until it stops breathing.
Take mouse out and verify depth of anesthesia by absence of response to toe pinch.
Pour ~30mL of the ice-cold oxygenated Brain Perfusion Solution into reservoir of cardiac perfusion system just prior to dissection, fill tubing, and stop when about 20mL of solution is left in the reservoir to be used for perfusion. Continue oxygenating the rest of the Brain Perfusion Solution4On ice.
Perform mouse dissection.
Insert the cardiac perfusion needle into left ventricle. Cut the right atrium with scissors.
Start the flow of perfusion system.
Perfuse about 15mL of perfusion solution (~0h 1m 0s) until the effluent is clear. Success of perfusion can be assessed by internal organs changing color to a lighter shade.
Fill a petri dish On ice with ~ 15mL of Brain Perfusion Solution.
Decapitate the mouse, Extract brain:
Cut skin above skull from caudal to rostral end and peel away to expose skull.
Make a midline cut with scissors in the skull towards the eye sockets. Use fine Graefe forceps to break and peel pieces of skull away from midline, exposing the brain.
Make a coronal cut with scissors between olfactory bulbs and the rest of the brain, and another cut between the brain and spinal cord to mobilize the brain.
Use a spatula to slightly lift the brain from the skull, use scissors to cut optical tracts, and finally extract the mobilized brain from the skull with a spatula (hippocampal tool) and push it into the Petri dish with the Brain Perfusion Solution.
Make a coronal cut between cerebellum and the rest of the brain, trying to make it as flat and perpendicular to the rostro-caudal axis of the brain as possible.
Lift the brain from the solution with a curved spatula and gently dab with filter paper to dry, especially the flat coronal aspect which will be glued down to the vibratome stage.
Put a little drop of super glue on the vibratome stage and spread it with a cotton tip to an area slightly larger than the brain.
Put the brain down on the stage area covered with glue, coronal aspect (now being the caudal end) down, rostral end up.
Mount the vibratome stage with the glued brain in the inner vibratome bath. About 0h 0m 10s after the brain attachment, fill the vibratome bath with the remaining oxygenated Brain Perfusion Solution to completely cover the brain in the bath.
Rotate the stage of the vibratome with the brain to orient ventral (hypothalamic side) towards and dorsal cortex away from the blade.
Make sure the oxygenation line is inserted into vibratome bath and is turned on throughout the brain slicing.
Lower the vibratome blade into cutting position.
Using vibratome control pad, move the stage and blade as needed to perform the first cut close to the rostral end of the brain (facing up in the bath).
Start cutting 300 µm thick slices (vibratome settings: speed: 0.2 mm/sec, amplitude: 1 mm), observing the anatomical cues until the target area is reached. The level of bregma+1.0mm is reached approximately when left and right parts of corpus callosum meet in the middle.
After reaching the target area, cut two consecutive 300 µm thick slices to be collected.
Pour all 9mL of Hibernate A media from the "dissection" tube into the second empty Petri dish 4On ice.
Transfer two target slices with a spatula from the vibratome bath into Petri dish with Hibernate A media.
Using 27G needles attached to 1mL syringes used as cutting tools, dissect the target brain areas from slices. Further cut dissected pieces in two, to produce tissue pieces about 1x1 mm in size.
Collect tissue pieces with a glass pipette, transfer them to the "harvest" tube with Hibernate A media, and place the tube 4On ice.
Prepare digestion solution : add 80U of stock papain suspension to the "papain" tube with 4mL of media (for a final concentration of 20U/ml; calculate the volume of papain to get80U beforehand based on the activity of specific batch, usually 65µLto 85µL). Mix by inversion and place in 34°C water bath. Check and mix by inversion every minute until the media is no longer cloudy (~0h 4m 0s).
During "papain" incubation, place the "harvest" tube with tissue pieces into 34°C water bath.
Using 0.22-µm syringe filter, filter papain solution into a "digestion" culture flask.
Using glass pipette, transfer tissue pieces from "harvest" tube into "digestion" culture flask with papain. Shake "digestion" flask to make sure tissue pieces are not clumped together but are floating separately to ensure proper digestion.
Incubate "digestion" flask in a shaking water bath at 34°C with shaking at 150rpm.
Meanwhile, prepare media solutions for trituration:
Prepare DNAse solution: add 500µL of Hibernate A media to a vial with DNAse solid, gently but thoroughly mix to dissolve completely (do not vortex, DNAse is sensitive to shear). Transfer 500µL of dissolved DNAse to the "DNAse" tube with 5.5mLof Hibernate A media (final volume 6mL, final concentration of DNAse0.1mg/mL). Invert gently to mix (do not vortex).
Prepare BSA solution: weigh 160 mg of bovine serum albumin solid and add to the "bsa" tube with 2 ml of Hibernate A (final concentration: 8% BSA). Vortex for 30 sec. Using 0.22 µm syringe filter, filter BSA solution into another "bsa filtered" tube. Transfer 1mL of filtered BSA to the "bsa centrifugation" tube.
Place 30μm Miltenyi SmartStrainer onto "cell suspension" tube to collect and filter cell suspension.
When 0h 30m 0s digestion is done, transfer tissue pieces from "digestion" to the "trituration" tube using glass pipette. Invert once and let tissue pieces settle. Aspirate almost all media with glass pipette, leaving only tissue pieces.
Add 3mL of the trituration media ("DNAse" tube) to the "trituration" tube with tissue pieces. Triturate with the largest pipette (0.9 mm) 10 times (draw large volume to collect all tissue pieces with each trituration stroke) over approximately 0h 0m 30s. Wait 0h 4m 0s for tissue pieces to settle. Use the second-largest (0.7 mm tip opening) glass pipette to transfer 2mL from the top of the cell suspension onto the cell strainer on top of "cell suspension" tube.
Add 2mL of trituration media to the "trituration" tube. Triturate 10 times with the second-largest 0.7 mm pipette. Wait 0h 3m 0s for pieces to settle. Use the third (0.5mm) glass pipette to transfer the top 2mL to the cell strainer/"cell suspension" tube.
Add 1mLof trituration media to the "trituration" tube. Triturate 5 times with the third 0.5 mm pipette. Wait 0h 2m 0sfor tissue pieces to settle. Use the fourth 0.3 mm glass pipette to transfer all the remaining solution from the "trituration" to the cell strainer/"cell suspension" tube.
Remove cell strainer from the "cell suspension" tube and layer all the solution from this tube onto the BSA layer in the "BSA centrifugation" tube using a glass pipette.
Centrifuge the "BSA centrifugation" tube1000rpm. Using an Eppendorf 5810R swinging bucket centrifuge, this corresponds to 67 rcf. Set "break" setting to '0', i.e. no breaking.
After centrifugation is done, carefully aspirate almost all media from the tube, leaving about 50µL of solution above the cell pellet.
Add 950µL of Hibernate A media to cell pellet. Resuspend cells with the glass pipette (gently pipette up and down 10 times).
Transfer all (~1mL) of the cell suspension from the "bsa centrifugation" tube to a new 2mLEppendorf LoBind tube.
Centrifuge at 300rcf in a small tabletop centrifuge (Eppendorf 5424: 1787 rpm) at 34Room temperature.
Carefully remove supernatant to leave ~50µLof the solution, avoiding disrupting the cell pellet (it won't be visible) and creating bubbles.
Using glass pipette, carefully resuspend the cell pellet in the remaining ~50µL of solution (pipette up and down about 10 times, avoid creating bubbles). This is the final cell suspension used for 10X library preparation. Place and store tube 34On ice until starting the 10X single cell protocol.
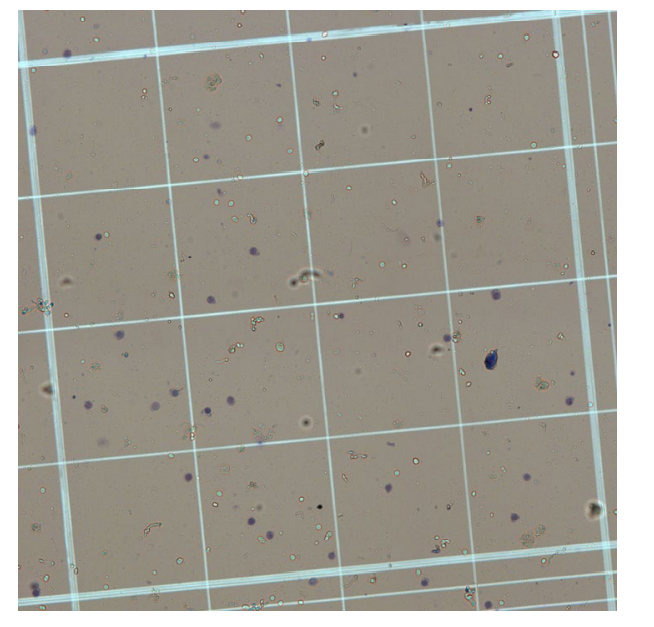
To determine the volume of solution to use for library preparation, determine cell concentration by counting cells using a hemocytometer. Transfer a 10µLaliquot from the final cell suspension to a new2mL LoBind tube. Add 10µLof Trypan Blue stain. Mix gently with a pipette. Pipette 10µL into a hemocytometer chamber. Count the number of live (transparent) and dead (blue) cells under the microscope. In case of harvesting primary and secondary motor areas from two 300 µm thick slices (4 tissue pieces ~1x2 mm in size), the expected cell concentration is ~1,000-2,000 cells/µl, expected viability is ~85%. To recover 10,000 cells in 10X protocol, the optimal concentration is 700-1,200 cells/µl. If obtained cell concentration is much higher than that, dilute cell suspension to the desired concentration by adding appropriate volume of Hibernate A media.
Proceed with the 10x Genomics Single Cell Protocol.
Expected results
In case of harvesting primary and secondary motor cortex areas from two 300 µm thick slices (4 tissue pieces ~1x2 mm in size), the expected cell concentration is ~1,000-2,000 cells/µl, expected viability is ~85%.
Figure 1 shows a representative image of the cell suspension loaded in hemocytometer for counting.