Protocol for Genomic DNA Extraction and Sequencing Library Preparation from Phage Stock
Shuborno Islam, Rathindranath Kabiraj, Al Amin, Sadnane Hussain Pranto, Shantu Dey, Apurba Rajib Malaker, Deb Purna Keya, Arif Mohammad Tanmoy, Samir K Saha, Yogesh Hooda, Senjuti Saha
Abstract
Whole Genome Sequencing (WGS) is an essential technique for understanding the genetic composition of organisms. This protocol describes a cost-effective method for propagating phages from stock, extracting DNA, and preparing sequencing libraries. These libraries are then sequenced using the Illumina iSeq100.
Before start
All pipettes, tips and centrifuge tubes in the assay must be sterile and DNase/RNase-free. To prevent contamination, filtered tips are required and should be replaced after each reagent or sample is added.
-
All workbench, hood surfaces, and supplies should be cleaned and disinfected regularly using 10% bleach and then 70% ethanol before and after completing an assay.
-
Before beginning each assay, each component must be thawed and briefly centrifuged. Avoid repeated freeze-thaw cycles.
-
Dispose of waste in compliance with institutional and local regulations.
Steps
Phage Propagation and Purification
Phage Propagation
Spotting of phages, co-cultures preparation, and DLA lawning
Melt soft TSA (0.7% agar) and aliquot 4 mL molten TSA in a screw-cap tube. Equilibrate the temperature between 56°C - 60°C to avoid agar solidification and damaging bacterial cells.
Following the Double-Layer Agar (DLA) method, add 200 µL of overnight grown Salmonella Typhi BRD948 culture to the 4 mL molten TSA tube and pour it on a TSA plate (1.5% agar). Let the plate solidify for 30 - 60 minutes at room temperature.
Make dilutions of the phage stock (10-2, 10-4, 10-6) and spot 2 µL from each dilution including the undiluted phage stock (100) on the prepared plate of Salmonella Typhi BRD948 culture ( step 1.2 ). Incubate the plates for 16-18 hours at 37°C.
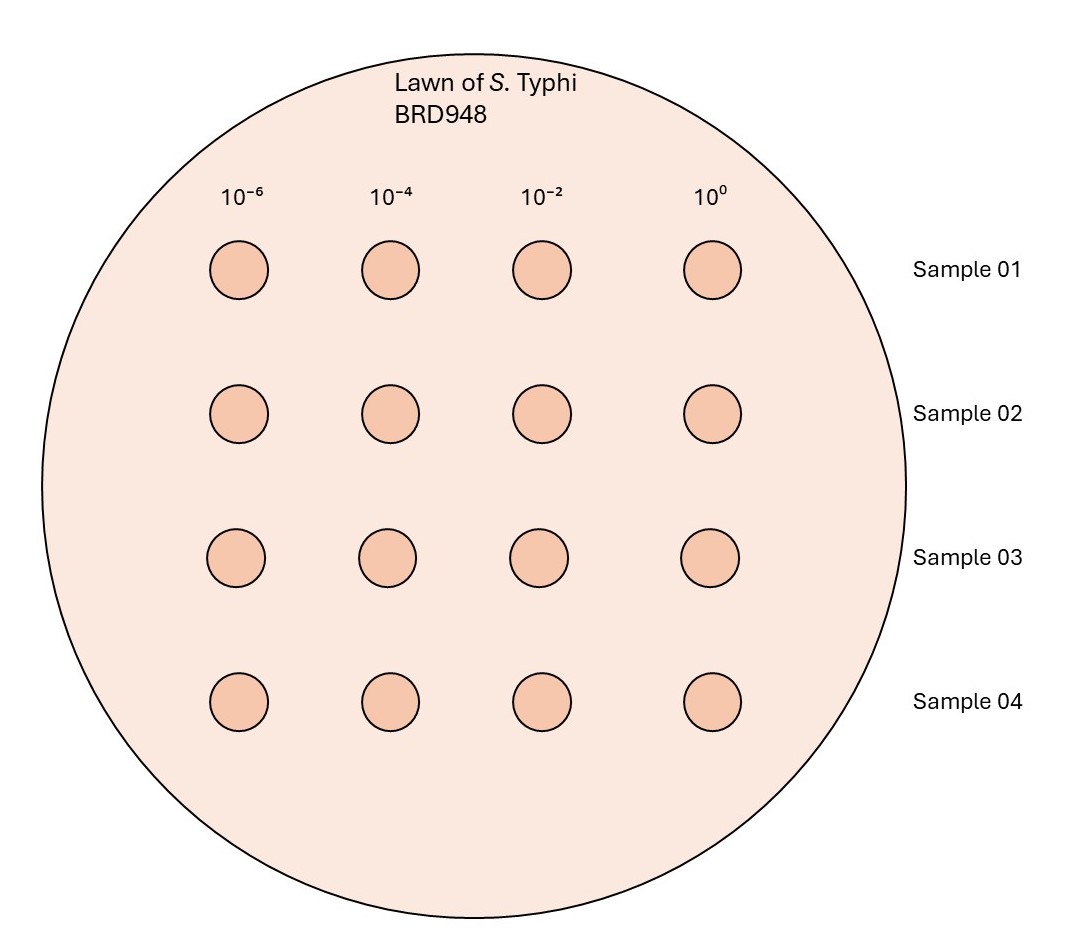
After incubation, observe the plate for lysis formation and record the number of plaques.
The dilution that creates an uncountable number of plaques is denoted as TNTC (Too numerous to count) around the spotted zone, but not a clear zone, is the targeted dilution needed for the next step. This is in between a single plaque-forming spot and a full lysed-forming spot (This dilution is required to form "webbed" confluent lysed plates).
Prepare a Co-Culture by adding 50 µL phage volume from targeted dilution to 200 µL Salmonella Typhi BRD948 overnight liquid culture. Incubate for 20 minutes at room temperature for host-phage attachment.
Add the Co-Culture (250 µL) into a 4 mL soft TSA tube (molten and equilibrated at 56°C - 60°C) and pour on a TSA plate (1.5% agar). Incubate the plates at 37°C for 16-18 hours.
After incubation, observe the plates for "webbed" confluent Lysed Plates.
Purification of Propagated Phages
Upon observation of the "webbed" confluent lysed plate, pour 4 mL of chilled LB (4°C) over the plate followed by incubation at 4°C for 4 hours.
Dislodge the top agar (using a sterile glass scrapper or a bent tip) and transfer 2 mL of the mixture into a new microcentrifuge tube using a pipette.
Add 4 drops of chloroform, vortex for 20 seconds, and rest for 10 minutes at room temperature.
Centrifuge at 10000 rcf for 10 minutes at room temperature.
Transfer 1 - 1.5 mL supernatant into a fresh microcentrifuge tube without disturbing the pellet. Label the tube and keep it at 4°C for downstream experiments.
Extraction of Phage genomic DNA
Removal of Bacterial DNA and RNA
Prepare enzyme mix containing 50 µL DNA Digestion Buffer, 1 µL DNase I (1U/µL), and 0.5 µL RNase A (20mg/mL). Add the enzyme mix to 450 µL propagated phage from step 2.5 . Incubate at 37°C for 1.5 hours. Do not shake or vortex .
After incubation, add 20 µL 0.5 M EDTA (final concentration 20 mM) to deactivate the enzymes. Incubate at room temperature for 10 minutes.
Digestion of Phage Protein
Add 1.25 µL Proteinase K (20 mg/mL). Incubate at 56°C for 1.5 hours. Do not shake or vortex .This is "Phage Lysate".
Phage gDNA purification
Add 200 µL AL buffer to 200 µL of the Phage Lysate from step 4.1 and vortex the mixture. Incubate the mixture at 70°C for 10 minutes.
Analyze the DNA using 0.8% agarose gel by loading 5 µL DNA sample to check the quality of the extracted genomic DNA.
The eluted phage DNA can be stored at -20°C until library preparation.
Let the tube cool to room temperature, add 200 µL ice-cold ethanol (99%), and vortex the mixture.
Transfer the mixture into the spin column, and centrifuge at 8000 rcf for 1 minute.
Discard the filtrate, add 500 µL AW1 buffer into the spin column, and centrifuge at 8000 rcf for 1 minute.
Discard the filtrate, add 500 µL AW2 buffer into the spin column, and centrifuge at 14000 rcf for 3 minutes.
Discard the filtrate and centrifuge for 1 minute at 14000 rpm to avoid carryover.
Transfer the spin column to a microcentrifuge tube and add 30 µL AE buffer. Incubate for 5 minutes at room temperature and centrifuge for 2 minutes at 8000 rcf to elute the DNA.
Add the eluted volume into the spin column again, and centrifuge for 2 minutes at 8000 rcf to elute a higher concentration of phage DNA.
Measure the DNA concentration using Qubit Fluorometer or Nanodrop (described below).
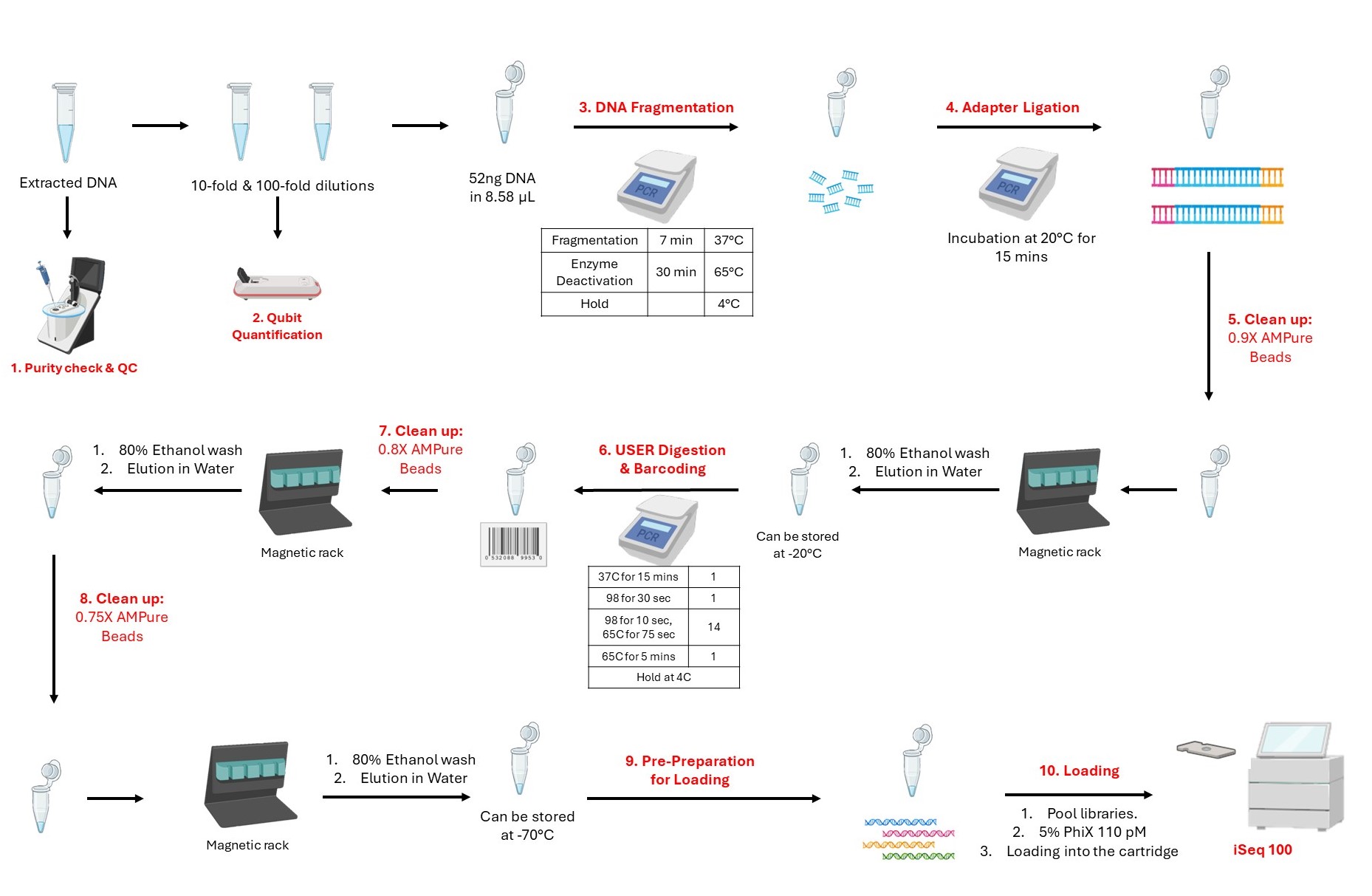
Workflow of Phage Genome Sequencing
Quantification and Normalization
Normalization before library preparation ensures an equivalent representation of the sample amount for downstream reactions, reducing biases from sequencing depth and technical variations for accurate genomic analysis. The eluted phage DNA is serially diluted for accurate concentration measurement on Qubit (quantification). Based on the concentrations, the amount of DNA is normalized to have ~52 ng of DNA in 8.58 µL volume. The Qubit™ 1X dsDNA High Sensitivity (HS) and Broad Range (BR) Assay Kits do Quantification for double-stranded DNA (dsDNA).
Bring the reagents and buffer to room temperature 30 minutes before the experiment.
| A | B | C |
|---|---|---|
| DNA Quantification (100-fold) | DNA Quantification (10-fold) | DNA Quantification (1-fold) |
| X | Y | Z |
| - | ||
| Volume for Normalization (100-fold) | Volume for Normalization (10-fold) | Volume for Normalization (1-fold) |
| 52/X | 52/Y | 52/Z |
| - | ||
| Water to make 8.58 µL | Water to make 8.58 µL | Water to make 8.58 µL |
| 8.58 - 52/X | 8.58 - 52/Y | 8.58 - 52/Z |
Volume calculation for Normalization
Prepare working solutions (reagents and buffer mix) for both standards and eluted phage DNA.
The final volume in each tube must be 100 µL. For each standard, 95 µL of the working solution is required while 99 µL is required for each DNA sample.
For Standards, take 95 µL of the working solution in 2 tubes. Add 5 µL of each Qubit standard to appropriate tubes, vortex, and spin them shortly.
For samples, take 99 µL of the working solution in N tubes ( N is the number of phage samples ). Add 1 µL of 100-fold diluted DNA to appropriate working solution tubes; vortex, and spin them shortly.
Incubate the tubes in the dark for 2 minutes.
Measure the standards and samples in the Qubit Fluorometer under the High Sensitivity Range
Calculate the concentration of 10-fold diluted samples and/or 1-fold tubes from the Qubit results.
Repeat the quantification step ( step 7.5 ) with 10-fold diluted tubes if the results for 100-fold diluted tubes read “Too Low.”
Calculate the volume of DNA and nuclease-free water required to make a solution of 52 ng of DNA in 8.58 µL volume. Select the volume and dilution factors that can be used for normalization.
Library Preparation and Phage Genome Sequencing
Fragmentation and End Prep
Prepare master mix by adding the following reagents to a 0.2 mL PCR tube on ice rack.
| A | B |
|---|---|
| Reagent | Volume (µL) |
| NEBNext Ultra II FS Reaction Buffer (YELLOW) | 2.31 |
| NEBNext Ultra II FS Enzyme mix (YELLOW) | 0.66 |
| Total | 2.97 |
Master mix calculation for fragmentation
Aliquot 2.97 µL of the master mix in each tube containing the normalized DNA (8.58 µL).
Total reaction volume: 11.55 µL
Vortex and spin
Thermocycler program:
| A | B | C |
|---|---|---|
| PCR Profile | ||
| Step | Time | Temperature |
| Fragmentation | 7 minutes | 37°C |
| Enzyme Deactivation | 30 minutes | 65°C |
| Hold | ∞ | 4°C |
| Lid: 105°C |
Adapter Ligation
Combine the following components in a 0.2 mL PCR tube to prepare the master mix (without the adapters):
Adapters were pre-diluted using a buffer.
| A | B | C |
|---|---|---|
| Reagent | Volume (µL) | Remarks |
| NEBNext Ultra II Ligation Master Mix (RED) | 9.9 | Master mix |
| NEBNext Ligation Enhancer (RED) | 0.33 | |
| Total | 10.23 | |
| - | ||
| NEBNext Adapter for illumina (1:100) | 0.825 | Check step 9.3 |
Master mix calculation for Adapter ligation
Aliquot 10.23 µL of the Adapter ligation master mix directly to the Fragmentation reaction mixture from step 8.2 (11.55 µL).
Add 0.825 µL adapter separately to avoid the formation of adapter dimers.
Total: 22.6 µL
Briefly vortex and spin the reaction mix
Incubate at 20°C for 15 minutes in a thermocycler with the heated LID OFF.
Clean-up of Adapter-ligated DNA and Size Selection (0.9X wash)
This step eliminates small untagged DNA fragments and short adapter sequences.
Prepare 80% EtOH.
Remove the tube from the magnetic rack. Add 6.95 µL of nuclease-free water.
Pipette well to mix and incubate for 2 minutes at room temperature off the magnetic rack.
Place on a magnetic rack again until the solution is clear (~2 minutes).
Transfer 4.95 µL of the supernatant (Elution) to a clean nuclease-free PCR tube.
Checkpoint: Checkpoint: Samples can be stored at -20 °C overnight and library preparation should be resumed the next day.
Use 0.9x ratio of beads - to - total volume of sample.
Add 20.34 µL of AMPure XP beads (0.9x) to Adaptor Ligated products. Mix well by pipetting.
Incubate for 5 miutesn at room temperature.
Place samples on a magnetic rack and incubate for 5 minutes on the rack.
Carefully remove the supernatant, without disturbing the beads.
Add 200 µL of 80% Ethanol to each sample in the magnetic rack. Incubate at room temperature for 30 seconds then remove Ethanol.
Repeat step 10.7 once more and carefully remove any residual ethanol with a 10 µL pipette tip.
Air dry the beads for 5 - 10 minutes while the tubes are on the magnetic rack with cap open.
USER Digestion and Barcoding
This step uses the USER enzyme to cleave the adaptor at the Uracil region, creating a gap for index primers (barcodes) to bind. In this step, libraries of each sample are tagged with a unique barcode sequence for sample identification which undergoes PCR enrichment.
Combine the following components in a 0.2 mL sterile PCR tube to prepare the master mix.
| A | B | C |
|---|---|---|
| Reagent | Volume (µL) | Remarks |
| USER Enzyme (WHITE) | 0.99 | Master mix |
| NEBNext Ultra II Q5 master mix (BLUE) | 8.25 | |
| Total | 9.24 | |
| - | ||
| 5 µM pre-mixed Forward & reverse index primers | 3.3 | Check step 11.2 |
Master mix calculation for USER Digestion and Barcoding
Aliquot 9.24 µL master mix directly to the adaptor-ligated sample tube to the purified adapter-ligated DNA from step 10.13 .
Add 3.3 µL (5 µM pre-mixed forward and reverse) index primers separately to each sample tubes.
Total volume: 17.49 µL
Briefly vortex and spin the reaction mix.
In a thermocycler run the following program with the heated lid on for PCR enrichment.
Thermocycler program:
| A | B |
|---|---|
| Thermocycler | Cycles |
| 37 °C for 15 minutes | 1 |
| 98 °C for 30 seconds | 1 |
| 98 °C for 10 seconds | 14 |
| 65 °C for 75 seconds | |
| 65 °C for 5 minutes | 1 |
| Hold at 4°C | ∞ |
| LID: 105°C |
Clean-up of Adapter-ligated DNA and Size Selection (0.8X wash)
This step washes the barcoded DNA by eliminating products < 100 bp.
Prepare 80% EtOH.
Remove tube from magnetic rack. Add 14.52 µL of nuclease free water.
Pipette well to mix and incubate for 2 minutes at room temperature off the magnetic rack.
Place on a magnetic rack again until the solution is clear (~2 minutes).
Remove 13.2 µL of the supernatant (Elution) and transfer to a clean nuclease-free PCR tube.
Use 0.8x ratio of beads -to -total volume of sample.
Add 13.99 µL of AMPure XP beads (0.8x) to barcoded products. Mix well by pipetting.
Incubate for 5 minutes at room temperature.
Place samples on a magnetic rack and incubate for 5 minutes on the rack.
Carefully remove the supernatant.
Add 200 µL of 80% Ethanol to each sample in the magnetic rack. Incubate at room temperature for 30 seconds then remove Ethanol.
Repeat step 12.7 once for a total of two washes. Carefully remove any residual ethanol with a 10 µL pipette tip.
Air dry the beads for 5-10 minutes while on the magnetic rack with the lid open.
Clean-up of Adapter-ligated DNA and Size Selection (0.75X wash)
This step washes the barcoded DNA by eliminating products < 200 bp.
Prepare 80% EtOH.
Remove tube from magnetic rack. Add 11.9 µL of nuclease free water.
Pipette well to mix and incubate for 2 minutes at room temperature off the magnetic rack.
Place on a magnetic rack again until the solution is clear (~2 minutes).
Remove 9.9 µL of the supernatant (Elution) and transfer to a clean nuclease-free PCR tube.
Checkpoint: _Checkpoint: Samples can be stored at -70 °C._
Use 0.75x ratio of beads -to -total volume of sample.
Add 9.9 µL of AMPure XP beads (0.75x) to barcoded products. Mix well by pipetting.
Incubate for 5 minutes at room temperature.
Place samples on a magnetic rack and incubate for 5 minutes on the rack.
Carefully remove the supernatant.
Add 200 µL of 80% Ethanol to each sample in the magnetic rack. Incubate at room temperature for 30 seconds then remove the Ethanol.
Repeat step 13.7 once for a total of two washes. Carefully remove any residual ethanol with a 10 µL pipette tip.
Air dry the beads for 5-10 minutes while on the magnetic rack with the lid open.
Loading on iSeq100
Quantify each of the libraries and pool 7-10 ng of the libraries into one tube.
Calculate the volume needed for desired loading concentration of the libraries (110 pM) and dilute the pooled tube accordingly (optional).
Prepare 5% 110 pM working PhiX.
Follow the Illumina protocol to thaw and prepare the iSeq 100 cartridge.
Load 20 µL diluted library with 5% PhiX in library-loading well of the cartridge.
Insert the flow cell into the cartridge and load it into the sequencer following the instructions on the instrument.