Proteome-Derived Peptide Libraries for Deep Specificity Profiling of N-terminal Modification Reagents
Haley N. Bridge, Haley N. Bridge, Amy M. Weeks, Amy M. Weeks
2-pyridinecarboxaldehyde
N terminomics
protein N terminus
proteome-derived peptide libraries
subtiligase
Abstract
Protein and peptide N termini are important targets for selective modification with chemoproteomics reagents and bioconjugation tools. The N-terminal ⍺-amine occurs only once in each polypeptide chain, making it an attractive target for protein bioconjugation. In cells, new N termini can be generated by proteolytic cleavage and captured by N-terminal modification reagents that enable proteome-wide identification of protease substrates through tandem mass spectrometry (LC-MS/MS). An understanding of the N-terminal sequence specificity of the modification reagents is critical for each of these applications. Proteome-derived peptide libraries in combination with LC-MS/MS are powerful tools for profiling the sequence specificity of N-terminal modification reagents. These libraries are highly diverse, and LC-MS/MS enables analysis of the modification efficiencies of tens of thousands of sequences in a single experiment. Proteome-derived peptide libraries are a powerful tool for profiling the sequence specificities of enzymatic and chemical peptide labeling reagents. Subtiligase, an enzymatic modification reagent, and 2-pyridinecarboxaldehyde (2PCA), a chemical modification reagent, are two reagents that have been developed for selective N-terminal peptide modification and can be studied using proteome-derived peptide libraries. This protocol outlines the steps for generating N-terminally diverse proteome-derived peptide libraries and for applying these libraries to profile the specificity of N-terminal modification reagents. Although we detail the steps for profiling the specificity of 2PCA and subtiligase in Escherichia coli and human cells, these protocols can easily be adapted to alternative proteome sources and other N-terminal peptide labeling reagents. © 2023 The Authors. Current Protocols published by Wiley Periodicals LLC.
Basic Protocol 1 : Generation of N-terminally diverse proteome-derived peptide libraries from E. coli
Alternate Protocol : Generation of N-terminally diverse proteome-derived peptide libraries from human cells
Basic Protocol 2 : Characterizing the specificity of 2-pyridinecarboxaldehyde using proteome-derived peptide libraries
Basic Protocol 3 : Characterizing the specificity of subtiligase using proteome-derived peptide libraries
INTRODUCTION
Proteome-derived peptide libraries are powerful tools for profiling the sequence specificity of enzymatic and chemical labeling reagents (Fig. 1). Profiling labeling reagents using proteome-derived peptide libraries provides a more comprehensive, cost-effective, and biologically relevant view of the sequence specificity compared to using chemically synthesized libraries (Keller & Schilling, 2010; Schilling & Overall, 2008; Schilling et al., 2011). Typically, proteome-derived peptide libraries require several steps to protect reactive groups, including lysine and cysteine side chains and N-terminal ⍺-amines (Schilling et al., 2011). Protection of these groups alters their chemical properties and can affect whether enzymes and chemical reagents are able to label them. This protocol outlines how to generate peptide libraries with unblocked lysine side chains and N termini for use in profiling N-terminal modification reagents (Fig. 1A).
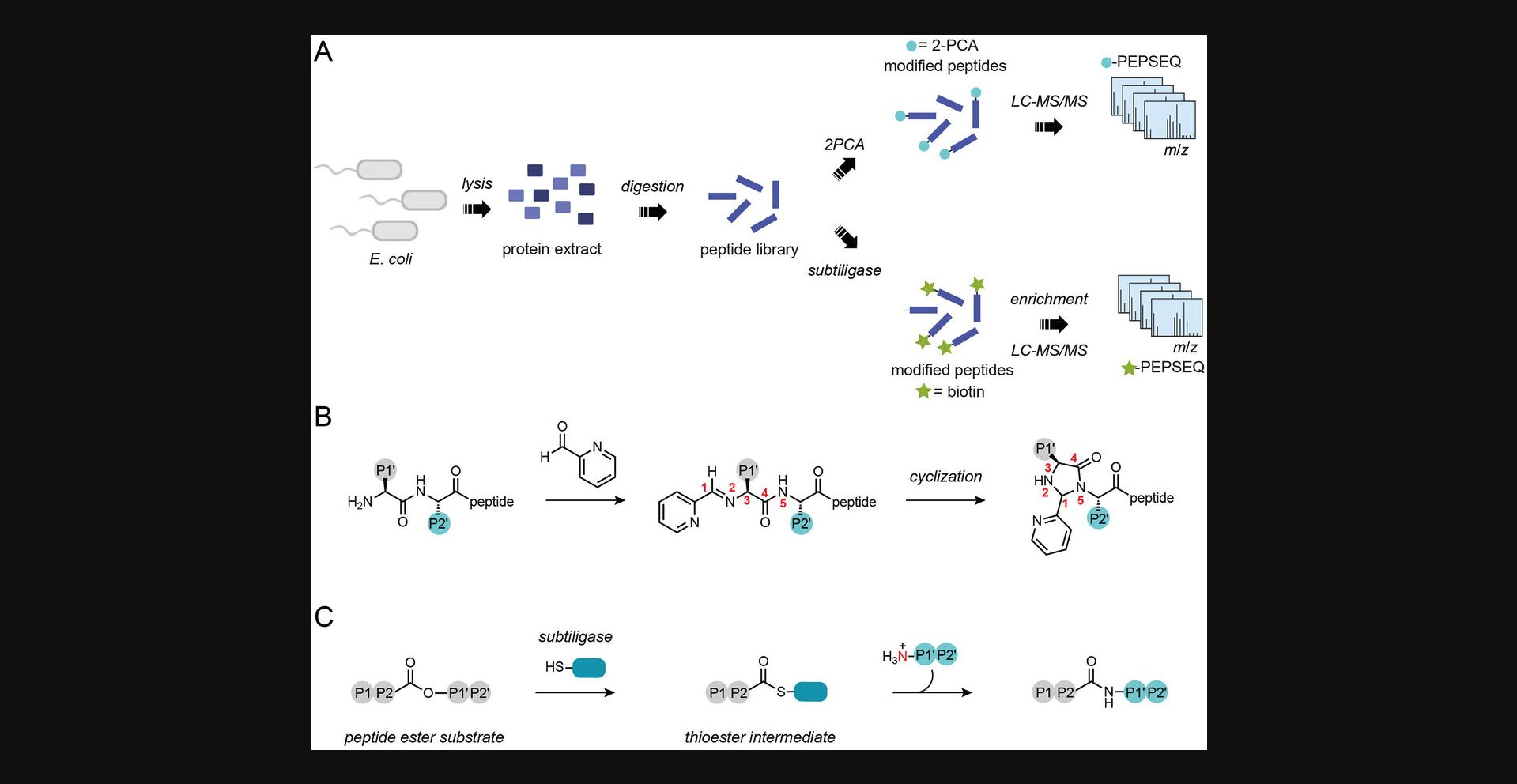
N-terminal modification reagents are important tools in the fields of protein bioconjugation and chemoproteomics (Griswold et al., 2019; Mahrus et al., 2008; Rosen & Francis, 2017; Weeks & Wells, 2018). In protein bioconjugation, targeting the N terminus enables site-specific single modification of proteins with payloads such as fluorophores, affinity handles, and drug molecules. Although there are many approaches to modifying the N terminus, they have variable selectivity and inherent sequence biases (MacDonald et al., 2015; Rosen & Francis, 2017). It is therefore important to define these properties for each reagent before selecting one for a particular application. In chemoproteomics, N-terminal modification reagents enable the selective capture of protein N termini for global study of proteolytic cleavage events (Fig. 1B and 1C; Griswold et al., 2019; Mahrus et al., 2008). Because the proteases under study may have their own sequence preferences, it is important to understand N-terminal modification reagent specificity to disentangle it from the specificity of the protease of interest.
Enrichment-based methods for N-terminal chemoproteomics, or N terminomics, rely on the ability to selectively biotinylate protein N termini while avoiding modification of other biological amines (Luo et al., 2019). Following biotinylation, the biotin affinity handle can be used to selectively isolate N-terminal peptides for analysis with tandem mass spectrometry (LC-MS/MS). One method, known as subtiligase N terminomics, uses the engineered peptide ligase subtiligase to selectively modify protein N termini with a biotinylated peptide (Fig. 1C; Mahrus et al., 2008; Weeks & Wells, 2019). N-terminal specificity is conferred by the molecular recognition properties of the subtiligase protein scaffold, which recognizes N-terminal α-amines but not lysine side chains (Weeks & Wells, 2019). Subtiligase N terminomics has been applied extensively to study caspase cleavages during apoptosis, providing molecular insights into the events that lead to programmed cell death (Agard et al., 2012; Mahrus et al., 2008; Shimbo et al., 2012). Proteomic Identification of Ligation Sites (PILS), a method based on subtiligase modification of proteome-derived peptide libraries, revealed that subtiligase specificity matches the specificity of caspases very well (Weeks & Wells, 2018). However, PILS also revealed that N-terminal peptides with acidic residues in the first two positions are poor substrates for subtiligase. PILS was subsequently deployed for engineering subtiligase variants that overcome this sequence limitation, demonstrating the utility of proteome-derived peptide libraries for informing chemoproteomics studies.
A second N terminomics method, Chemical enrichment Of Protease Sites (CHOPS), uses a biotinylated derivative of the N-terminally selective reagent 2-pyridinecarboxaldehyde (2PCA) for selective modification of protein N termini (Griswold et al., 2019; MacDonald et al., 2015). The specificity of 2PCA reagents for the N terminus is conferred by the chemical mechanism of modification (Fig. 1B). The first step involves a reversible condensation between the N-terminal α-amine and the aldehyde of 2PCA to form an imine (MacDonald et al., 2015). In principle, this step can also occur with lysine ε-amines. However, selectivity arises from a second, irreversible step in which the neighboring amide in the peptide backbone attacks the imine to form a stable cyclic imidazolidinone. Because lysine side chains do not have an appropriately positioned amide, this cyclization reaction cannot occur and 2PCA only stably modifies N-terminal α-amines. Importantly, N termini in which proline is the second residue also lack a neighboring nucleophilic amide and cannot be modified efficiently by 2PCA reagents. Deep profiling of 2PCA specificity with proteome-derived peptide libraries experimentally confirmed this bias and also revealed a less substantial bias against 2PCA modification of N-terminal glycine residues (Bridge et al., 2023). Despite these well-defined biases, 2PCA nonetheless has very broad sequence specificity and is thus useful as a modification reagent for N terminomics. The 2PCA-based method CHOPS has been applied to identify proteolytic neo-N termini that arise from cleavage by dipeptidyl peptidases 8 and 9 (DPP8 and DPP9), shedding light on how these enzymes regulate the Nlrp1 inflammasome (Griswold et al., 2019). More recently, another 2PCA-based strategy, Chemical enrichment Of Protease sites with Purchasable, Elutable Reagents (CHOPPER), has been developed to eliminate the need for chemical synthesis of CHOPS probes by introducing a click-chemistry-based biotinylation step following N-terminal modification with alkyne-modified 2PCA (Bridge et al., 2023). CHOPPER has been applied to apoptotic proteolysis, leading to the discovery of many previously unknown caspase cleavage sites.
Both 2PCA and subtiligase are powerful tools for N terminomics, but each method has key advantages and limitations. Both modification strategies have broad N-terminal specificity as determined by deep specificity profiling using proteome-derived peptide libraries, making them well suited for global sequencing of proteolytic neo-N termini (Bridge et al., 2023; Weeks & Wells, 2018). This contrasts with other methods that have been explored for modification of cellular N termini, such as application of the transpeptidase sortase (Swee et al., 2015). Efficient sortase modification requires one or more N-terminal glycine residues, making this enzyme poorly suited for proteome-scale N-terminal modification. Both subtiligase N terminomics and 2PCA-based methods target only unblocked N termini and not those that are acetylated, methylated, myristoylated, or otherwise blocked, making these reagents most useful for studying unblocked N termini that arise from proteolysis. This limits their utility for the study of N termini that bear post-translational modifications, although subtiligase has been used to detect increases in the abundance of unblocked N termini upon knockdown of N-acetyltransferase enzymes (Yi et al., 2011). Both 2PCA reagents and subtiligase enable enrichment of N-terminal peptides for identification of N-terminal sequences with single-amino-acid resolution by LC-MS/MS. Although most polypeptide molecules are linear and contain exactly one N terminus, certain regulatory modifications, such as ubiquitination, SUMOylation, and NEDDylation, generate branched protein structures that contain more than one N terminus (Swatek & Komander, 2016). Proteolytic removal of these modifications leaves an unblocked di-glycine N terminus linked to the modified protein by an isopeptide bond. Standard 2PCA-based and subtiligase-based workflows do not provide information on such branched structures.
Treatment of N-terminally diverse proteome-derived peptide libraries with a reagent of interest followed by analysis by LC-MS/MS enables the assessment of tens of thousands of peptide sequences as potential modification targets, providing a deep profile of sequence and site specificity (Bridge et al., 2023; Weeks & Wells, 2018). This article outlines how to generate N-terminally diverse proteome-derived peptide libraries and how to apply them to define the sequence specificity of N-terminal modification reagents (Fig. 1). Although protocols are provided for specificity characterization of 2-pyridinecarboxaldehyde (2PCA; MacDonald et al., 2015) and subtiligase (Weeks & Wells, 2018), the Strategic Planning section describes considerations for planning experiments with other N-terminal modification reagents. Basic Protocol 1 describes a method to generate peptide libraries from the E. coli proteome. The Alternate Protocol describes a method to generate peptide libraries from the human proteome. Basic Protocol 2 describes how to apply proteome-derived peptide libraries for specificity profiling of 2PCA, a chemical reagent for N-terminal modification. Basic Protocol 3 describes how to apply proteome-derived peptide libraries for specificity profiling of subtiligase, an enzymatic N-terminal modification reagent.
STRATEGIC PLANNING
Before beginning Basic Protocol 1, it is important to consider which digest protease(s) are best suited for characterization of the reagent of interest. Protease specificity is typically described using the Schechter and Berger nomenclature (Schechter & Berger, 1967), in which residues on the nonprime side (N-terminal to scissile bond) are denoted with a P and numbered outward and residues on the prime side (C-terminal to the scissile bond) are denoted with Pʹ and numbered outward. The specificity of trypsin, chymotrypsin, and GluC is determined by the P1 residue (N-terminal to the scissile bond). Trypsin, chymotrypsin, and GluC all recognize characteristic P1 residues, resulting in depletion of certain amino acids at the N termini of peptides. For example, if a proteome-derived peptide library is generated with trypsin, which cleaves C-terminal to Lys and Arg (P1 = K or R), Lys and Arg will not be well represented at the N termini of the resultant peptides but will be primarily found at the C termini. In most cases, modification reagents should be characterized using multiple libraries generated with proteases of orthogonal specificity to ensure maximum sequence coverage. It is important to note that certain amino acids (Pro, Cys, and Trp) will be unavoidably underrepresented due to their low abundance in the proteome. It is also important to keep in mind that Cys residues in libraries generated according to Basic Protocol 1 are carbamidomethylated.
Basic Protocol 2 describes a method for profiling the specificity of an N-terminal modification reagent without enrichment of the modified peptides, whereas Basic Protocol 3 describes a method in which modified peptides are enriched before analysis. Enrichment refers to the inclusion of a workflow for selective isolation of modified peptides before analysis by LC-MS/MS. Before choosing between these approaches for profiling a new modification reagent, several key properties of the reagent should be considered. The first consideration is whether the reagent under study can be modified to enable enrichment by derivatization with biotin or another affinity handle. In the absence of an enrichment handle, only Basic Protocol 2 can be used. If prior knowledge is available about how broad or specific the reagent is expected to be, this can inform which protocol should be chosen. Reagents that modify >10% of the sequences in the library can be profiled with Basic Protocol 2, as this method is likely to lead to the identification of thousands of modified peptides even in the absence of enrichment. However, if the extent of modification is lower, Basic Protocol 3 may be better suited for specificity profiling as the enrichment step will decrease sample complexity and increase sampling depth for modified peptides.
When profiling a new N-terminal modification reagent, there are several details of both Basic Protocol 2 and Basic Protocol 3 that should be carefully considered. The first is the buffer to be used for the reaction. In general, the buffer that supports the reaction and/or enzymatic activity should be chosen. Most buffers are compatible with proteome-derived peptide libraries. If the buffer used contains detergents, polymers, or other molecules that cannot easily be separated from peptides using C18 desalting, the SP3 desalting strategy outlined in Basic Protocol 2 should be used.
Basic Protocol 1: GENERATION OF N-TERMINALLY DIVERSE PROTEOME-DERIVED PEPTIDE LIBRARIES FROM Escherichia coli
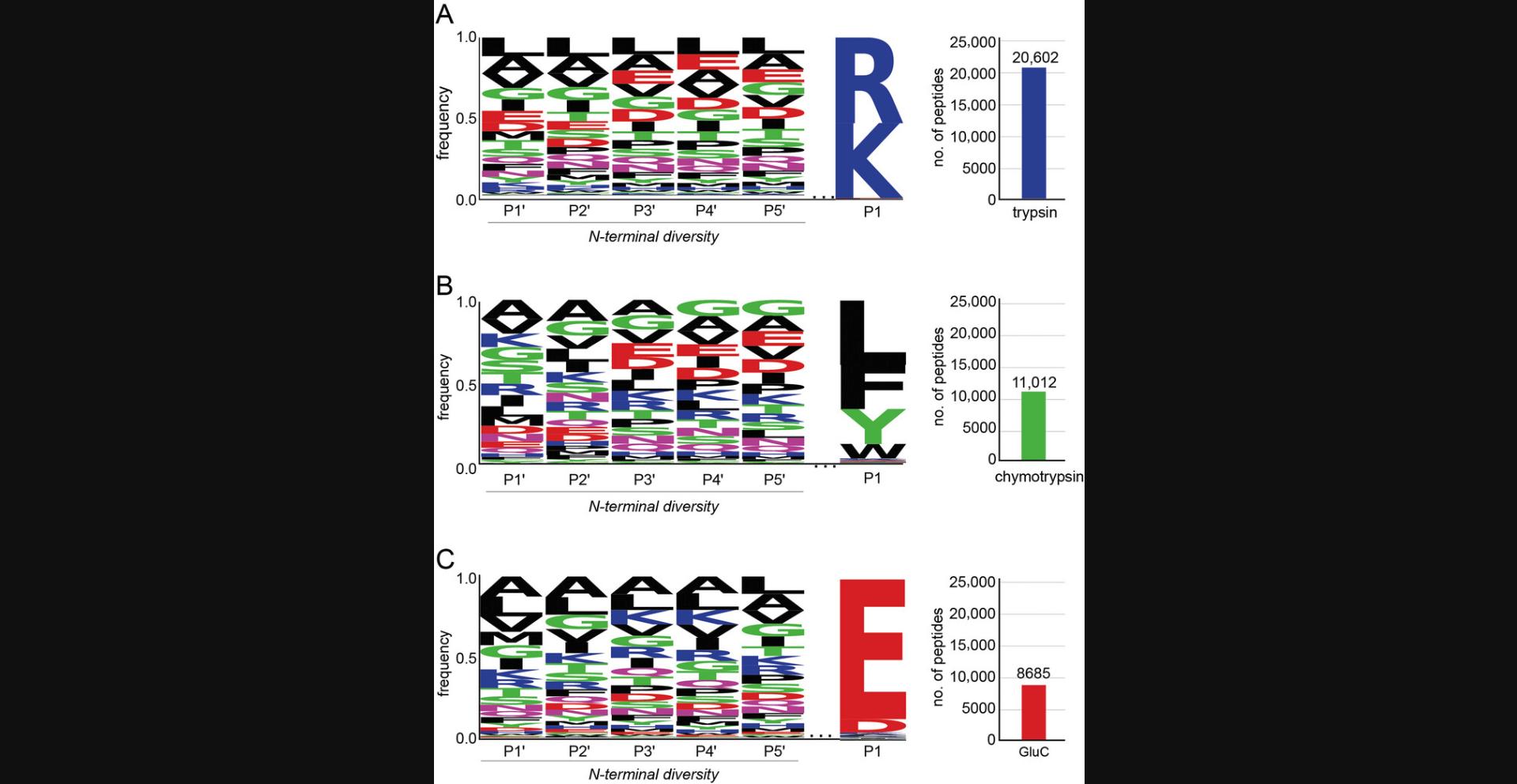
This protocol describes how to generate N-terminally diverse peptide libraries from the E. coli proteome. Cells are lysed and proteins are extracted and digested with the desired digest protease(s). Peptide libraries generated according to this protocol will contain thousands of peptides with a known conserved sequence at the C-terminus and diverse sequences at the N-terminus (Fig. 2).
Materials
-
Luria-Bertani (LB) Broth
-
Escherichia coli XL10 (or other E. coli strain)
-
Phosphate-buffered saline (PBS; see recipe)
-
E. coli lysis buffer (see recipe)
-
1 M HEPES, pH 7.5 (Fisher Scientific, cat. no. AC172570010)
-
1 M dithiothreitol (DTT; Goldbio, cat. no. DTT100)
-
500 mM iodoacetamide (Sigma-Aldrich, cat. no. I1149-5G; make fresh and protect from light)
-
100% (w/v) trichloroacetic acid (TCA; Sigma-Aldrich, cat. no. T6399)
-
Methanol (Fisher Scientific, cat. no. A456-4), prechilled to –20°C
-
20 mM sodium hydroxide (NaOH; Sigma-Aldrich, cat. no. 415413-500ML)
-
Bicinchoninic acid (BCA) protein assay kit (Thermo Scientific, cat. no. 23228)
-
Sequencing-grade trypsin (Promega, cat. no. V5113), GluC (Promega, cat. no. V1651), or chymotrypsin (Promega, cat no. V1061)
-
SDS-PAGE gels (Invitrogen, cat. no. NP0321BOX)
-
1 M phenylmethanesulfonylfluoride (PMSF) in DMSO (make fresh before use)
-
8 M guanidine hydrochloride (Chem-Impex, cat. no. 00152)
-
Trifluoroacetic acid (TFA; Optima LC-MS grade; Fisher, cat. no. A116)
-
Acetonitrile (Optima LC-MS grade; Fisher, cat. no. A955)
-
Water (Optima LC-MS grade; Fisher, cat. no. W64)
-
0.1% TFA in water (use LC-MS-grade chemicals)
-
0.1% TFA/50% acetonitrile in water (use LC-MS-grade chemicals)
-
0.1% formic acid in water (use LC-MS-grade chemicals)
-
Ultrapure Milli-Q water
-
Baffled culture flask that can accommodate 1 L (such as a 2.8-L Fernbach flask, Sigma-Aldrich, cat. no. CLS44242XL)
-
Shaker-incubator, 37°C
-
Refrigerated centrifuge capable of operating at 10,000 × g
-
50-ml conical tubes
-
Homogenizer, microfluidizer, or French press (such as the Avestin EmulsiFlex-C3, Avestin, cat. no. C321220)
-
Probe ultrasonicator (such as the Qsonica Q700, Fisher Scientific, cat. no. 15-338-281)
-
Rocker or rotisserie mixer (such as the Labnet Mini Lab Roller, Labnet, cat. no. H5500)
-
BCA protein assay kit (Thermo Scientific, cat. no. 23228)
-
Chemical fume hood
-
Benchtop microcentrifuge
-
Microplate reader suitable for reading absorbance at 562 nm (such as the Tecan Infinite M200)
-
Incubator, 37°C
-
Apparatus to run SDS-PAGE gel
-
pH strips (EMD Millipore, cat. no. 1.09535.0001)
-
Waters Sep-Pak C18 cartridge, 360 mg sorbent (Fisher Scientific, cat. no. 50-818-645)
-
1.5-ml microcentrifuge tubes (Axygen, cat. no. MCT-150-L-C)
-
Vacuum centrifuge (e.g., SpeedVac)
-
Ultra-low-temperature (–80°C) freezer
-
Dionex UltiMate 3000 RSLCnano system (Thermo Scientific)
-
Acclaim PepMap RSLC column (Thermo Scientific, cat no. 164942)
-
Orbitrap Exploris 480 Mass Spectrometer (Thermo Scientific)
-
Computer with the following minimum requirements: 2 GHz processor, 2 GB RAM, video card and monitor capable of 1280 × 1024 resolution, screen resolution of 96 dpi, 1 TB available on C drive, New Technology File System format)
-
Proteome Discoverer 2.4 analysis software (Thermo Scientific)
Growth and harvesting the proteome source
1.Using a sterile inoculation loop or sterile wooden stick,, inoculate 1 L of autoclaved LB medium in a 2.8-L flask with E. coli XL10.
2.Grow the E. coli for 14-16 hr at 37°C, shaking at 200 rpm.
3.Harvest the cells by centrifugation for 15 min at 5000 × g , 4°C.
4.Pour off the supernatant into a liquid waste container and retain the cell pellet.
5.Resuspend the cell pellet in 50 ml PBS by pipetting up and down with a serological pipet.
6.Weigh an empty 50-ml conical tube and note the weight.
7.Transfer the resuspended cells into the 50-ml conical tube.
8.Centrifuge 5 min at 5000 × g , 4°C.
9.Pour off the supernatant into a liquid waste container and retain the cell pellet.
10.Determine the pellet mass by subtracting the mass of the empty 50-ml conical tube from the mass of the tube containing the cell pellet.
11.Resuspend the cell pellet in 50 ml E. coli lysis buffer.
12.Lyse the cells by three passes through a homogenizer, microfluidizer, or French press at 15,000 psi.
13.Centrifuge 30 min at 10,000 × g , 4°C.
14.Transfer the clarified supernatant to a fresh 50-ml conical tube. Discard the insoluble pellet.
15.Add the appropriate volume of 1 M HEPES to the supernatant to achieve a final concentration of 100 mM HEPES. Save a sample to measure the protein concentration using the BCA protein assay kit according to the manufacturer's instructions using a microplate reader.
First reduction and alkylation
16.Proceed with 50 ml of the supernatant from step 15.Add 250 µl of 1 M DTT to achieve a final concentration of 5 mM and incubate at room temperature on a rocker or rotisserie mixer for 30 min.
17.Add 1 ml of 500 mM iodoacetamide to achieve a final concentration of 10 mM and incubate for 60 min at room temperature in the dark on a rocker or rotisserie mixer.
18.Quench any unreacted iodoacetamide by adding 250 µl of 1 M DTT to achieve an accumulated final DTT concentration of 10 mM. Incubate at room temperature for 15 min.
Protein isolation
19.Using the BCA assay results from step 15, transfer aliquots of the protein extract from step 18 containing 10 mg protein into 15-ml conical tubes.
20.Precipitate protein adding 100% (w/v) TCA to a final concentration of 15% (w/v) TCA.
21.Incubate the precipitated protein samples at –20°C for a minimum of 2 hr. After 2 hr, continue to step 22 or use this as a pause point.
22.Thaw the precipitated protein samples.
23.Centrifuge 30 min at 10,000 × g , 4°C.
24.Pour off the supernatant into a liquid waste container and retain the protein pellet. Centrifuge briefly to collect residual supernatant at the bottom of the tube. Use a pipet to carefully remove residual traces of supernatant.
25.Overlay the pellet with 200 µl of –20°C methanol to wash the pellet.
26.Holding the tube upright, carefully remove the methanol with a pipet.
27.Repeat steps 25 and 26.
28.After removing the second methanol wash from all of the samples, use a fresh pipet tip to carefully remove any visible traces of methanol.
29.Let the pellet air dry for 20 min.
30.Add 5 ml of 20 mM NaOH to the protein pellet to achieve an assumed protein concentration of 2 mg/ml (based on aliquoting 10 mg per tube in step 19).
31.Use probe ultrasonication (20% amplitude, 10 cycles of 5 s on/5 s off) to help dissolve the pellet.
32.Add 1 M HEPES, pH 7.5, to a final concentration of 200 mM.
33.Centrifuge 30 min at 10,000 × g , 4°C.
34.Transfer the supernatant to a fresh 15-ml conical tube. Discard the insoluble pellet.
35.Determine the protein concentration and total protein mass using the BCA assay method. Save a 100 µl pre-digestion sample.
36.Digest with chymotrypsin, GluC, trypsin, or other suitable protease. Use a protease-to-proteome ratio of 1:100 (wt/wt) and incubate 16-20 hr at 37°C.
37.Run 10 µg of protein from the sample saved in step 35 and 10 µg of the digested protein sample on an SDS-PAGE gel. No bands above 10 kDa should be visible in the digested sample.
38.Add 1 M PMSF in DMSO to a final concentration of 1 mM to inhibit protease digestion. Invert several times to mix and heat the sample at 95°C to completely inactivate the protease.
39.Add 8 M guanidine hydrochloride to a final concentration of 1 M.
40.Centrifuge 30 min at 10,000 × g , 4°C.
Second reduction and alkylation
41.Transfer the supernatant to a fresh 15-ml conical tube. Discard the insoluble pellet.
42.Add DTT to a final concentration of 5 mM and incubate at 37°C for 1 hr.
43.Add iodoacetamide to a final concentration of 10 mM and incubate at 37°C for 1 hr in the dark.
44.Add DTT to a final concentration of 15 mM and incubate at 37°C for 10 min to quench excess iodoacetamide.
Desalting peptide libraries using C18 solid-phase extraction
45.Prepare the peptide library for desalting by acidifying the sample to a pH <3 by adding 100% TFA to a final concentration of 5% (v/v).
46.Condition a Sep-Pak cartridge by pushing 3 ml acetonitrile through the cartridge dropwise using a 3-ml syringe. Collect the flowthrough in a waste breaker.
47.Equilibrate the cartridge by pushing 3 ml of 0.1% TFA through dropwise using a 3-ml syringe. Collect the flowthrough into a waste beaker.
48.Repeat step 46.
49.Load 4 mg of the peptide library onto the equilibrated C18 cartridge using a syringe. Collect flowthrough into waste beaker or save if desired.
50.Equilibrate the cartridge by pushing 3 ml of 0.1% TFA through dropwise using a 3 ml syringe. Collect the flowthrough into a waste beaker.
51.Repeat step 50.
52.Elute the peptide library by pushing 1.5 ml of 80% acetonitrile/20% water through the cartridge. Collect the eluted peptide into a 1.5-ml microcentrifuge tube.
53.Repeat step 52.
54.Use a vacuum centrifuge (e.g., SpeedVac) to remove acetonitrile from the peptide libraries. Concentrate the peptide library until the volume is reduced to half of the original volume (750 µl in each tube).
55.Add 500 µl LC-MS-grade water and concentrate the peptide library until the volume has again been reduced to 750 µl.
56.Repeat step 55 two additional times.
57.Pool the concentrated elution fractions. Determine the final peptide concentration by BCA assay.
58.Adjust the concentration of the peptide libraries to 2 mg/ml using LC-MS-grade water. Store peptide libraries in water in 50- to 100-µl aliquots at –80°C.
LC-MS/MS analysis of proteome-derived peptide libraries
59.Dilute a sample of proteome-derived peptide library to a concentration of 0.1 mg/ml in 0.1% formic acid.
60.Analyze peptides by LC-MS/MS.
| Parameter | Setting |
|---|---|
| Source | Nano-ESI |
| Ion transfer tube temperature | 325°C |
| Positive spray voltage | 2000 V |
| Full scan mass range | 300-1200 m/z |
| Full scan parameters | |
| Orbitrap resolution | 60,000 at 200 m/z |
| Scan range | 300-1200 m/z |
| RF lens | 40% |
| Normalized AGC target | 300% |
| Maximum injection time | Auto |
| Intensity threshold | 5 × 103 |
| Charge state | 2-6 |
| Dynamic exclusion | 20 s; precursor mass tolerance ±10 ppm |
| Top-N MS2 | 20 |
| ddMS2 parameters | |
| Isolation window | 1.4 m/z |
| Collision energy mode | Fixed |
| Collision energy type | Normalized |
| HCD collision energy | 30% |
| Orbitrap resolution | 15,000 at 200 m/z |
| Scan range mode | Define first mass |
| First mass | 110 m/z |
| AGC target | Standard |
| Maximum injection time | 22 ms |
61.Analyze raw files using Proteome Discoverer software. Search data using the human or E. coli SwissProt database, depending on the proteome used to generate the library. Select the appropriate protease specificity and set it to full cleavage with up to 2 missed cleavages. For data collected on a Thermo Orbitrap Exploris, set the precursor ion mass tolerance to 10 ppm and fragment ion mass tolerance to 0.02 Da. Set cysteine carbamidomethylation as a static modification. Set acetylation, methionine loss, and methionine loss plus acetylation as dynamic N-terminal protein modifications.
Alternate Protocol: GENERATION OF N-TERMINALLY DIVERSE PROTEOME-DERIVED PEPTIDE LIBRARIES FROM HUMAN CELLS
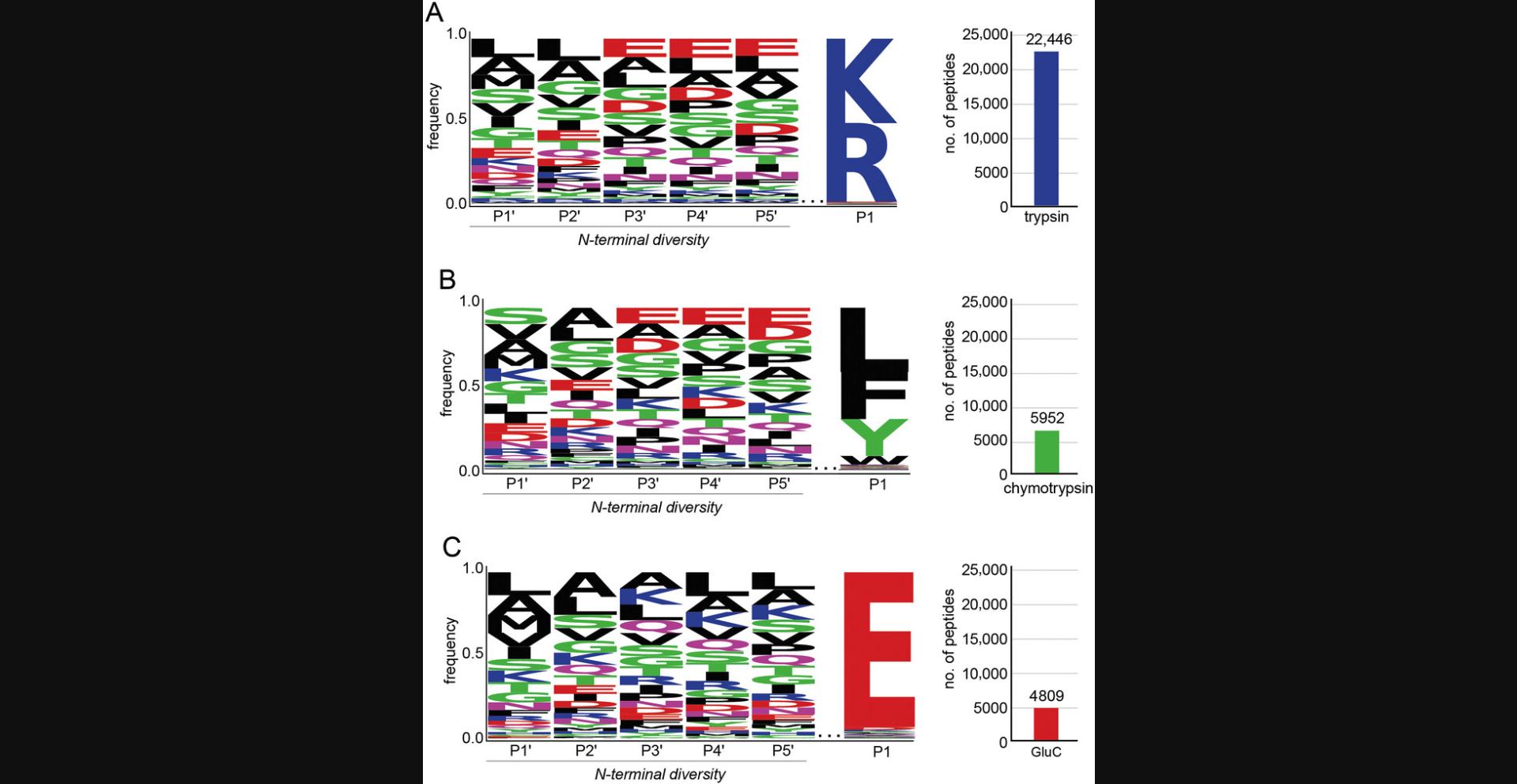
This protocol is performed instead of Basic Protocol 1 if human-derived N-terminally diverse peptide libraries are desired. Peptide libraries derived from human sources will have post-translational modifications that E. coli -derived peptide libraries do not have. Human-derived peptide libraries can be used as a complement to E. coli -derived peptide libraries, or on their own. Successful completion of this protocol will result in thousands of N-terminally diverse human peptides with a conserved C-terminal sequence (Fig. 3).
Additional Materials (also see Basic Protocol 1)
-
HEK293T cells (ATCC, cat. no. CRL-3216, RRID:CVCL_0063)
-
Complete DMEM (see recipe)
-
Phosphate-buffered saline (PBS; VWR, cat. no. 45000-448)
-
Versene (Thermo Scientific, cat. no. 15040066)
-
Mammalian cell lysis buffer (see recipe)
-
Laminar-flow hood (BSC Class II)
-
225-cm2 cell culture flasks
-
5% CO2, 37°C tissue culture incubator
-
Vacuum aspirator
-
Tissue culture microscope
Growing and harvesting the proteome source
1.Grow one 225-cm2 flask of HEK293T cells in complete DMEM medium to 90% confluence in a 5% CO2, 37°C tissue culture incubator, with 85% humidity.
2.Remove the medium using a vacuum aspirator, being careful not to disturb the cells.
3.Carefully wash the cells with 20 ml PBS. Remove the PBS with a vacuum aspirator.
4.Add 10 ml Versene to detach the cells. Incubate in 5% CO2, 37°C tissue culture incubator for 15 min, occasionally tapping the sides of flask to help cells detach.
5.Transfer the Versene cell suspension to a 15-ml conical tube.
6.Centrifuge cells 5 min at 300 × g , 4°C. Remove the supernatant with a vacuum aspirator.
7.Resuspend cells gently in 10 ml PBS.
8.Centrifuge resuspended cells 5 min at 300 × g , 4°C. Remove the supernatant with a vacuum aspirator.
9.While step 8 is running, aliquot 800 µl of mammalian cell lysis buffer into a 1.5-ml microcentrifuge tube and preheat to 95°C.
10.Resuspend the cell pellet in the preheated lysis buffer from step 9.Heat at 95°C for 10 min in a 1.5-ml microcentrifuge tube.
11.Complete the lysis by probe ultrasonication at 20% amplitude using 10 cycles of 5 s on/5 s off.
12.Centrifuge the lysate for 10 min at 20,000 × g , 4°C, in a benchtop microcentrifuge.
13.Transfer the clarified supernatant to a fresh 2.0-ml microcentrifuge tube. Discard the insoluble pellet.
14.Continue with Basic Protocol 1 starting from step 19 (protein isolation).
Basic Protocol 2: CHARACTERIZING THE SPECIFICITY OF 2-PYRIDINECARBOXALDEHYDE USING PROTEOME-DERIVED PEPTIDE LIBRARIES
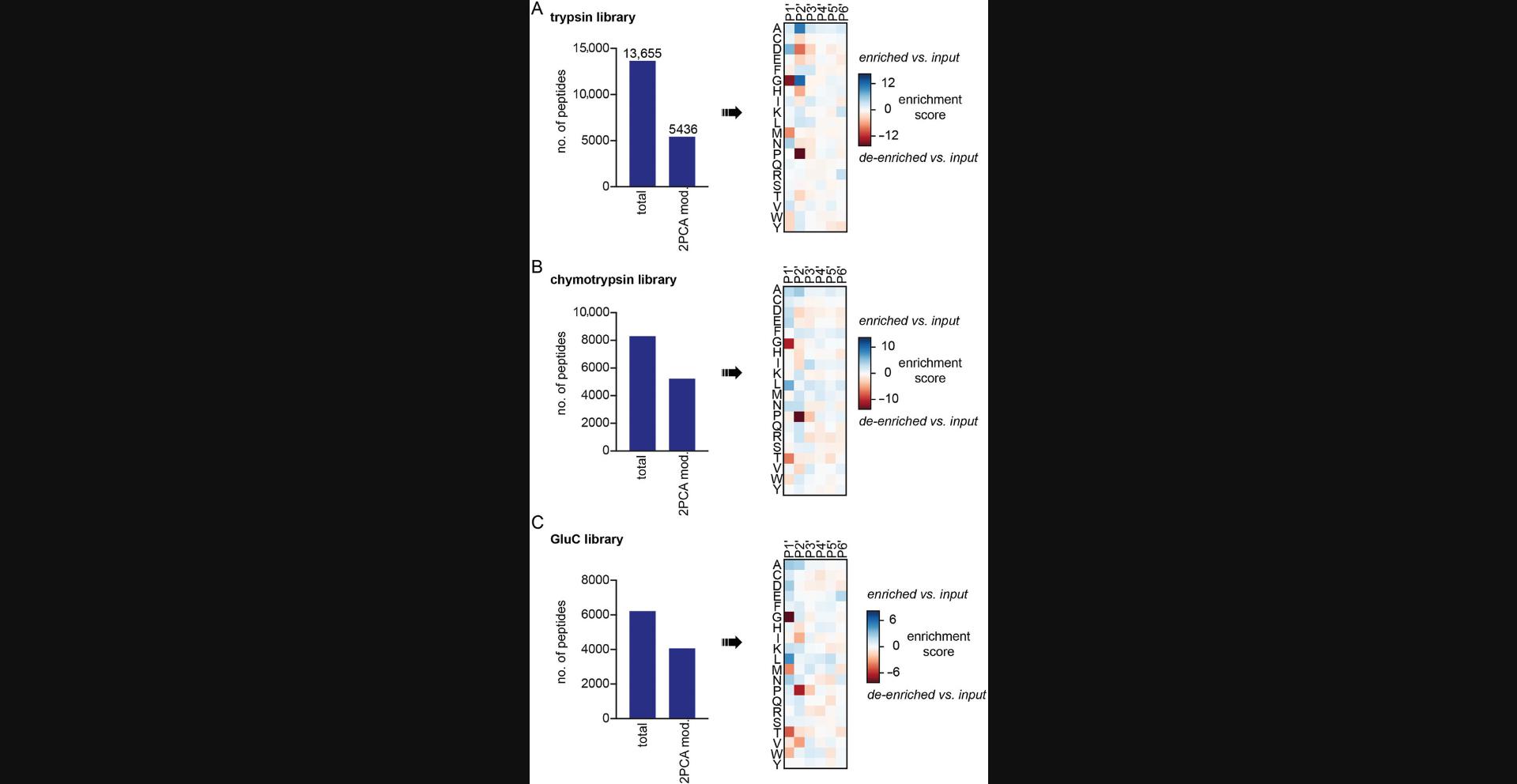
This protocol is performed to profile the labeling specificity of a chemical N-terminal labeling reagent: 2-pyridinecarboxaldehyde (2PCA; MacDonald et al., 2015). Proteome-derived N-terminally diverse peptide libraries are incubated with 2PCA to label the N termini. The labeled peptide sequences are then identified by LC-MS/MS and the labeling preference is analyzed (Fig. 4).
Materials
-
Ultrapure Milli-Q water
-
400 mM sodium phosphate, pH 7.5
-
2 mg/ml proteome-derived peptide library (from Basic Protocol 1)
-
500 mM 2-pyridinecarboxaldehyde (2PCA; Sigma-Aldrich, cat. no. P62003-100G) in Milli-Q water
-
Water (Optima LC-MS grade; Fisher, cat. no. W64)
-
Acetonitrile (Optima LC-MS grade; Fisher, cat. no. A955)
-
0.1% formic acid in water (use LC-MS-grade chemicals)
-
1.5- and 2-ml microcentrifuge tubes (Axygen, cat. no. MCT-150-L-C)
-
Incubator (37°C)
-
Sera-Mag Speedbeads Carboxyl magnetic beads, hydrophobic (Cytiva, cat. no. 65152105050250)
-
Sera-Mag Speedbeads Carboxyl magnetic beads, hydrophilic (Cytiva, cat. no. 45152105050250)
-
Vortex
-
Magnetic stand (such as MagRack 6, Cytiva, cat. no. 28-9489-64)
-
Benchtop microcentrifuge
-
Water bath sonicator
-
Thermomixer C (Eppendorf)
-
Spectrophotometer (such as a Nanodrop microvolume spectrophotometer)
-
Dionex UltiMate 3000 RSLCnano system (Thermo Scientific, cat. no. ULTIM3000RSLCNANO)
-
Acclaim PepMap RSLC column (Thermo Scientific, cat no. 164942)
-
Orbitrap Exploris 480 Mass Spectrometer (Thermo Scientific)
-
Proteome Discoverer 2.4 software (Thermo Scientific)
Labeling of proteome-derived peptide libraries with 2-PCA
1.Prepare a 100-µl reaction mixture in a 1.5-ml microcentrifuge tube by combining 27.5 µl Milli-Q water, 12.5 of µl 400 mM sodium phosphate, pH 7.5, 50 µl of 2 mg/ml peptide library, and 10 µl of 500 mM 2PCA. Mix well.
2.Allow the reaction to proceed for the desired length of time (2-24 hr for 2PCA) at 37°C.
Sample desalting and cleanup
3.Desalt the samples using the single-pot, solid-phase-enhanced sample-preparation (SP3) method. Aliquot 20 µl hydrophilic Sera-Mag Speedbeads and 20 µl hydrophobic Sera-Mag Speedbeads into the same 2-ml microcentrifuge tube. Vortex briefly to mix.
4.Place the tube on a magnetic stand for 2 min to collect the beads. Aspirate the supernatant.
5.Remove the tube from the magnetic stand. Add 1 ml water and vortex for 10 s to mix.
6.Centrifuge for 2 s to collect liquid from the cap and sides of the tube. Place on a magnetic stand for 2 min to collect the beads. Aspirate the supernatant.
7.Repeat steps 5 and 6 two additional times.
8.Aliquot 1900 µl of LC-MS-grade acetonitrile into a 2-ml microcentrifuge tube. Add the 2PCA reaction mixture to the tube.
9.Transfer the acetonitrile/peptide mixture from step 8 to the tube containing the washed beads from step 7.Vortex immediately.
10.Incubate samples on a Thermomixer at 1000 rpm at room temperature for 10 min.
11.Centrifuge for 2 s to collect liquid on the sides of the tube and cap. Place the tube on a magnetic stand for two minutes to collect the beads. Aspirate the supernatant.
12.Wash the beads by adding 1 ml LC-MS-grade acetonitrile. Vortex immediately, and then place on a Thermomixer at 1000 rpm at room temperature for 30 s.
13.Centrifuge for 2 s to collect liquid on the sides of the tube and cap. Place the tube on a magnetic stand for two minutes to collect the beads. Aspirate the supernatant.
14.Repeat steps 12 and 13 two additional times.
15.Air dry the beads for 2 min to ensure that no acetonitrile remains.
16.Elute the peptides by adding 100 µl LC-MS-grade water. Sonicate the samples in a water bath sonicator for 1 min, and then place the tube in a Thermomixer at room temperature at 1000 rpm for 5 min.
17.Centrifuge for 2 s to collect liquid from the sides of the tube and cap. Place the tube on a magnetic stand for 2 min.
18.Transfer the supernatant, which contains the eluted peptides, to a clean 1.5-ml microcentrifuge tube.
Quantification and analysis of peptides
19.Quantify the peptides by measuring the absorbance at 280 nm on a Nanodrop spectrophotometer.
20.Dilute the sample to a concentration of 0.1 mg/ml in 0.1% formic acid.
21.Analyze 5 µl of 0.1 mg/ml sample by LC-MS/MS.
22.Analyze raw files using Proteome Discoverer software. Search data using the human or E. coli SwissProt database, depending on the proteome used to generate the library. Select the appropriate protease specificity and set it to full cleavage with up to 2 missed cleavages. For data collected on a Thermo Orbitrap Exploris, set the precursor ion mass tolerance to 10 ppm and fragment ion mass tolerance to 0.02 Da. Set cysteine carbamidomethylation (+57.0215 Da) as a static modification. Set acetylation (+42.0106 Da), methionine loss (–131.0404 Da), and methionine loss plus acetylation (–89.0299 Da) as dynamic N-terminal protein modifications. Set 2PCA (+89.0265 Da) as a dynamic modification at peptide N termini.
Basic Protocol 3: CHARACTERIZING THE SPECIFICITY OF SUBTILIGASE USING PROTEOME-DERIVED PEPTIDE LIBRARIES
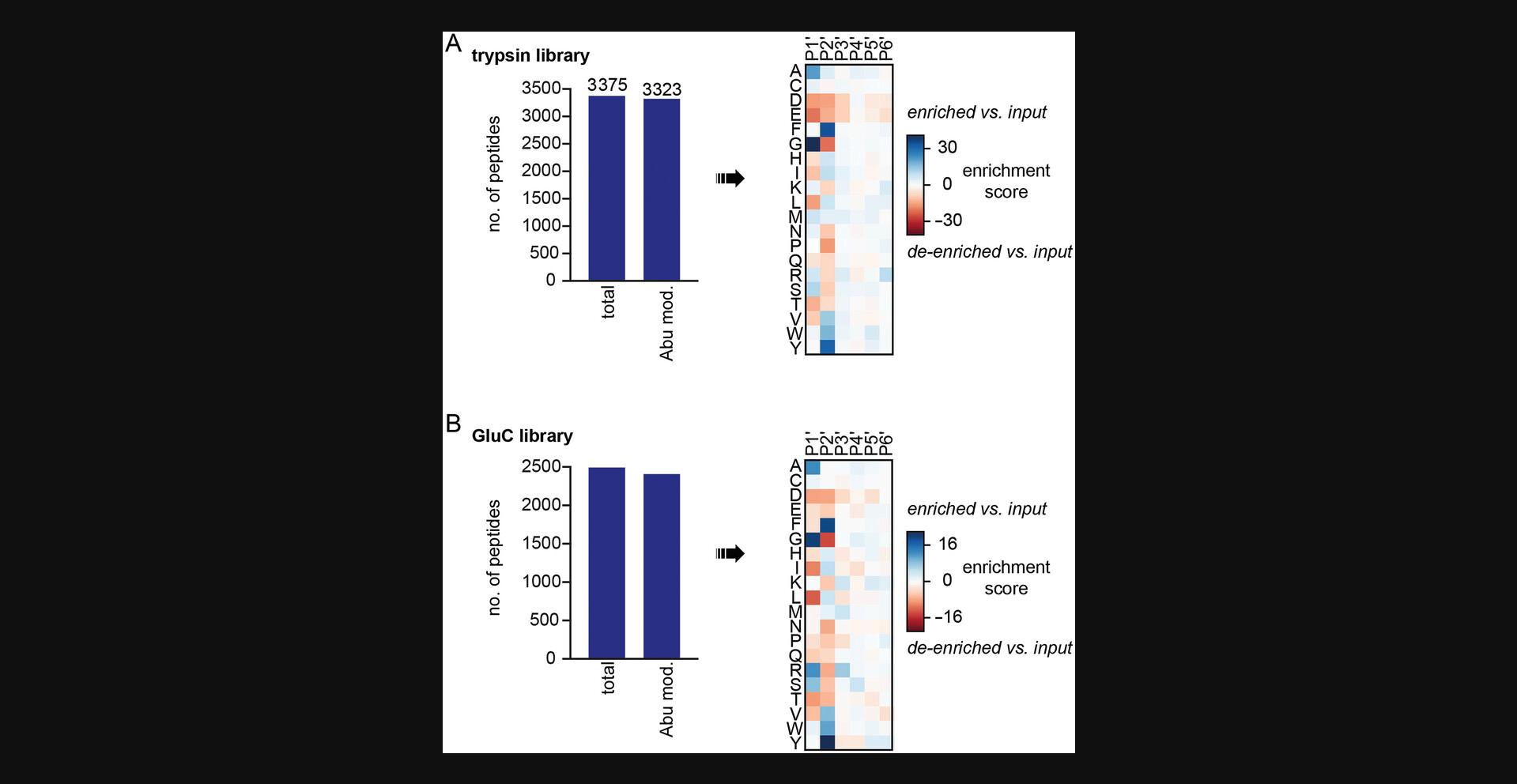
This protocol describes how to profile the specificity of subtiligase using proteome-derived peptide libraries. Detailed protocols for purification of subtiligase and synthesis of its substrate have been previously published in another Current Protocols paper (Weeks & Wells, 2020). Libraries are modified with a biotinylated peptide ester subtiligase substrate (Tev Ester 6) that also contains a tobacco etch virus (TEV) protease cleavage site and an aminobutyric acid (Abu) mass tag (Weeks & Wells, 2018, 2020). Subtiligase-modified peptides are enriched on neutravidin resin and then selectively eluted using TEV protease, leaving Abu at the N terminus of each subtiligase-modified peptide. Analysis of the eluted peptides by LC-MS/MS reveals the sequences of the labeled peptides, enabling the user to identify the sequence preferences and biases of the subtiligase variant tested. This protocol is expected to result in the enrichment and identification of thousands of peptides from each peptide library (Fig. 5).
Materials
-
Milli-Q water
-
1 M tricine, pH 8 (Bio-Rad, cat. no. 1610713)
-
2 mg/ml trypsin, GluC, and/or chymotrypsin proteome-derived peptide libraries (see Basic Protocol 1)
-
20 mM Tev Ester 6 (TE6) in DMSO (Weeks & Wells, 2020)
-
Subtiligase or subtiligase mutant, 50 µM stock solution (Weeks & Wells, 2020)
-
8 M and 4 M guanidine hydrochloride (GdnHCl; ChemImpex, cat. no. 00152)
-
High-capacity Neutravidin Agarose resin (Thermo, cat. no. 29202 or 29204)
-
100 mM ammonium bicarbonate (Sigma-Aldrich, cat. no. 09830-500G; make fresh on day of use)
-
1 M DTT (Goldbio, cat. no. DTT100)
-
TEV protease (New England BioLabs, cat. no. P8112S, or purified in-house)
-
Acetonitrile (Optima LC-MS grade; Fisher, cat. no. A955)
-
0.1% TFA (use LC-MS-grade water [Fisher, cat. no. W64] to prepare)
-
0.1% formic acid (use LC-MS-grade chemicals to prepare)
-
0.1% TFA/50% acetonitrile (use LC-MS-grade chemicals to prepare)
-
1.5-ml microcentrifuge tubes (Axygen, cat. no. MCT-150-L-C)
-
Microcentrifuge
-
Rotisserie mixer (such as the Labnet Mini Lab Roller, Labnet, cat. no. H5500)
-
1-ml-volume spin columns (Pierce Snap-Cap, Thermo, cat. no. 69725)
-
Vacuum centrifuge (e.g., SpeedVac)
-
SOLA HRP Cartridges (Thermo, cat. no. 60109-001) or similar reverse-phase desalting devices
-
Spectrophotometer (such as a Nanodrop microvolume spectrophotometer)
Labeling peptide libraries
1.Prepare a 100-µl reaction mixture by combining 37 µl Milli-Q water, 10 µl of 1 M tricine, pH 8, 50 µl of 2 mg/ml peptide library, and 1 µl of 20 mM TE6.
2.Initiate the reaction by adding 2 µl of 50 µM subtiligase or variant. Mix well by pipetting up and down several times.
3.Allow the reaction to proceed for 1 hr at room temperature.
4.Add 100 µl of 8 M GdnHCl to achieve a final concentration of 4 M GdnHCl. Vortex to mix.
Enrichment of biotinylated peptides
5.Prepare High-Capacity NeutrAvidin Agarose resin for enrichment. Invert bottle several times to generate a uniform slurry, and then aliquot 250 µl slurry into a 1.5-ml reaction tube for each sample.
6.Centrifuge resin for 2 min at 500 × g , room temperature, in a microcentrifuge. Aspirate the supernatant.
7.Resuspend resin in 500 µl of 4 M GdnHCl.
8.Centrifuge resin for 2 min at 500 × g , room temperature, in a microcentrifuge. Aspirate the supernatant.
9.Repeat steps 7 and 8 two more times.
10.Dilute the reaction mixture from step 4 with 100 µl of 4 M GdnHCl. Add the mixture to the prepared NeutrAvidin resin from step 9.
11.Place the tubes on a rotisserie or end-over-end mixture and incubate 30 min-20 hr.
12.Centrifuge resin for 2 min at 500 × g , room temperature, in a microcentrifuge. Aspirate the supernatant.
13.Resuspend resin in 500 µl of 4 M GdnHCl. Vortex briefly to mix.
14.Centrifuge resin for 2 min at 500 × g , room temperature, in a microcentrifuge. Aspirate the supernatant.
15.Repeat steps 13 and 14 two more times.
16.Add 500 µl 100 mM ammonium bicarbonate to the resin. Vortex briefly to mix.
17.Centrifuge resin for 2 min at 500 × g , room temperature, in a microcentrifuge. Aspirate the supernatant.
18.Repeat steps 16 and 17 two more times.
19.Resuspend the resin in 250 µl 100 mM ammonium bicarbonate. The total volume of buffer and slurry is now 375 µl.
20.Add 1 µl 1 M DTT to achieve a final concentration of 2.7 mM DTT.
21.Add 5 µl of 2 mg/ml (10 µg total) TEV protease. Incubate at room temperature on a rotisserie or end-over-end mixer for 2-6 hr at room temperature or for 16-20 hr at 4°C.
22.Centrifuge resin 2 min at 500 × g , room temperature, to collect droplets that may have accumulated on the cap.
23.Resuspend resin in the supernatant, and transfer to a snap-cap spin column. Place column in a clean 1.5-ml microcentrifuge tube.
24.Centrifuge column 2 min at 500 × g , room temperature. Save flowthrough, which contains the eluted N-terminal peptides.
25.Wash resin once with 125 µl of 100 mM ammonium bicarbonate. Combine wash with the flowthrough from step 24.
26.Dry sample in a vacuum centrifuge.
27.Resuspend pellet in 50-100 µl of 5% TFA to precipitate the TEV protease. Incubate on ice for 10 min. Add 1 µl solution to a strip of pH paper to check that the pH is below 3.
28.Centrifuge using a benchtop microcentrifuge 10 min at 21,000 × g , 4°C, to pellet precipitated TEV protease.
29.Transfer supernatant to a fresh 1.5-ml microcentrifuge tube.
30.Desalt the sample with a SOLA HRP Cartridge according to the manufacturer's instructions.
31.Dry the peptide in a vacuum centrifuge.
32.Resuspend dried peptides in 12-20 µl of 0.1% formic acid.
Quantification and analysis of peptides
33.Quantify the peptides by measuring the absorbance at 280 nm on a Nanodrop spectrophotometer.
34.Dilute the sample to a concentration of 0.1 mg/ml in 0.1% formic acid.
35.Analyze 5 µl of sample by LC-MS/MS.
36.Analyze raw files using Proteome Discoverer software. Search data using the human or E. coli SwissProt database, depending on the proteome used to generate the library. Select the appropriate protease specificity and set it to full cleavage with up to 2 missed cleavages. For data collected on a Thermo Orbitrap Exploris, set the precursor ion mass tolerance to 10 ppm and fragment ion mass tolerance to 0.02 Da. Set cysteine carbamidomethylation (+57.0215 Da) as a static modification. Set acetylation (+42.0106 Da), methionine loss (–131.0404 Da), and methionine loss plus acetylation (–89.0299 Da) as dynamic N-terminal protein modifications. Select Abu (+85.05276 Da) as a dynamic modification at peptide N termini.
REAGENTS AND SOLUTIONS
Use ultrapure water (ddH2O) for all solutions and protocol steps unless another solvent is specified.
Complete DMEM
- Dulbecco's modified eagle medium (DMEM) with high glucose (Cytiva, cat. no. SH30243.02)
- 100× penicillin-streptomycin (Cytiva, cat. no. SV30010)
- Fetal bovine serum (FBS; Cytiva, cat. no. SH30396.03)
Supplement high-glucose DMEM with 1× penicillin-streptomycin and 10% (v/v) FBS. Supplemented medium can be stored up to 1 year at 4°C.
E. coli lysis buffer
- 10 mM HEPES, pH 7.5 (checked with a pH meter)
- 0.5 mM EDTA
- 1 mM PMSF
Buffer without PMSF can be prepared and stored at 4°C for up to 1 year. 1 M PMSF in DMSO should be added to a final concentration of 1 mM PMSF immediately before use as PMSF is unstable in water.
Mammalian cell lysis buffer
- 100 mM Tris·Cl, pH 8.5 (checked with a pH meter)
- 6 M guanidine hydrochloride
- 5 mM TCEP
- 10 mM chloroacetamide
Buffer without TCEP and chloroacetamide can be prepared and stored up to 1 year at 4°C. TCEP and chloroacetamide should be added immediately before use. TCEP and chloroacetamide are preferred because they can be used for simultaneous reduction and alkylation without loss of cysteine alkylation efficiency.
COMMENTARY
Background Information
Proteome-derived peptide libraries are a powerful tool for profiling the sequence specificity of enzymatic and chemical labeling reagents (Bridge et al., 2023; Weeks & Wells, 2018). Proteome-derived peptide libraries provide a more comprehensive, cost-effective, and biologically relevant view of sequence specificity in comparison with synthetic peptide libraries. These libraries may be modified to block lysine side chains and peptide N termini for studies of protease specificity (Schilling et al., 2011) or can be generated with lysine side chains and peptide N termini left unblocked to study enzymes and reagents that target these sites (Bridge et al., 2023; Weeks & Wells, 2018). Proteome-derived peptide libraries first emerged as a tool for characterizing protease specificity (Schilling & Overall, 2008). More recently, they have been applied to characterize the specificity of the designed peptide ligase subtiligase (Weeks & Wells, 2018) and the N-terminally specific modification reagent 2PCA (Bridge et al., 2023). In principle, proteome-derived peptide libraries are well suited for characterizing the specificity of any reagent or enzyme that accepts peptides as substrates and can be generated from any proteomic sample.
We have recently applied proteome-derived peptide libraries to characterize the specificity of 2PCA, a chemical reagent that specifically modifies the ⍺-amine of protein and peptide N termini but not the ε-amine of lysine side chains (Bridge et al., 2023). The specificity of 2PCA had been previously evaluated using a small synthetic peptide library with the sequence XADSWAG, where X was varied to every amino acid (MacDonald et al., 2015). This small peptide library did not allow assessment of the effect of varying the second residue, which participates in the modification reaction, nor could pairwise combination of residues at different positions be evaluated. By profiling 2PCA specificity with proteome-derived peptide libraries, we were able to assess the effect of varying each amino acid at the first six positions of a potential 2PCA modification substrate as well as the effect of pairwise combinations of residues at different positions on 2PCA modification efficiency. Notably, we observed that although peptides with glycine in the first position are modified with low efficiency, the efficiency can be rescued when the amino acid at the second position is glycine, alanine, or lysine. This pairwise interaction is typical of what is often observed in enzymes with multiple subsites for substrate recognition but had been previously overlooked as a feature of 2PCA.
Here, we detail the preparation of peptide libraries with both peptide N termini and lysine side chains unblocked, which were required for deep profiling of 2PCA specificity. In previous studies, the most widely used proteome-derived peptide libraries have had both lysine side chains and peptide N termini blocked by N-terminal dimethylation (Schilling & Overall, 2007; Schilling et al., 2011). Our protocol omits this step, enabling specificity profiling of reagents that target these groups. We provide example protocols for profiling both chemical and enzymatic N-terminal modification reagents with or without enrichment of modified substrates. For broad-specificity reagents, enrichment of modified substrates may not be required, whereas for reagents that modify only a small percentage of the library, enrichment may be needed.
Beyond profiling the specificity of N-terminal modification reagents, the libraries described here have many other potential applications. They are suitable as substrates of other enzymes and chemical reagents that act on peptides, such as reagents that target specific amino acid side chains or enzymes that modify specific sites or motifs. Libraries generated here can also be further modified to enable enrichment-based characterization of other enzymes. For example, we used proteome-derived peptide libraries to optimize conditions for complete N-terminal modification while leaving lysine side chains unblocked. The N-terminally blocked libraries that we generated were then used to profile the specificity of several subtilisin/kexin-type proprotein convertases, a group of proteases that recognize lysine as part of their consensus cleavage motif (Bridge et al., 2023). Many other applications in profiling specificity of kinases, phosphatases, acetyltransferases, and other enzymes can also be envisioned.
Critical Parameters
Protease digestion
Successful generation of proteome-derived peptide libraries depends on complete digestion with the protease of choice. Protease digestion should occur for a minimum of 16 hr at the appropriate temperature and pH optimal for the protease of choice. The digest should be checked on an SDS-PAGE gel to confirm that no bands above 10 kDa are visible. When proteases other than those used here are used in library generation, it is important to use buffer conditions compatible with maximal activity of the protease. It is also critical to consider specificity biases in the library that may arise from properties of the protease. For example, trypsin does not accept proline in the position C terminal to the cleavage site, and proline will therefore not be well represented in the N-terminal position of libraries generated with trypsin.
Protein resolubilization pH
Resolubilization of the proteins will occur most efficiently in a pH range of 7-8 (Basic Protocol 1, steps 20-32). A pH outside this range will result in incomplete resolubilization of the proteins, leading to a decrease in overall peptide yield. Additionally, the solution containing resolubilized protein may have a low pH incompatible with protease digestion.
Selection of buffer conditions for N-terminal modification reagent specificity profiling
For the specificity profiling reactions described in Basic Protocols 2 and 3, it is important to buffer the solution to the desired pH and to use buffer conditions compatible with the reagent or enzyme of interest. 2PCA labeling should be performed at a pH between 7.5 and 8.5 for optimal activity.
Troubleshooting
Table 2 lists common problems encountered during Basic Protocol 1 and possible solutions. Table 3 lists common problems encountered during 2PCA specificity profiling and possible solutions. Table 4 lists common problems encountered during subtiligase specificity profiling and possible solutions.
| Problem | Possible causes | Solutions |
|---|---|---|
| Proteins do not dissolve after TCA precipitation | pH is too low | Check that the pH is 7.0-8.0. |
| Protein solution is too concentrated | Add buffer to achieve an estimated protein concentration of ≤2 mg/mL. | |
| Low number of peptides identified by LC-MS/MS | Incomplete digestion with protease | Check the digested proteins by SDS-PAGE gel to verify that no bands >10 kDa remain, or extend protease digestion time. |
| Inefficient de-salting | Ensure the pH of the peptide sample is below 3 before it is applied to C18 resin and that the resin is equilibrated properly. | |
| LC-MS/MS TIC shows evenly spaced repeats | Polymer contamination in the sample | Ensure all de-salting solutions are made using LC-MS-grade reagents. Use non-autoclaved plastics. |
| Problem | Possible causes | Solutions |
|---|---|---|
| Few or no peptides identified by LC-MS/MS | Inefficient de-salting | Ensure the pH of the peptide sample is <3 before it is applied to C18 resin and that the resin is equilibrated. properly. |
| Contamination with polymer or detergent | Use only non-autoclaved plastics. Do not store buffers in glassware washed with detergents. |
| Problem | Possible causes | Solutions |
|---|---|---|
| Low modification efficiency | pH of peptide library/reaction mixture is too low |
Ensure that the pH of the peptide library is close to neutral using pH paper. Use a high concentration of tricine in the reaction to maintain pH at 8 |
| No Abu-tagged peptides detected by LC-MS/MS | Many possible causes: |
TEV cleave and desalt a sample of the reaction mixture before enrichment to test whether peptides are modified. Check Proteome Discoverer search parameters to ensure the Abu mass modification is correct. |
| LC-MS/MS TIC shows evenly spaced repeats | Polymer contamination in the sample | Ensure that all de-salting solutions are made using LC-MS-grade reagents. Use non-autoclaved plastics. |
Understanding Results
Anticipated results from Basic Protocol 1 are shown in Figure 2. For a peptide library generated from E. coli cells by trypsin digestion, ∼20,000 peptides can be identified by LC-MS/MS. E. coli libraries generated by digestion with chymotrypsin or GluC typically have 5000-10,000 peptides identified by LC-MS/MS. Example data that can be obtained from performing the Alternate Protocol is shown in Figure 3. Libraries generated from HEK293T cells and trypsin typically have >20,000 peptides identified by LC-MS/MS. Digestion with GluC or chymotrypsin will result in 4000-8000 peptides. Peptide library sequences can be analyzed by generating a sequence logo. Peptides should have a conserved C-terminal sequence corresponding to the protease used and a diverse sequence at the N-terminal residues.
Anticipated results from Basic Protocol 2 are shown in Figure 4. Labeling a tryptic peptide library with 2PCA should result in >15,000 total peptides identified by LC-MS/MS. The fraction of peptides with 2PCA at the N terminus will vary depending on the 2PCA concentration used. Peptides labeled with 2PCA are expected to exhibit a reduced preference for peptides that contain glycine at the P1 position. No peptides with proline at the P2 position should be labeled.
Anticipated results for Basic Protocol 3 are shown in Figure 5. Thousands of peptides are expected to be identified, and >80% of peptides are expected to be modified at their N-termini with aminobutyric acid (Abu).
Time Considerations
Basic Protocol 1 and the Alternate Protocol will take 3-5 days once the cell cultures are ready to harvest. The protocol up to the digestion step can be performed in 1 day if the TCA precipitation is carried out for 2 hr. On the second day, the proteome-derived peptide libraries can be desalted and concentrated. On the third day, the peptide libraries can be analyzed by LC-MS/MS. The protocol may take two additional days if pause points are utilized.
Basic Protocol 2 requires 1-2 days, depending on the desired length of 2PCA labeling.
Basic Protocol 3 requires 1-2 days, depending on the length of the incubation with TEV protease. Note that the preparation of subtiligase and TEV ester 6 required 2-3 days each and are described in another article (Weeks & Wells, 2020).
For all Basic Protocols, individual LC-MS/MS runs take 90 min each.
Acknowledgments
Research in the Weeks lab is supported by an NIH Director's New Innovator Award (1DP2GM149548-01), a David and Lucille Packard Fellowship for Science and Engineering, and a Career Award at the Scientific Interface from the Burroughs Wellcome Fund (1017065; to A.M.W.). H.N.B is supported in part by a William R. and Dorothy E. Sullivan Wisconsin Distinguished Graduate Fellowship.
Author Contributions
Haley N. Bridge : Data curation, formal analysis, writing—original draft, writing—review and editing; Amy M. Weeks : Conceptualization, data curation, formal analysis, funding acquisition, methodology, project administration, writing—original draft, writing—review and editing.
Conflict of Interest
The authors declare no conflict of interest.
Open Research
Data Availability Statement
No new data were generated in the preparation of this manuscript. Figures showing anticipated results were generated from data previously published in Bridge, Frazier, and Weeks (2023) and Weeks and Wells (2018).
Literature Cited
- Agard, N. J., Mahrus, S., Trinidad, J. C., Lynn, A., Burlingame, A. L., & Wells, J. A. (2012). Global kinetic analysis of proteolysis via quantitative targeted proteomics. Proceedings of the National Academy of Sciences , 109(6), 1913–1918. https://doi.org/10.1073/pnas.1117158109
- Bridge, H. N., Frazier, C. L., & Weeks, A. M. (2023). An expanded 2-pyridinecarboxaldehyde (2PCA)-based chemoproteomics toolbox for probing protease specificity. BioRxiv , https://doi.org/10.1101/2023.02.12.528234
- Griswold, A. R., Cifani, P., Rao, S. D., Axelrod, A. J., Miele, M. M., Hendrickson, R. C., Kentsis, A., & Bachovchin, D. A. (2019). A chemical strategy for protease substrate profiling. Cell Chemical Biology , 26(6), 901–907. e6. https://doi.org/10.1016/j.chembiol.2019.03.007
- auf dem Keller, U., & Schilling, O. (2010). Proteomic techniques and activity-based probes for the system-wide study of proteolysis. Biochimie , 92(11), 1705–1714. https://doi.org/10.1016/j.biochi.2010.04.027
- Luo, S. Y., Araya, L. E., & Julien, O. (2019). Protease substrate identification using N-terminomics. ACS Chemical Biology , 14(11), 2361–2371. https://doi.org/10.1021/acschembio.9b00398
- MacDonald, J. I., Munch, H. K., Moore, T., & Francis, M. B. (2015). One-step site-specific modification of native proteins with 2-pyridinecarboxyaldehydes. Nature Chemical Biology , 11(5), 326–331. https://doi.org/10.1038/nchembio.1792
- Mahrus, S., Trinidad, J. C., Barkan, D. T., Sali, A., Burlingame, A. L., & Wells, J. A. (2008). Global sequencing of proteolytic cleavage sites in apoptosis by specific labeling of protein N termini. Cell , 134(5), 866–876. https://doi.org/10.1016/j.cell.2008.08.012
- Rosen, C. B., & Francis, M. B. (2017). Targeting the N terminus for site-selective protein modification. Nature Chemical Biology , 13(7), 697–705. https://doi.org/10.1038/nchembio.2416
- Schechter, I., & Berger, A. (1967). On the size of the active site in proteases. I. Papain. Biochemical and Biophysical Research Communications , 27(2), 157–162. https://doi.org/10.1016/s0006-291x(67)80055-x
- Schilling, O., Huesgen, P. F., Barré, O., auf dem Keller, U., & Overall, C. M. (2011). Characterization of the prime and non-prime active site specificities of proteases by proteome-derived peptide libraries and tandem mass spectrometry. Nature Protocols , 6(1), 111–120. https://doi.org/10.1038/nprot.2010.178
- Schilling, O., & Overall, C. M. (2007). Proteomic discovery of protease substrates. Current Opinion in Chemical Biology , 11(1), 36–45. https://doi.org/10.1016/j.cbpa.2006.11.037
- Schilling, O., & Overall, C. M. (2008). Proteome-derived, database-searchable peptide libraries for identifying protease cleavage sites. Nature Biotechnology , 26(6), 685–694. https://doi.org/10.1038/nbt1408
- Shimbo, K., Hsu, G. W., Nguyen, H., Mahrus, S., Trinidad, J. C., Burlingame, A. L., & Wells, J. A. (2012). Quantitative profiling of caspase-cleaved substrates reveals different drug-induced and cell-type patterns in apoptosis. Proceedings of the National Academy of Sciences , 109(31), 12432–12437. https://doi.org/10.1073/pnas.1208616109
- Swatek, K. N., & Komander, D. (2016). Ubiquitin modifications. Cell Research , 26(4), 399–422. https://doi.org/10.1038/cr.2016.39
- Swee, L. K., Lourido, S., Bell, G. W., Ingram, J. R., & Ploegh, H. L. (2015). One-step enzymatic modification of the cell surface redirects cellular cytotoxicity and parasite tropism. ACS Chemical Biology , 10(2), 460–465. https://doi.org/10.1021/cb500462t
- Weeks, A. M., & Wells, J. A. (2018). Engineering peptide ligase specificity by proteomic identification of ligation sites. Nature Chemical Biology , 14(1), 50–57. https://doi.org/10.1038/nchembio.2521
- Weeks, A. M., & Wells, J. A. (2019). Subtiligase-catalyzed peptide ligation. Chemical Reviews , 120(6), 3127–3160. https://doi.org/10.1021/acs.chemrev.9b00372
- Weeks, A. M., & Wells, J. A. (2020). N-terminal modification of proteins with subtiligase specificity variants. Current Protocols in Chemical Biology , 12(1), e79. https://doi.org/10.1002/cpch.79
- Yi, C. H., Pan, H., Seebacher, J., Jang, I.-H., Hyberts, S. G., Heffron, G. J., Vander Heiden, M. G., Yang, R., Li, F., Locasale, J. W., Sharfi, H., Zhai, B., Rodriguez-Mias, R., Luithardt, H., Cantley, L. C., Daley, G. Q., Asara, J. M., Gygi, S. P., Wagner, G., … Yuan, J. (2011). Metabolic regulation of protein N-alpha-acetylation by Bcl-xL promotes cell survival. Cell , 146(4), 607–620. https://doi.org/10.1016/j.cell.2011.06.050

