Programmable Gene Knockdown in Diverse Bacteria Using Mobile-CRISPRi
Amy B. Banta, Amy B. Banta, Ryan D. Ward, Ryan D. Ward, Jennifer S. Tran, Jennifer S. Tran, Emily E. Bacon, Emily E. Bacon, Jason M. Peters, Jason M. Peters
Bacillus subtilis
biofuels
conjugation
CRISPR-Cas9
CRISPRi
Escherichia coli
ESKAPE pathogens
functional genomics
Listeria monocytogenes
Zymomonas mobilis
Abstract
Facile bacterial genome sequencing has unlocked a veritable treasure trove of novel genes awaiting functional exploration. To make the most of this opportunity requires powerful genetic tools that can target all genes in diverse bacteria. CRISPR interference (CRISPRi) is a programmable gene-knockdown tool that uses an RNA-protein complex comprised of a single guide RNA (sgRNA) and a catalytically inactive Cas9 nuclease (dCas9) to sterically block transcription of target genes. We previously developed a suite of modular CRISPRi systems that transfer by conjugation and integrate into the genomes of diverse bacteria, which we call Mobile-CRISPRi. Here, we provide detailed protocols for the modification and transfer of Mobile-CRISPRi vectors for the purpose of knocking down target genes in bacteria of interest. We further discuss strategies for optimizing Mobile-CRISPRi knockdown, transfer, and integration. We cover the following basic protocols: sgRNA design, cloning new sgRNA spacers into Mobile-CRISPRi vectors, Tn 7 transfer of Mobile-CRISPRi to Gram-negative bacteria, and ICE Bs1 transfer of Mobile-CRISPRi to Bacillales. © 2020 The Authors.
Basic Protocol 1 : sgRNA design
Basic Protocol 2 : Cloning of new sgRNA spacers into Mobile-CRISPRi vectors
Basic Protocol 3 : Tn 7 transfer of Mobile-CRISPRi to Gram-negative bacteria
Basic Protocol 4 : ICE Bs1 transfer of Mobile-CRISPRi to Bacillales
Support Protocol 1 : Quantification of CRISPRi repression using fluorescent reporters
Support Protocol 2 : Testing for gene essentiality using CRISPRi spot assays on plates
Support Protocol 3 : Transformation of E. coli by electroporation
Support Protocol 4 : Transformation of CaCl2-competent E. coli
INTRODUCTION
CRISPRi (Fig. 1A) is a contemporary gene-perturbation strategy with substantial advantages over classic genetic approaches (e.g., gene deletions, transposon mutagenesis). First, CRISPRi is programmable (Qi et al., 2013): to specify a gene for knockdown, one need only alter the first 20 nucleotides (nt) of the sgRNA (known as the “spacer”) to match the target gene (provided that there is a nearby protospacer-adjacent motif, or PAM; see Strategic Planning). This programmability makes it straightforward to construct single-gene knockdowns or knockdown libraries targeting defined sets of genes (Liu et al., 2017; Peters et al., 2016) or all genes in the genome (Lee et al., 2019; Rousset et al., 2018; Wang et al., 2018; Yao et al., 2020). Because the sgRNA spacer uniquely identifies the target gene, spacers can act as barcodes to obtain counts of individual knockdown strains and measure their fitness in pooled growth experiments. Second, CRISPRi is inducible and titratable, and therefore can be used to target essential genes (Li et al., 2016; Peters et al., 2016). The timing and extent of CRISPRi knockdown can be controlled by expressing the sgRNA and dcas9 genes from inducible promoters. Further, knockdown gradients can be achieved by expressing CRISPRi components from constitutive promoters of differing strengths (Qu et al., 2019), by using truncated spacers (Vigouroux, Oldewurtel, Cui, Bikard, & Teeffelen, 2018), or by systematically mutating the sgRNA to introduce mismatches between the spacer and target gene that reduce knockdown efficacy to a predictable extent (Hawkins et al., 2020). Finally, CRISPRi can be multiplexed to target several genes in the same cell by cloning arrays of sgRNAs with different spacers (Ellis, Kim, & Machner, 2020; Peters et al., 2016; Reis et al., 2019). These advantages suggest that CRISPRi will become a common tool for bacterial functional genomics in the near future.
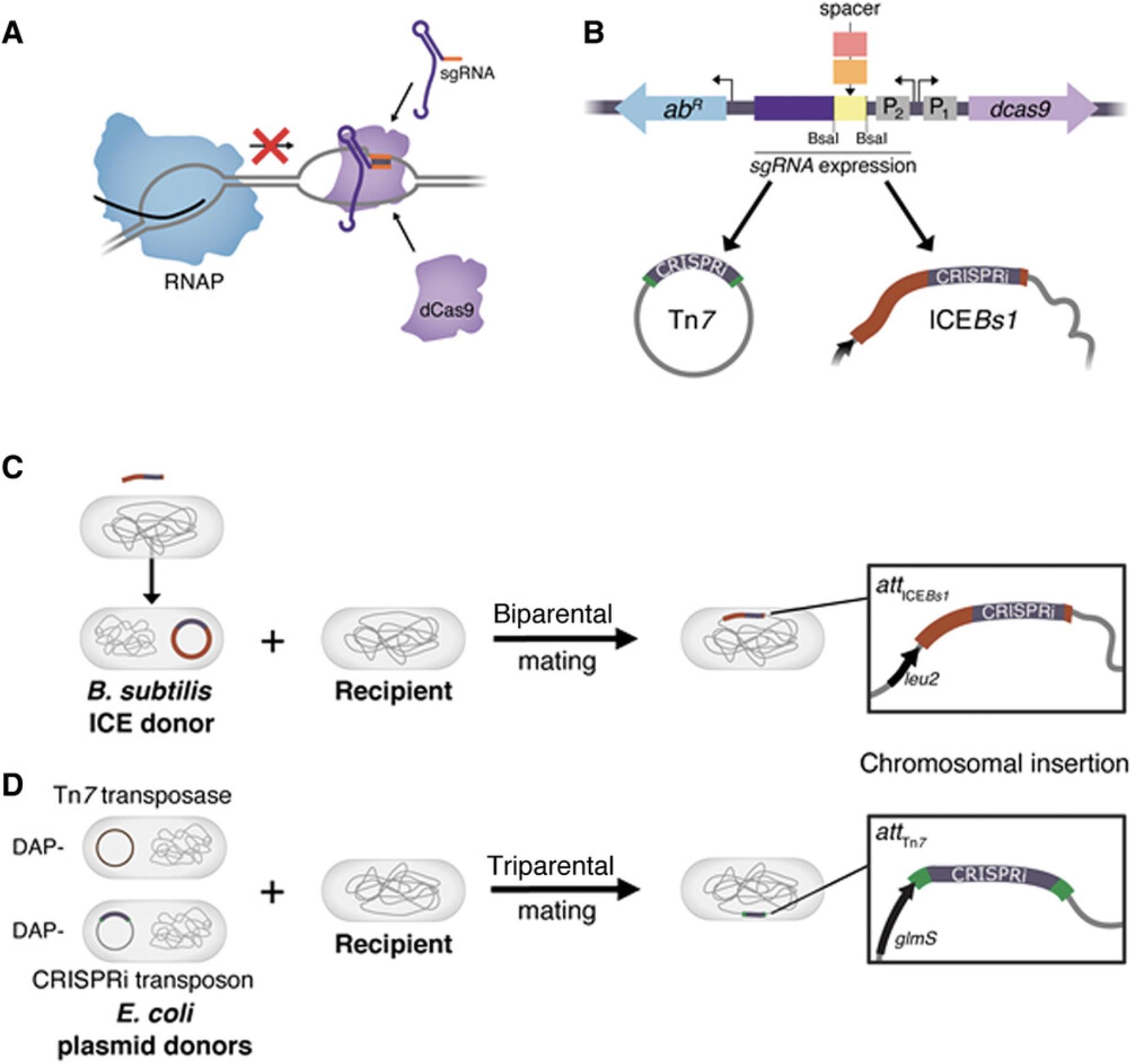
Despite the utility of CRISPRi, its widespread adoption in bacteria has been limited by a lack of generalizable systems. To facilitate the use of CRISPRi in diverse bacteria, we developed Mobile-CRISPRi—a suite of modular vectors that transfer by conjugation and stably integrate into the genomes of recipient bacteria (Peters et al., 2019). All Mobile-CRISPRi vectors contain (1) dcas9 , (2) an sgRNA with either a targeting spacer or restriction sites for cloning new spacers, and (3) an antibiotic resistance marker for selection in recipient bacteria (Fig. 1B). The modularity of Mobile-CRISPRi enables facile cloning of sgRNA libraries. There are two general sets of Mobile-CRISPRi vectors: ICE Bs1 -based vectors for Gram-positive bacteria related to Bacillus subtilis (i.e., order Bacillales) that integrate downstream of the leu2 tRNA (Fig. 1C), and Tn 7 -based vectors for Gram-negative bacteria that integrate downstream of the glmS gene (Fig. 1D). Tn 7 Mobile-CRISPRi vectors transfer from a diaminopimelic acid (DAP)-dependent donor strain of Escherichia coli to recipient strains of interest via the RP4 conjugation machinery and require a helper plasmid for integration (also transferred from E. coli) that expresses the Tn 7 transposition genes (tnsABCD ; triparental mating). ICE Bs1 Mobile-CRISPRi vectors are first integrated into the genome copy of ICE Bs1 in a B. subtilis donor strain using natural competence transformation and are then transferred to recipient strains of interest through induction of the ICE Bs1 conjugation machinery (biparental mating). Integration of Mobile-CRISPRi downstream of glmS or leu2 does not impart a fitness defect in the strains tested (although Tn 7 insertion might not be neutral in all species, and expression of dcas9 may be toxic in some bacteria; see Commentary).
Mobile-CRISPRi transfer, integration, and knockdown efficiencies vary by recipient strain for reasons that are sometimes but not always understood (Banta, Enright, Siletti, & Peters, 2020; Peters et al., 2019; Qu et al., 2019). To rapidly assess Mobile-CRISPRi knockdown efficacy, we developed “test” vectors for both the Tn 7 and ICE Bs1 systems that contain either an mRFP or an sfGFP gene (encoding monomeric red fluorescent protein or superfolder green fluorescent protein, respectively) and an sgRNA targeting mRFP or sfGFP that allows knockdown to be easily measured in live cells using a fluorometer or flow cytometer. The modular nature of Mobile-CRISPRi enables straightforward knockdown optimization approaches, such as replacing the sgRNA and dcas9 promoters or dcas9 ribosome-binding site with native regulatory sequences from the recipient bacterium (see Commentary). Thus far, we have demonstrated Mobile-CRISPRi transfer/integration and knockdown in several Gram-negative (γ-Proteobacteria: E. coli , Enterobacter cloacae , Enterobacter aerogenes , Pseudomonas aeruginosa , Klebsiella pneumoniae , Vibrio casei , Acinetobacter baumannii , Salmonella enterica , and Proteus mirabilis [Peters et al., 2019]; α-Proteobacteria: Zymomonas mobilis [Banta et al., 2020]) and Gram-positive species (Firmicutes: B. subtilis , Listeria monocytogenes , Staphylococcus aureus , and Enterococcus faecalis [Peters et al., 2019]).
Here, we describe the following basic protocols: (1) sgRNA design for selecting and computationally optimizing guide spacers (Basic Protocol 1), (2) cloning of new sgRNA spacers into Mobile-CRISPRi vectors to target specific genes for knockdown (Basic Protocol 2), (3) Tn 7 transfer of Mobile-CRISPRi to Gram-negative bacteria via conjugation with E. coli donors (Basic Protocol 3), and (4) ICE Bs1 transfer of Mobile-CRISPRi to Bacillales via conjugation with a B. subtilis donor (Basic Protocol 4). We also provide the following support protocols: (1) quantification of CRISPRi repression using fluorescent reporters that measure knockdown in live cells (Support Protocol 1), (2) testing for gene essentiality using CRISPRi spot assays on plates to examine the effects of CRISPRi gene knockdown on plating efficiency (Support Protocol 2), (3) transformation of E. coli by electroporation (Support Protocol 3), and (4) transformation of CaCl2-competent E. coli to facilitate cloning sgRNA spacers and create donor strains for Mobile-CRISPRi mating (Support Protocol 4).
Biosafety caution
CAUTION : Follow all biosafety requirements relevant to the microorganism under investigation. See Burnett et al. (2009) for more information.
STRATEGIC PLANNING
Before beginning CRISPRi experiments in a new strain, one must first design sgRNA spacers against target genes (see Basic Protocol 1). In general, desirable sgRNAs are on target (have only one binding site in the genome) and have high efficacy (although lower-efficacy guides may be useful for targeting essential genes; Hawkins et al., 2020; Vigouroux et al., 2018). To design on-target guides, we use a strategy developed by the Weissman and Gross labs that takes into account the DNA-binding preferences of the dCas9-sgRNA complex to score guide specificity (Gilbert et al., 2014; Peters et al., 2019). For instance, noncomplementarity between the sgRNA and a target site proximal to the PAM (the sgRNA “seed” region) has a strong negative effect on binding, whereas PAM-distal mismatches (e.g., 15-20 nt away from the PAM) have less impact on binding and may result in off-target effects. To identify and eliminate off-target guides, we strongly recommend designing sgRNA spacers for the entire genome at once (see Basic Protocol 1). If no closed genome sequence exists for the strain of interest, it is not possible to predict off-target sgRNA activity; therefore, gene-knockdown phenotypes must be confirmed by multiple guides. Because bacterial genomes are relatively small, most sgRNA spacers have high specificity (i.e., a maximum specificity score of 39 in our code below). sgRNA knockdown efficacy is complex and the subject of ongoing research (Calvo-Villamañán et al., 2020), although some general rules have emerged. The clearest rule is that guides targeting the nontemplate strand of the gene (antisense, or “anti” in our code below) have much higher efficacy than template-targeting guides (Bikard et al., 2013; Qi et al., 2013); the mechanistic underpinnings of this effect are unknown. Early observations suggested that sgRNAs targeting the 5′ ends of genes are more efficacious, but subsequent studies have shown that guide efficacy is constant across genes on average (Cui et al., 2018; Rousset et al., 2018). Nonetheless, we tend to prefer spacers that target toward the 5′ ends of genes because the effects of the CRISPRi transcription block on translation are unclear. Targeting promoters with CRISPRi can also be effective, but we prefer to target genes because of the lack of promoter location data in the majority of bacteria and the fact that spacer distribution is more limited in intergenic regions (i.e., there are fewer NGG PAM sequences in AT-rich promoter regions).
Basic Protocol 1: sgRNA DESIGN
This protocol describes how to design an exhaustive list of 20-nucleotide sgRNA spacers using a command-line interpreter. The process is streamlined and should be modified by changing the environment variable “ACC_NO” to specify a chromosomal accession number. Steps 1 and 2 are only required in the first setup and should be omitted afterwards.
Materials
- Linux- or Unix-compatible computer with Internet access and the following software:
- Conda (https://docs.conda.io/projects/conda/en/latest/)
- GitHub (https://github.com/)
- Excel or other spreadsheet management software
Download dependencies for sgRNA design script
1.Prepare a conda environment, “sgrna_design”, to host the sgRNA design scripts. It is only necessary to perform environment creation once.
- conda create -n sgrna_design -c bioconda
bowtie=1.2.3' biopython pysam entrez-direct gitpython>3'
2.Retrieve the “sgrna_design” project from GitHub, then move into the newly created directory. It is only necessary to perform project download once.
- git clone https://github.com/ryandward/sgrna_design.git && cd sgrna_design
Activate Conda environment
3.Activate the environment to load script dependencies This is required every time a new terminal window is opened.
- conda activate sgrna_design
Obtain genome sequence data from NCBI using the accession number
4.If not known, consult the NCBI Nucleotide Database (see Internet Resources) to locate the accession number corresponding to the chromosome of interest, and adjust the environment variable “ACC_NO” accordingly. This example uses the E. coli MG1655 chromosome “U00096.3”; this number will serve as the template for the names of all other files.
- ACC_NO="U00096.3"
5.Retrieve and save the GenBank chromosome file (.gb file)—here automatically named U00096.3.gb. This command also issues a warning if NCBI returns an empty response and may be run multiple times as needed.
- efetch -db nuccore -format gb -id
{ACC_NO}.gb && file ${ACC_NO}.gb | grep -iq ascii && echo "File contains data, continue." || echo "Empty file, retry this step."
Run sgRNA design script
6.Run the main python script, producing a tab-separated variable file corresponding to the accession number followed by "_sgrna". Upon successful completion, this command also confirms the name of the results file—here "U00096.3_sgrna.tsv".
- ./build_sgrna_library.py --input_genbank_genome_name
{ACC_NO}_sgrna.tsv && echo "Output saved as ${ACC_NO}_sgrna.tsv"
These results can be accessed by navigating to the present working directory and viewed using a spreadsheet utility, text editor, or command-line interpreter tool (e.g., cat).
Basic Protocol 2: CLONING NEW sgRNA SPACERS INTO MOBILE-CRISPRi VECTORS
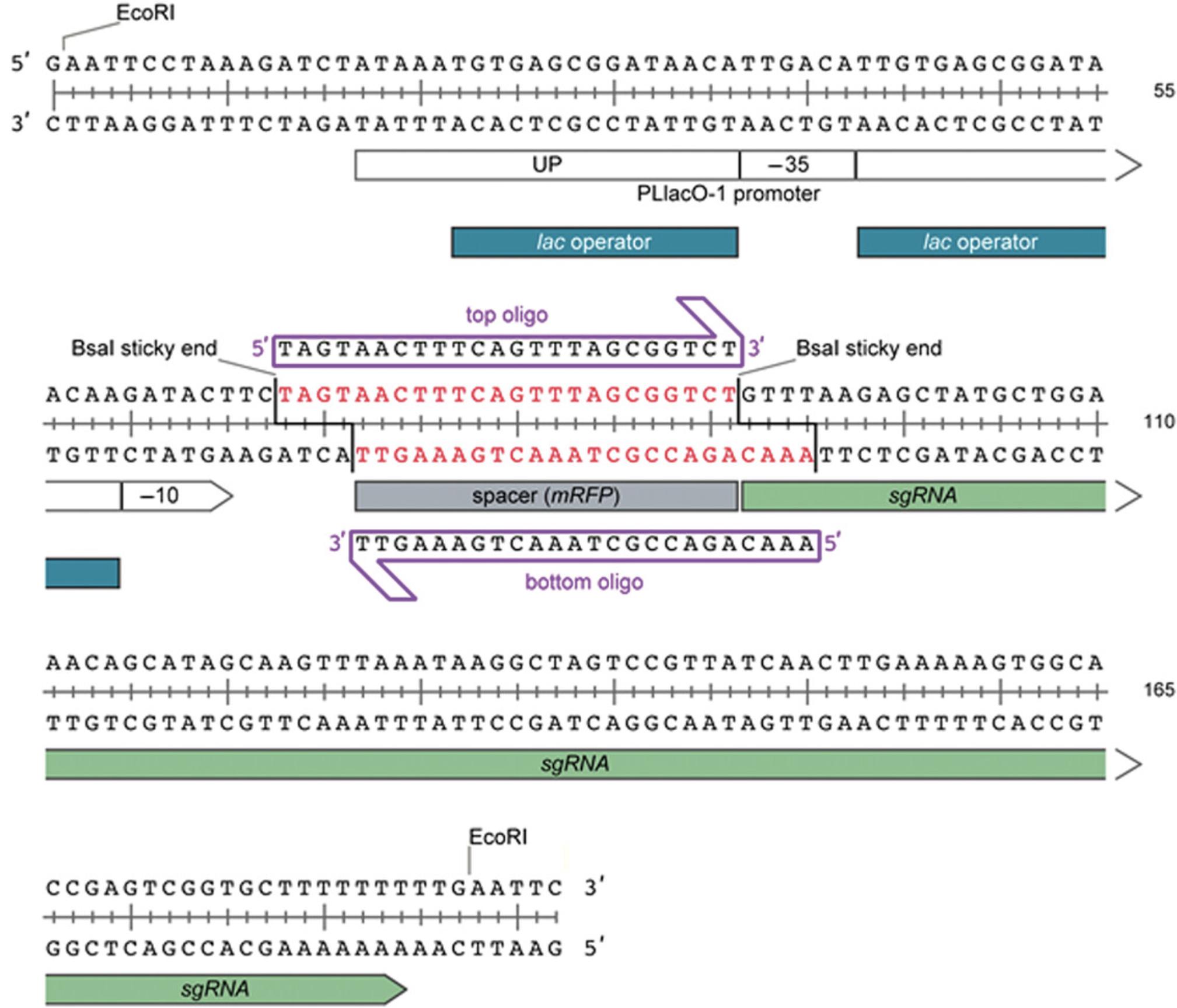
This protocol describes the construction of Mobile-CRISPRi vectors encoding an sgRNA spacer targeting a gene of interest. Two oligonucleotides are designed such that when annealed, they form the desired sgRNA spacer sequence with overhangs enabling ligation into a BsaI-digested Mobile-CRISPRi plasmid (Fig. 2). The resulting plasmid, which contains the entire Mobile-CRISPRi system encoding the sgRNA, dCas9, and antibiotic resistance marker between Tn 7 transposon ends, can be used as a donor for transposition of the system into the recipient att Tn7 site.
Materials
- Mobile-CRISPRi plasmid DNA (see Table 1)
- BsaI-HF-v2 restriction enzyme and 10× CutSmart buffer (NEB R3733)
- DNA spin-purification kit (e.g., NEB T1030 or Zymo Research D4003)
- Custom oligonucleotides (Top and Bottom, each 100 µM; see Basic Protocol 1, Strategic Planning, and Fig. 2)
- T4 DNA ligase with 10× buffer (e.g., NEB M0202)
- 1 mM ATP (e.g., Thermo R0441, diluted 100×)
- 100 mM dithiothreitol (DTT; see recipe)
- Electrocompetent E. coli cells (pir + strain, BW25141; Support Protocol 3)
- LB agar with 100 µg/ml ampicillin agar plates
- Liquid LB medium with 100 µg/ml ampicillin
- Plasmid DNA miniprep kit (e.g., Thermo GeneJet kit K0502)
| JMP no. | Addgene no. | Rep. | Antibiotic res.a | sgRNA prom. | sgRNA | dCas9 prom. | dCas9 | Comment |
|---|---|---|---|---|---|---|---|---|
| ICEBs1 system | ||||||||
| pJMP1333 | 119268 | mRFP | ampR, kanR | Pveg | mRFP (NT1) | Pxyl/tet | Spy::3Xmyc | mRFP “test” (– sgRNA) |
| pJMP1335 | 119269 | mRFP | ampR, kanR | None | None | Pxyl/tet | Spy::3Xmyc | mRFP “test” (+ sgRNA) |
| pJMP1337 | 119270 | None | ampR, kanR | Pveg | None (BsaI) | Pxyl/tet | Spy::3Xmyc | For cloning new sgRNAs |
| Tn7 system | ||||||||
| pJMP1039 | 119239 | NA | ampR, N/A | N/A | N/A | N/A | N/A | Expresses Tn7 transposase |
| pJMP1183 | 119254 | mRFP | ampR, kanR | PLlacO1 | mRFP (NT1) | PLlacO1 | Spy::3Xmyc | mRFP “test” (+ sgRNA) |
| pJMP1185 | 119255 | mRFP | ampR, kanR | None | None | PLlacO1 | Spy::3Xmyc | mRFP “test” (– sgRNA) |
| pJMP1187 | 119256 | mRFP | ampR, kanR | PLlacO1 | mRFP (NT1) | PLlacO1 | HSA Spy::3Xmyc | mRFP “test” (+ sgRNA) |
| pJMP1189 | 119257 | mRFP | ampR, kanR | None | None | PLlacO1 | HSA Spy::3Xmyc | mRFP “test” (– sgRNA) |
| pJMP1339 | 119271 | None | ampR, kanR | PLlacO1 | None (BsaI) | PLlacO1 | HSA Spy::3Xmyc | For cloning new sgRNAs |
| pJMP1354 | 119275 | None | ampR, tmpR | PLlacO1 | None (BsaI) | PLlacO1 | HSA Spy::3Xmyc | For cloning new sgRNAs |
| pJMP1356 | 119276 | None | ampR, cmR | PLlacO1 | None (BsaI) | PLlacO1 | HSA Spy::3Xmyc | For cloning new sgRNAs |
| pJMP1358 | 119277 | None | ampR, spcR | PLlacO1 | None (BsaI) | PLlacO1 | HSA Spy::3Xmyc | For cloning new sgRNAs |
| pJMP1360 | 119278 | None | ampR, gentR | PLlacO1 | None (BsaI) | PLlacO1 | HSA Spy::3Xmyc | For cloning new sgRNAs |
| pJMP2754 | 160666 | sfGFP | ampR, gentR | PLlacO1 | None (BsaI) | PLlacO1 | Spy::3Xmyc | sfGFP “test” (– sgRNA) |
| pJMP2774 | 160667 | sfGFP | ampR, gentR | PLlacO1 | Gmc6 (sfgfp) | PLlacO1 | Spy::3Xmyc | sfGFP “test” (+ sgRNA) |
| pJMP2782 | 160668 | None | ampR, gentR | PLlacO1 | None (BsaI) | PLlacO1 | Spy::3Xmyc | For cloning new sgRNAs |
| pJMP2820 | 160669 | sfGFP | ampR, cmR | PLlacO1 | None (BsaI) | PLlacO1 | Spy::3Xmyc | sfGFP “test” (– sgRNA) |
| pJMP2822 | 160670 | sfGFP | ampR, kanR | PLlacO1 | None (BsaI) | PLlacO1 | Spy::3Xmyc | sfGFP “test” (– sgRNA) |
| pJMP2824 | 160671 | sfGFP | ampR, spcR | PLlacO1 | None (BsaI) | PLlacO1 | Spy::3Xmyc | sfGFP “test” (– sgRNA) |
| pJMP2832 | 160672 | sfGFP | ampR, cmR | PLlacO1 | Gmc6 (sfgfp) | PLlacO1 | Spy::3Xmyc | sfGFP “test” (+ sgRNA) |
| pJMP2834 | 160673 | sfGFP | ampR, kanR | PLlacO1 | Gmc6 (sfgfp) | PLlacO1 | Spy::3Xmyc | sfGFP “test” (+ sgRNA) |
| pJMP2836 | 160674 | sfGFP | ampR, spcR | PLlacO1 | Gmc6 (sfgfp) | PLlacO1 | Spy::3Xmyc | sfGFP “test” (+ sgRNA) |
| pJMP2844 | 160675 | None | ampR, cmR | PLlacO1 | None (BsaI) | PLlacO1 | Spy::3Xmyc | For cloning new sgRNAs |
| pJMP2846 | 160676 | None | ampR, kanR | PLlacO1 | None (BsaI) | PLlacO1 | Spy::3Xmyc | For cloning new sgRNAs |
| pJMP2849 | 160677 | None | ampR, spcR | PLlacO1 | None (BsaI) | PLlacO1 | Spy::3Xmyc | For cloning new sgRNAs |
- a
Resistance(s) of recipient E. coli. ampR, ampicillin resistance; cmR, chloramphenicol resistance; gentR, gentamycin resistance; kanR, kanamycin resistance; spcR, spectinomycin resistance; tmpR, trimethoprim resistance.
-
Rep., reporter; prom., promoter. All plasmids have the R6K ori (replicate only in pir+ strains); gmc6 encodes sfGFP sgRNA (CATCTAATTCAACAAGAATT); mRFP (NT1) encodes mRFP sgRNA (AACTTTCAGTTTAGCGGTCT); HSA Spy::3Xmyc dCas9 is human codon optimized, “None (BsaI)” sgRNA has a BsaI cloning site for insertion of new sgRNA encoding DNA; PLlacO1 promoters are regulated by lacIq; and Pxyl/tet promoter is regulated by TetR.
Prepare BsaI-digested Mobile-CRISPRi plasmid
1.Digest 1 µg plasmid DNA by setting up the following reaction mixture:
| Component | Volume |
|---|---|
| 10× CutSmart buffer (NEB) | 5 µl |
| Plasmid DNA (100 ng/µl) | 10 µl |
| BsaI-HF-v2 (NEB) | 1 µl |
| Deionized H2O | 34 µl |
| Total | 50 µl. |
2.Incubate 2-8 hr at 37°C to digest.
3.Incubate 20 min at 80°C to heat inactivate.
4.Purify the digested plasmid using a DNA spin-purification kit according to the manufacturer's protocol. Elute in 10 µl.
Anneal oligonucleotides
5.Set up each oligonucleotide annealing reaction:
| Component | Volume |
|---|---|
| 10× CutSmart Buffer (NEB) | 5 µl |
| Oligonucleotide-Top (100 µM) | 1 µl |
| Oligonucleotide-Bottom (100 µM) | 1 µl |
| dH2O | 43 µl |
| Total | 50 µl. |
6.Incubate 5 min at 95°C to denature DNA.
7.Cool ∼15 min to room temperature to anneal oligonucleotides.
8.Dilute 1:40 in deionized water.
Ligate BsaI-digested plasmid and annealed oligonucleotides
9.Set up each ligation:
| Component | Volume |
|---|---|
| 10× T4 DNA ligase buffer | 1 µl |
| 1 mM ATP | 1 µl |
| 100 mM DTT | 1 µl |
| T4 DNA ligase | 0.5 µl |
| 1:40 diluted annealed oligonucleotides (step 8) | 2 µl |
| 50 ng BsaI-digested plasmid (step 4) | X µl |
| Deionized water | X µl |
| Total | 10 µl. |
10.Incubate 1-2 hr at room temperature to ligate.
11.Incubate 15 min at 65°C to heat inactivate.
Transformation
12.Follow Support Protocol 3 to transform 1 µl of ligation mix into E. coli BW25141 and select on LB-ampicillin agar plates.
Extract plasmid DNA and confirm sequence
13.Pick an isolated colony into 5 ml LB + ampicillin medium in a culture tube and incubate cultures ∼16 hr at 37°C, shaking at 250 rpm or on a roller drum.
14.Pellet cells from entire 5-ml culture and extract plasmid DNA using a kit following the manufacturer's instructions.
15.Sequence the region of the plasmid near the insertion site to confirm the insertion and the fidelity of the cloning.
Basic Protocol 3: Tn7 TRANSFER OF MOBILE-CRISPRi TO GRAM-NEGATIVE BACTERIA
This protocol transfers the Tn 7 -based Mobile-CRISPRi system into the chromosome of a Gram-negative bacterium of interest. E. coli donor strains have a chromosomal copy of the RP4 transfer machinery to mobilize the Tn 7 plasmids. A plasmid with a Tn 7 transposon carrying CRISPRi components and a second plasmid encoding Tn 7 transposition genes are transferred to recipient bacteria by triparental mating. In the recipient, transposition proteins integrate the CRISPRi system into the recipient genome downstream of the glmS gene. Selection on plates lacking DAP eliminates the DAP-dependent E. coli donors, whereas R6K ori plasmids are lost because they cannot replicate in recipient cells that lack the pir gene. Antibiotic selection results in retention of only the recipients with an integrated CRISPRi system. Once integrated into the chromosome, the Mobile-CRISPRi system is stable without further antibiotic selection in all organisms tested so far. This method has been used with a variety of recipient γ-Proteobacteria, including Acinetobacter , Enterobacter , Escherichia , Klebsiella , Proteus , Pseudomonas , Salmonella , Shewanella , and Vibrio , as well as the α-Proteobacterium Zymomonas , and is likely to be adaptable to a wider range of bacteria.
Materials
-
Plasmid DNA: Mobile-CRISPRi-encoding plasmids (Tn 7 version; Table 2)
-
LB medium, with additions of diaminopimelic acid (DAP) and/or ampicillin, as appropriate
-
LB agar plates, with appropriate additions, prewarmed to 37°C
-
Culture medium and agar plates suitable for growth of the recipient bacterium (with addition of antibiotics, as appropriate, depending on Mobile-CRISPRi-encoded resistance)
-
1× phosphate-buffered saline (PBS; e.g., Fisher BP399500, diluted 1:10)
-
Cellulose filters (e.g., MF-Millipore HAWG01300)
-
Tweezers, sterilized in ethanol
| Strain | JMP no. | Genotype | Selection | Comment | Ref. |
|---|---|---|---|---|---|
| E. coli BW25141 | sJMP146 | ∆(araD-araB)567, ∆lacZ4787(::rrnB-3), ∆(phoB-phoR)580, λ-, galU95, ∆uidA3::pir+, recA1, endA9(Δins)::FRT, rph-1, ∆(rhaD-rhaB)568, hsdR514 | LB |
E. coli pir+ “cloning strain” |
Datsenko & Wanner (2000) |
| E.coli WM6026 | sJMP424 | lacIq, rrnB3, DElacZ4787, hsdR514, DE(araBAD)567, DE(rhaBAD)568, rph-1 att-lambda::pAE12-del (oriR6K/cat::frt5), Δ4229(dapA)::frt(DAP–), Δ(endA)::frt, uidA(ΔMluI)::pir(wt), attHK::pJK1006::Δ1/2(ΔoriR6K-cat::frt5, ΔtrfA::frt) | LB + DAP | E. coli pir+ “mating strain” | Blodgett et al. (2007) |
Prepare Mobile-CRISPRi donor strains for Tn7 transposition
1.To make E. coli donor strains, transfer Mobile-CRISPRi-encoding plasmids (Tn 7 version) to E. coli mating strain (WM6026) following Support Protocol 3 or 4.Select on LB + 300 µM DAP + 100 µg/ml ampicillin.
2.Prepare stock strains in medium + 15% glycerol and store at −80°C.
Tn7 transposition of Mobile-CRISPRi
3.Streak strains onto agar plates from −80°C stocks to obtain isolated colonies. For E. coli donor strains, use LB + 300 µM DAP + 100 µg/ml ampicillin agar plates and incubate ∼14-18 hr at 37°C. For recipient strain, use appropriate medium and culture conditions.
4.Inoculate liquid cultures from a single colony and grow to saturation. For E. coli donor strains, use 5 ml LB + 300 µM DAP + 100 µg/ml ampicillin medium in a culture tube and incubate ∼14-18 hr at 37°C with aeration (shaking at 250 rpm or on drum roller). For recipient strain, use appropriate medium, culture conditions, and culture volume to obtain between 1- and 10-fold the number of recipient cells compared to the E. coli donor cultures (i.e., equivalent or excess recipient cells).
5.Add 700 µl LB (or recipient-appropriate medium) to a microcentrifuge tube. Add 100 µl of each of the three cultures for a total volume of 1 ml. Centrifuge 2 min at 7000 × g , room temperature.
6.Remove supernatant by pipetting and resuspend pellet in 1 ml of LB (or recipient-appropriate medium). Centrifuge 2 min at 7000 × g.
7.Repeat the above wash step.
8.Remove supernatant and resuspend pellet in 30 µl LB (or recipient-appropriate medium). Pipet the cells onto a cellulose filter placed on a prewarmed LB + 300 µM DAP plate (or recipient-appropriate/compromise medium). Incubate for ∼2-24 hr at 37°C (or recipient appropriate temperature).
9.Using ethanol-sterilized tweezers, transfer the filter to a microcentrifuge tube containing 200 µl 1× PBS. Vortex ∼15 s to dislodge cells from the filter and resuspend in the buffer.
10.Plate cells on agar medium that selects for the Mobile-CRISPRi transposon-encoded antibiotic resistance gene and recipient (e.g., LB + kanamycin) without DAP. Initially, plate several volumes to determine optimal amount of your recipient organism to plate to obtain isolated colonies.
11.Stock strains in medium + 15% glycerol and store at −80°C.
12.Once recipient strains have been generated, CRISPRi knockdown can be tested by targeting fluorescent proteins (Support Protocol 1) or by observing reduced plating efficiency upon targeting of essential genes (Support Protocol 2).
Basic Protocol 4: ICEBs1 TRANSFER OF MOBILE-CRISPRi TO BACILLALES
This protocol describes how to integrate the Mobile-CRISPRi system into the chromosome of a bacterium of interest using the B. subtilis integrative and conjugative element (ICE Bs1 , or conjugative transposon; Table 3). This protocol has been used with members of the Bacillales Firmicutes (e.g., Bacillus subtilis , Staphylococcus aureus , Listeria monocytogenes , and Enterococcus faecalis).
| Strain | sJMP no. | Antibiotic res.a | Selectionb | Description/reference |
|---|---|---|---|---|
| B. subtilis with ICE | sJMP251 | cmR, spcR strS | LB with 6 µg/ml chloramphenicol | B. subtilis ICE-containing (cmR) donor strain |
| B. subtilis with Mobile-CRISPRi containing ICE | NAc | kanR, strS | LB with 7.5 µg/ml kanamycin | B. subtilis mating strain with Mobile CRISPRi (kanR) in ICE element |
| B. subtilis with Mobile-CRISPRi-containing ICE | sJMP274 | kanR, strS | LB with 7.5 µg/ml kanamycin | B. subtilis mating strain with “test” Mobile CRISPRi (kanR), mRFP, +sgRNA in ICE element |
| B. subtilis with Mobile-CRISPRi-containing ICE | sJMP275 | kanR, strS | LB with 7.5 µg/ml kanamycin | B. subtilis mating strain with “test” Mobile CRISPRi (kanR), mRFP, -sgRNA in ICE element |
| Bacillus subtilis CAL89 | sJMP210 | kanS, strR | LB with 100 µg/ml streptomycin | Recipient strain (select for ICE(kan) on LB + 7.5 µg/ml kanamycin + 100 µg/ml streptomycin) |
| Enterococcus faecalis | sJMP382 | kanS, strR | BHI with 100 µg/ml streptomycin | Recipient strain (select for ICE(kan) on BHI agar + 100 µg/ml kanamycin + 100 µg/ml streptomycin) |
| Listeria monocytogenes | sJMP17 | kanS, strR | BHI with 100 µg/ml streptomycin | Recipient strain (select for ICE(kan) on BHI agar + 50 µg/ml kanamycin + 100 µg/ml streptomycin) |
| Staphylococcus aureus | sJMP18 | kanS, strR | TSB with 100 µg/ml streptomycin | Recipient strain (select for ICE(kan) on TSB agar + 50 µg/ml kanamycin + 100 µg/ml streptomycin) |
- a
Resistance (s) of recipient E. coli. cmR, chloramphenicol resistance; kanR, kanamycin resistance; spcR, spectinomycin resistance; strR, streptomycin resistance.
- b
See Reagents and Solutions for LB, BHI, and TSB medium and stock solution recipes.
- c
To be created with a Mobile-CRISPRi plasmid (ICE version) expressing an sgRNA targeting your gene of interest using Basic Protocol 2. The B. subtilis strains are being made available through the Bacillus Genetic Stock Center (http://www.bgsc.org/); other strains are available from the ATCC.
Materials
-
MC medium (see recipe) with 6 µg/ml chloramphenicol
-
B. subtilis containing a chloramphenicol-marked ICE Bs1 element and IPTG-inducible rapI (sJMP251; Table 3)
-
BMK (competence) medium (see recipe) + 6 µg/ml chloramphenicol
-
Plasmid DNA (Mobile-CRISPRi-encoding plasmids; ICE version)
-
LB, BHI, and/or TSB medium (see recipes)
-
Agar plates with strain-specific medium (LB, BHI, or TSB) and antibiotic (chloramphenicol, kanamycin, and/or streptomycin; see Table 3 for appropriate concentration)
-
50% (v/v) glycerol
-
1 M IPTG
-
Spizizen's medium and agar plates (see recipe)
-
125-ml flask
-
1-ml deep 96-well plates, sterile (e.g., USA Scientific 1896-1000)
-
AeraSeal sterile breathable film (Sigma A9224)
-
Microplate orbital shaker (e.g., GeneMate MP4)
-
Analytical filter funnels (100 ml) with cellulose nitrate (CN) filters (47 mm, 0.2 µm, sterile; e.g., Nalgene 145-0020) and vacuum source.
Create ICE donor strains by integrating Mobile-CRISPRi (ICEBs1 version) into the Bacillus subtilis chromosomal ICEBs1 by natural competence
1.Inoculate 3 ml MC medium + 6 µg/ml chloramphenicol in a sterile culture tube with a single colony of B. subtilis containing a chloramphenicol-marked ICE Bs1 element and IPTG-inducible rapI (sJMP251), and incubate ∼12-16 hr at 37°C with aeration (shaking at 250 rpm or on a roller drum).
2.Dilute the overnight culture to OD600 = 0.1 in 30 ml BMK (competence) medium + 6 µg/ml chloramphenicol in a 125-ml flask and incubate at 37°C with shaking (250 rpm) until OD600 ∼1.5.
3.Mix 120 μl culture with ≥100 ng (∼1-5 μl) plasmid DNA in a deep 96-well plate, cover with breathable film, and incubate 10 min at 37°C without shaking, then 2 hr with shaking (900 rpm) on a microplate shaker.
4.Plate cells on LB agar + 7.5 µg/ml kanamycin and incubate 16-24 hr at 37°C. Plate several amount to obtain isolated colonies and/or restreak for isolation on a new selection plate.
5.Prepare stock strains in LB + 7.5 µg/ml kanamycin +15% glycerol at −80°C.
ICEBs1 transfer of Mobile-CRISPRi from B. subtilis donor to recipient strains
6.Inoculate 3 ml LB + 3.75 µg/ml kanamycin with a single colony of B. subtilis ICE Bs1 -CRISPRi donor strain and 3 ml LB (or strain-specific rich medium) with a single colony of the recipient strain, and incubate until exponential phase (∼2 hr) at 37°C with aeration (shaking at 250 rpm or rotating on a drum roller).
7.Dilute the starter cultures to OD600 = 0.02 in 5 ml LB + 3.75 µg/ml kanamycin for donors or 15 ml LB (or recipient-specific rich medium) for recipients, and incubate until OD600 ∼0.2 (∼1 hr) at 37°C with aeration.
8.Add 5 µl 1 M IPTG to 5 ml donor cultures (1 mM IPTG) to induce rapI expression and continue to incubate all cultures 1 hr at 37°C with aeration.
9.Adjust OD600 of cultures to 0.9.For each mating, add 2.5 ml each of donor and recipient cultures to 5 ml Spizizen's medium in a 50-ml conical tube and vortex to mix.
10.Vacuum cell suspension through an analytical filter funnel to collect the cells on a CN filter, add 5 ml Spizizen's medium, and vacuum again to wash the filter.
11.Using flame-sterilized forceps, transfer the filter to a Spizizen's medium agar plate and incubate at 37°C for 3 hr.
12.Transfer each filter to a 50-ml conical tube containing 5 ml Spizizen's medium, and vortex to resuspend cells.
13.Plate on LB + kanamycin (7.5 µg/ml or strain-specific medium and kanamycin concentration) + streptomycin (100 µg/ml) agar plates to select for transconjugants. Adjust volume plated to obtain isolated colonies and/or restreak for isolation on a new selection plate.
14.Stock strains in LB + 15% glycerol at −80°C.
15.Once recipient strains have been generated, CRISPRi knockdown can be tested by targeting fluorescent proteins (Support Protocol 1) or by observing reduced plating efficiency upon targeting essential genes (Support Protocol 2).
Support Protocol 1: QUANTIFICATION OF CRISPRi REPRESSION USING FLUORESCENT REPORTERS
This protocol describes how to quantitatively test the function of the CRISPRi system. A fluorescent protein (e.g., mRFP or sfGFP) expression cassette is incorporated into a Mobile-CRISPRi construct expressing either an sgRNA targeting the fluorescent protein gene or a nontargeting control. Strains with these Mobile-CRISPRi constructs are grown with inducer, fluorescence is measured on a fluorometer, and fold repression can be calculated. This protocol is useful when initially testing and optimizing Mobile-CRISPRi in a new bacterium. (Reference publications for example data for this procedure: Banta et al., 2020; Peters et al., 2019.)
Materials
-
LB medium and plates (or medium appropriate for your bacterium)
-
IPTG (CRISPRi-Tn 7 version) or anhydrotetracycline (aTc; CRISPRi-ICE Bs1 version)
-
1× PBS (e.g., Fisher BP399500, diluted 1:10)
-
1-ml deep 96-well plates (e.g., USA Scientific 1896-1000) or culture tubes, sterile
-
Multichannel pipets (e.g., Rainin Pipet-lite multi 17013810 and 17013807)
-
AeraSeal sterile breathable film (Sigma A9224)
-
Microplate orbital shaker (e.g., GeneMate MP4)
-
Centrifuge capable of spinning 96-well plates or culture tubes (e.g., Eppendorf 5920R)
-
96-well clear-bottom black microplates (e.g., Corning C3631)
-
Microplate fluorometer (e.g., Tecan Infinite 200 PRO Mplex)
Construct strains with a fluorescent reporter to test CRISPRi knockdown
1.Transfer CRISPRi system with fluorescent reporter (see Table 1) to recipient strain according to Basic Protocol 3 (Tn 7 transfer to Gram-negative bacteria) or 4 (ICE Bs1 transfer to Bacillales).
Grow strains to test function of CRISPRi system
2.Add 300 µl LB (or appropriate medium) to wells of a deep 96-well plate. Inoculate wells with a single colony. Test three or four isolates per strain. Cover plate with sterile breathable film and incubate ∼16 hr at 37°C (or appropriate temperature for the cells) shaking at 900 rpm on a microplate shaker.
3.Serially dilute cultures 1:10,000 into 300 µl medium using a multichannel pipet as follows: dilute cultures 1:100 into LB (or appropriate medium), then dilute 1:100 again into LB (or appropriate medium) containing 0 or 1 mM IPTG (for CRISPRi-Tn 7 version) or 0 or 0.1 µg/ml aTc (for CRISPRi-ICE Bs1 version). Cover plate with sterile breathable film and incubate for ∼10 doublings at 37°C (or appropriate temperature) shaking at 900 rpm on a microplate shaker.
Measure fluorescence to determine efficacy of CRISPRi system
4.Spin down cultures in plates for 10 min at 4000 × g , room temperature. Decant medium, and resuspend pellet in an equal volume of 1× PBS (∼300 µl) by pipetting up and down ∼10 times.
5.Transfer 200 µl cells into a 96-well clear black-bottom microplate. In a fluorescence microplate reader, measure cell density (OD600) and fluorescence (excitation 485 nm, emission 510 nm for sfGFP; excitation 584 nm, emission 607 nm for mRFP).
6.Analyze knockdown efficacy by first calculating fluorescence/OD600 and then calculating a ratio of value of strains with and without the targeting sgRNA.
Support Protocol 2: TESTING GENE ESSENTIALITY USING CRISPRi SPOT ASSAYS ON PLATES
This protocol describes how to test for gene essentiality using CRISPRi. Serial dilutions of strains with Mobile-CRISPRi-encoding sgRNAs targeting genes of interest are spotted on agar plates with various concentrations of inducer. This protocol is useful when initially determining essentiality and for optimizing level of induction of Mobile-CRISPRi needed for partial knockdown for subsequent experiments.
Materials
-
LB medium and agar plates (or medium appropriate for your bacterium), with additions of DAP, IPTG, and/or other additives as appropriate
-
IPTG (CRISPRi-Tn 7 version) or anhydrotetracycline (aTc; CRISPRi-ICE version)
-
1-ml deep 96-well plates (e.g., USA Scientific 1896-1000) or culture tubes, sterile
-
AeraSeal sterile breathable film (Sigma A9224)
-
Sterile 96-well V-bottom microplates (e.g., Corning 3896)
-
Microplate orbital shaker (e.g., GeneMate MP4)
-
Multichannel pipets (e.g., Rainin Pipet-lite multi 17013810 and 17013807)
-
150 × 15-mm Petri plates (e.g., Falcon 1351058)
1.Construct strains with targeting and nontargeting (control) sgRNAs according to Basic Protocol 2 followed by either Basic Protocol 3 or 4.
2.Add 300 µl LB (or appropriate medium) to wells in a row of a deep 96-well plate. Inoculate wells with a single colony. Cover plate with sterile breathable film and incubate ∼16 hr at 37°C (or appropriate temperature), shaking at 900 rpm on a microplate shaker.
3.Using a multichannel pipet, add 90 µl LB medium to all wells of a sterile V-bottom 96-well microplate. Add 10 µl of each culture to the wells in row A, mix by pipetting 10 times, and serially dilute (10 µl culture + 90 µl LB medium) into rows B-H, mixing each time.
4.Prepare 150 × 15-mm petri plates with LB agar + 0, 10, 100, and 1000 µM IPTG (CRISPRi-Tn 7 version) or LB + 0, 0.0001, 0.001, and 0.1 µg/ml aTc (CRISPRi-ICE version). Prewarm plates and ensure that the plate surface is not wet.
5.Using a multichannel pipet, spot 3 µl of each dilution onto the plate. When spot has soaked in, invert plates and incubate ∼16 hr at 37°C.
6.Image and analyze culture growth on plates. Reduced growth compared to the control (nontargeting sgRNA) will indicate reduced fitness of strains in which CRISPRi targets essential genes. Using plates with a range of IPTG concentrations may aid in selecting an inducer concentration appropriate for partial or complete knockdown.
Support Protocol 3: TRANSFORMATION OF E. coli BY ELECTROPORATION
This protocol details the preparation of electrocompetent E. coli cells for transformation as well as the electroporation procedure. While either electrocompetent or chemically competent cells can be used for the protocols detailed here, electroporation is generally of higher efficiency and faster for small-scale experiments, but requires specialized equipment and is salt sensitive. In this protocol, electroporation is appropriate for transforming ligations into a cloning strain.
Materials
-
E. coli strains to be transformed
-
LB medium and agar plates (with selective antibiotics as necessary)
-
5% and 15% glycerol, autoclaved
-
SOC medium
-
2-L baffled flask, sterile
-
Spectrophotometer capable of measuring OD600 (e.g., Thermo Genesys 30)
-
Centrifuge able to spin 250-ml bottles and 1-ml conical tubes at 4°C (e.g., Eppendorf 5920R)
-
Disposable cuvettes (e.g., Fisher 14-955-127)
-
250-ml polypropylene centrifuge bottles (e.g., Nalgene 3120-0250)
-
15-ml conical centrifuge tubes, sterile
-
1.5-ml microcentrifuge tubes, sterile
-
Electroporation cuvettes, 0.1-cm gap (e.g., Fisher FB101)
-
Electroporator (e.g., Bio-Rad Gene Pulser Xcell or Bio-Rad Micropulser)
Preparation of electrocompetent E. coli cells
1.Streak strains onto LB agar plates from −80°C stocks to obtain isolated colonies. Incubate ∼14-18 hr at 37°C.
2.Place 5 ml LB medium in a culture tube, inoculate with a single colony, and incubate ∼14-18 hr at 37°C with aeration (shaking at 250 rpm or on a drum roller).
3.Place 500 ml LB medium in a 2-L baffled flask, inoculate with the 5 ml culture (starting OD600 ∼0.03), and incubate until mid-exponential phase (OD600 ∼0.3-0.4, ∼2 hr) at 37°C shaking at 250 rpm.
4.Swirl flask in a pan with an ice/water slurry for ∼5 min to quickly cool the culture.
5.Pellet the cells in 250-ml polypropylene bottles by centrifuging 10 min at 4000 × g , 4°C.
6.Pour off supernatant and, while holding bottle in ice, resuspend cells in 2 ml 5% glycerol and then add 250 ml 5% glycerol, mix gently by inversion, and pellet the cells by centrifuging 10 min at 4000 × g , 4°C.
7.Pour off supernatant, resuspend cells in 2 ml 5% glycerol and then add 125 ml 5% glycerol, mix gently by inversion, and pellet the cells by centrifuging 10 min at 4000 × g , 4°C.
8.Pour off supernatant, resuspend cells in 5 ml 15% glycerol, transfer all 10 ml to a single 15-ml centrifuge tube, and pellet the cells by centrifuging 10 min at 4000 × g , 4°C.
9.Pipet off supernatant, resuspend cells in 2.5 ml 15% glycerol, and, on ice, transfer 210-µl aliquots of cells to each tube (∼13-14 tubes, about four 50-µl transformations/tube) and freeze at −80°C.
Transformation of E. coli by electroporation
10.Thaw electrocompetent E. coli cells ∼5 min on ice.
11.Prewarm two or three selective plates (i.e., LB agar + appropriate antibiotics and additives) for each transformation to 37°C.
12.Transfers 50-µl aliquots of cells to 1.5-ml microcentrifuge tubes on ice.
13.Add up to 1 µl ligation or purified plasmid DNA to cells and transfer to a 0.1-cm-gap electroporation cuvette on ice.
14.Set electroporator to exponential decay pulse, 25 µF, 200 Ω, 1.8 kV (i.e., “Bacterial 1” preset on Bio-Rad Gene Pulser Xcell or “EC1” on Bio-Rad Micropulser).
15.Wipe cuvette dry with a Kimwipe, place in the holder, and then push the pulse button.
16.After completion of pulse, remove cuvette from the holder, add 800 µl SOC medium to the cuvette, mix by pipetting, and transfer cells and medium to a culture tube.
17.Incubate cultures 1 hr at 37°C shaking at 250 rpm or on a roller drum.
18.Plate transformation on two or three prewarmed selective plates to obtain isolated colonies. Incubate plates ∼16 hr at 37°C before proceeding with the rest of your experiment.
Support Protocol 4: TRANSFORMATION OF CaCl2-COMPETENT E. coli
This protocol details the preparation of CaCl2-competent E. coli cells for transformation as well as the heat-shock procedure for transforming these cells. This method requires less specialized equipment than transformation by electroporation and is of lower efficiency but can easily be adapted to be higher throughput when many strains need to be constructed at once, such as when transferring intact plasmids to a mating strain.
Materials
-
E. coli strains to be transformed
-
LB medium and agar plates (with selective antibiotics as necessary)
-
50 mM CaCl2 with 10 mM Tris, pH 7.5
-
50 mM CaCl2 with 10 mM Tris, pH 7.5, and 15% glycerol
-
SOC medium
-
Spectrophotometer capable of measuring OD600 (e.g., Thermo Genesys 30)
-
Disposable cuvettes (e.g., Fisher 14-955-127)
-
500-ml baffled flask, sterile
-
50-ml conical centrifuge tubes, sterile
-
Centrifuge able to spin 50-ml conical tubes at 4°C (e.g., Eppendorf 5920R)
-
15-ml conical centrifuge tubes, sterile
-
Sterile 96-well V-bottom microplates (e.g., Corning 3896)
-
1.5-ml microcentrifuge tubes, sterile
-
0.2-ml PCR strip tubes or 96-well PCR plates, sterile (for higher throughput transformation)
-
AeraSeal sterile breathable film (Sigma A9224)
-
Heat block or thermal cycler (for transformation in strip tubes or 96-well plates) or water bath (for transformation in microcentrifuge tubes)
-
Foil seals (e.g., Microseal “F” Foil Bio-Rad MSF1001)
-
Multichannel pipets (e.g., Rainin Pipet-lite multi 17013810 and 17013807)
-
Microplate orbital shaker (e.g., GeneMate MP4)
Preparation of chemically competent E. coli cells
1.Streak strains onto LB agar plates from −80°C stocks to obtain isolated colonies. Incubate ∼14-18 hr at 37°C.
2.Place 5 ml LB medium in a sterile culture tube, inoculate with a single colony, and incubate ∼14-18 hr at 37°C with aeration (shaking at 250 rpm or on a drum roller).
3.Place 100 ml LB in a sterile 500-ml baffled flask, inoculate with 1 ml culture (starting OD600 ∼0.03), and incubate until mid-exponential phase (OD600 ∼0.3-0.4, ∼2 hr) at 37°C shaking at 250 rpm.
4.Swirl flask in a pan with an ice/water slurry ∼5 min to quickly cool the culture.
5.Pellet the cells in sterile 50-ml conical centrifuge tubes by centrifuging 10 min at 4000 × g , 4°C.
6.Pour off supernatant and, while holding tubes in ice, resuspend cells in 25 ml 50 mM CaCl2/10 mM Tris, pH 7.5.Mix gently by inversion and place on ice for 15 min.
7.Pellet the cells by centrifuging 10 min at 4000 × g , 4°C.
8.Pour off supernatant and, while holding tubes in ice, resuspend cells from each tube in 3.3 ml 50 mM CaCl2/10 mM Tris, pH 7.5/15% glycerol, combine resuspended cells into one tube, and place on ice for 30 min.
9.For high-throughput transformations, transfer 35 µl per well to wells of two sterile 96-well PCR plates on ice, cover with a foil seal, and freeze at −80°C. Alternatively, transfer 650 µl cells per tube to 1.5-ml tubes (∼10-11, tubes each enough for ∼16 transformations [35 µl]) and freeze at −80°C.
Transformation of chemically competent E. coli
10.Thaw CaCl2 competent E. coli cells on ice ∼5 min and transfer 35-µl aliquots to sterile 0.2-ml PCR strip tubes or a 96-well PCR plate.
11.Add 1-2 µl plasmid DNA (>10 ng) to cells and gently pipet to mix. Close strip caps or cover 96-well plate with adhesive foil.
12.Hold on ice for 30 min, incubate for exactly 2 min in a 42°C heat block, and then hold on ice for 5 min.
13.Add 100 µl SOC medium to a sterile 96-well microplate. Using a multichannel pipet, transfer cells into plate with SOC and gently pipet to mix. Cover with breathable membrane and incubate 1 hr at 37°C, shaking at 900 rpm on a microplate shaker.
14.Spot transformation onto a prewarmed selective plate and streak out to obtain isolated colonies. Incubate plates ∼16 hr at 37°C before proceeding with the rest of your experiment.
REAGENTS AND SOLUTIONS
Prepare reagents, media, and solutions with purified water (e.g., Milli-Q). Sterilize all solutions. Some can be autoclaved (as indicated below) for 20-50 min, liquid cycle. For the others, filter sterilize through a 0.2-µm-pore-size filter: syringe filter (e.g., Fisher 09-719C), Steriflip (Sigma SCGP00525), or bottle-top filter (e.g., Nalgene 595-4520). In general, these media and solutions can be stored indefinitely at room temperature.
BMK (competence) medium
Combine all components (Table 4) and filter sterilize.
| Name | Formula | MW | Per liter | |
|---|---|---|---|---|
| Potassium phosphate dibasic | K2HPO4 | 174.2 | 10.7 g | Sigma P8281 |
| Potassium phosphate monobasic | KH2PO4 | 136.1 | 5.2 g | Sigma P0662 |
| D-(+)-Glucose | C₆H₁₂O₆ | 180.2 | 20 g | Sigma D9434 |
| Trisodium citrate dihydrate | Na3C6H5O7·2H2O | 294.1 | 0.88 g | Fisher S279-500 |
| Ferric ammonium citrate | C6H8FeNO7 | 262.0 | 0.022 g | Sigma F5879 |
| Potassium aspartate | C4H5K2NO4 | 209.3 | 2.5 g | Sigma A6558 |
| Magnesium sulfate heptahydrate | MgSO4·7H2O | 246.5 | 10 ml 1 M stocka (10 mM) | Sigma M1880 |
| Manganese chloride | MnCl2·4H2O | 197.9 | 0.5 ml 300 µM stocka (150 nM) | Sigma M3634 |
| L-Tryptophan | C11H12N2O2 | 204.2 | 40 mg | Fisher BP395-100 |
| Yeast extract | 0.5 g | BD 212750 |
- a
See Reagents and Solutions for stock solution recipes.
Brain-heart infusion (BHI) medium (BD 299070)
Dissolve 38 g/L in H2O and autoclave. For plates, add 15 g/l agar (e.g., BD 214530) before autoclaving.
DL-Dithiothreitol, 100 mM
Dissolve 0.154 g DTT (MW 154.3; Sigma D9779) in 9 ml dH2O, adjust volume to 10 ml, filter sterilize, aliquot, and store at −20°C.
Glucose, 20% (w/v)
Dissolve 200 g glucose ( Sigma D9434) in 800 ml dH2O, adjust volume to 1 liter, and filter sterilize.
Glycerol, 5% (v/v)
Add 50 ml glycerol (Fisher BP2291) to 950 ml dH2O and autoclave.
Glycerol, 50% (v/v)
Add 250 ml glycerol to 250 ml deionized H2O and stir ∼10 min to mix. Aliquot 100 ml/bottle and autoclave.
Lysogeny broth (LB, Lennox; BD 240230)
Dissolve 20 g/L in H2O and autoclave. For plates, add 15 g/l agar (e.g., BD 214530) before autoclaving.
Magnesium chloride (MgCl2), 1 M
Dissolve 20.3 g MgCl2·6H2O (Sigma M2670) in 80 ml deionized H2O, adjust volume to 100 ml, and autoclave or filter sterilize.
Manganese chloride (MnCl2), 300 mM
Dissolve 5.9 g MnCl2·2H2O (Sigma M3634) in 8 ml dH2O, adjust volume to 10 ml. Filter sterilize.
Manganese chloride (MnCl2), 300 µM
Dilute 300 mM MnCl2 1:1000 (10 µl in 10 ml sterile dH2O).
Magnesium sulfate (MgSO4), 1 M
Dissolve 24.6 g MgSO4·7H2O ( Sigma M1880) in 80 ml dH2O, adjust volume to 100 ml, and autoclave or filter sterilize.
MC medium
Combine all components (Table 5) and filter sterilize.
| Name | Formula | MW | Per liter | |
|---|---|---|---|---|
| Potassium phosphate dibasic | K2HPO4 | 174.2 | 10.7 g | Sigma P8281 |
| Potassium phosphate monobasic | KH2PO4 | 136.1 | 5.2 g | Sigma P0662 |
| D-(+)-Glucose | C₆H₁₂O₆ | 180.2 | 20 g | Sigma D9434 |
| Trisodium citrate dihydrate | Na3C6H5O7·2H2O | 294.1 | 0.88 g | Fisher S279-500 |
| Ferric ammonium citrate | C6H8FeNO7 | 262.0 | 0.022 g | Sigma F5879 |
| Casamino acids | 1.0 g | BD 223050 | ||
| Potassium glutamate monohydrate | C5H10KNO5·H2O | 203.2 | 2.2 g | Sigma G1501 |
| Magnesium sulfate heptahydrate | MgSO4·7H2O | 246.5 | 20 ml 1 M stocka (20 mM) | Sigma M1880 |
| Manganese chloride | MnCl2·4H2O | 197.9 | 1 ml 300 µM stocka (300 nM) | Sigma M3634 |
| L-Tryptophan | C11H12N2O2 | 204.2 | 20 mg | Fisher BP395-100 |
- a
See Reagents and Solutions for stock solution recipes.
Medium additives
Prepare additives (Table 6), filter sterilize each additive, and divide into aliquots before storage. Sterilized medium should be cooled below 55°C before these reagents are added.
| Reagent | Abbr. | [Stock] | Solvent | Storage | Product no. | Notes |
|---|---|---|---|---|---|---|
| Ampicillin | amp | 100 mg/ml | dH2O | –20°C | Fisher BP17605 | 1000× stock for E. coli |
| Anhydrotetracycline | aTc | 0.1 mg/ml | DMSO | –20°C | Sigma 37919 | use at 0.001-0.1 µg/ml |
| Chloramphenicol | cm | 20 mg/ml | 80% ethanol | –20°C | Sigma C1919 | 1000× stock for E. coli |
| Diaminopimelic acid | DAP | 30 mM (MW 190.2) | dH2O | 4°C | Sigma 33240 | 100× stock for dap– E. coli |
| Gentamycin | gent | 15 mg/ml | dH2O | –20°C | Sigma G1264 | 1000× stock for E. coli |
| Isopropyl β-D-1-thiogalactopyranoside | IPTG |
1 M (MW 238.3) |
dH2O | –20°C | Sigma I6758 | Light sensitive; use at 1-1000 µM |
| Kanamycin | kan | 30 mg/ml | dH2O | –20°C | Fisher BP906 | 1000× stock for E. coli |
| Spectinomycin | spec | 50 mg/ml | dH2O | –20°C | Sigma S4014 | 1000× stock for E. coli |
| Streptomycin | strep | 100 mg/ml | dH2O | –20°C | Sigma S1277 | 1000× stock for E. coli |
Potassium chloride (KCl), 1 M
Dissolve 7.5 g KCl (Sigma P9333) in 80 ml deionized H2O, adjust volume to 100 ml, and autoclave or filter sterilize.
SOC medium
Dissolve 4 g tryptone (BD 211705), 1 g yeast extract (BD212750), 0.1 g NaCl (Fisher S271), and 500 µl 1 M KCl (see recipe) in 200 ml H2O in a bottle with a stir bar. Autoclave to sterilize. When solution has cooled to below ∼60°C, add 1 ml sterile 1 M MgCl2 (see recipe) and 3.6 ml sterile 20% (w/v) glucose (see recipe) and stir to mix. Aliquot 10 ml/tube and store between −20°C and 25°C.
Spizizen's medium and agar plates
Combine all components listed in Table 7 and filter sterilize.
| Name | Formula | MW | Per liter | |
|---|---|---|---|---|
| Potassium phosphate dibasic | K2HPO4 | 174.2 | 14 g | Sigma P8281 |
| Potassium phosphate monobasic | KH2PO4 | 136.1 | 6 g | Sigma P0662 |
| Ammonium sulfate | (NH4)2SO4 | 132.1 | 2 g | Sigma A4418 |
| D-(+)-Glucose | C₆H₁₂O₆ | 180.2 | 5 g | Sigma D9434 |
| Trisodium citrate dihydrate | Na3C6H5O7·2H2O | 294.1 | 1 g | Fisher S279-500 |
| Magnesium sulfate heptahydrate | MgSO4·7H2O | 246.5 | 0.2 g | Sigma M1880 |
| L-Tryptophan | C11H12N2O2 | 204.2 | 50 mg | Fisher BP395-100 |
- a
For plates, mix 250 ml filter-sterile 2× medium with 250 ml autoclaved H2O + 7.5 g agar, cooled to 55°C, swirl to mix, and pour into 15 × 100-mm petri dishes (e.g., Corning 351029).
Tryptic Soy broth (TSB)
Dissolve 30 g/L TSB (BD 211825) in H2O and autoclave. For plates, add 15 g/liter agar (e.g., BD 214530) before autoclaving.
COMMENTARY
Background Information
CRISPR interference (CRISPRi) is a programmable gene-knockdown tool based on the clustered regularly interspaced short palindromic repeats (CRISPR) adaptive immune systems found in bacteria and archaea that restrict viral and plasmid DNA and RNA. Several types of CRISPR systems have been co-opted for use in CRISPRi (Qi et al., 2013; Specht, Xu, & Lambert, 2020; Zheng et al., 2019), but here we will focus on the commonly used Type II-A system from Streptococcus pyogenes (reviewed in Wright, Nuñez, & Doudna, 2016). In the native S. pyogenes CRISPR system, a CRISPR array containing spacers that designate target genes is transcribed into a pre-crRNA (CRISPR RNA), which is subsequently processed into individual crRNAs containing only one spacer. Processed crRNAs form a complex with a tracrRNA (trans-activating CRISPR RNA) and the CRISPR-associated nuclease Cas9; this complex is directed to target DNA by base-pairing between the crRNA spacer and a complementary DNA sequence in the target known as a protospacer. In addition to spacer-protospacer complementarity, Cas9 requires a short protospacer-adjacent motif (PAM; NGG for S. pyogenes) for DNA binding that prevents self-targeting of CRISPR arrays that lack this motif. The Cas9-tracr/crRNA complex binds to the PAM sequence, unzips the DNA duplex, anneals the crRNA and protospacer DNA, and then—if the spacer and protospacer match sufficiently—cleaves both strands of the DNA (Sternberg, Redding, Jinek, Greene, & Doudna, 2014). Doudna, Carpentier, and colleagues first showed that Cas9 is an RNA-guided endonuclease and simplified the natural dual RNA system by engineering a fused tracr/crRNA known as a single guide RNA (sgRNA; Jinek et al., 2012). Cas9 can thus be programmed to target new genes of interest simply by changing the 20-nt sgRNA spacer to match a protospacer with an adjacent PAM in the target DNA.
To repurpose CRISPR as a gene-knockdown technology, Qi and colleagues mutated the two nuclease active sites in Cas9, producing an inactive variant known as dCas9 (“dead Cas9”; Qi et al., 2013). dCas9 retains the ability to be directed to target genes via programmable sgRNAs but can no longer cut DNA. Instead, dCas9 inhibits transcription at the step of initiation or elongation by acting as a steric block to RNA polymerase in bacterial systems. The modest sequence requirements for CRISPRi repression—i.e., an NGG PAM sequence and adjacent spacer (Jinek et al., 2012)—and engineered Cas9 variants with altered- or relaxed-specificity PAM dependencies (Walton, Christie, Whittaker, & Kleinstiver, 2020) suggest that nearly all bacterial genes can be targeted by CRISPRi. CRISPRi systems have been established in many diverse bacteria and have primarily been used to phenotype individual essential genes in proof-of-principle work. However, CRISPRi has been increasingly valuable in targeting larger, defined sets of genes (e.g., essential genes; Liu et al., 2017; Peters et al., 2016) and in pooled phenotyping approaches at the genome scale for both model (Rousset et al., 2018; Wang et al., 2018) and nonmodel bacteria (Lee et al., 2019; Yao et al., 2020; reviewed in Vigouroux & Bikard, 2020).
Critical Parameters and Troubleshooting
Optimizing Mobile-CRISPRi transfer and integration
Distinguishing between Mobile-CRISPRi transfer and integration problems presents a challenge because both processes are required to obtain transconjugants. The efficiency of Mobile-CRISPRi transfer and integration varies by strain, such that two strains of the same species can produce vastly different numbers of transconjugants (e.g., P. aeruginosa PAO1 and PA14 differ by >100-fold; Peters et al., 2019). Cell surface features (e.g., capsules) as well as defense systems that destroy horizontally transferred DNA (e.g., restriction enzymes and other CRISPR systems) can reduce transfer and integration efficiency (Thomas & Nielsen, 2005). Extending the mating time to 24 hr may improve recovery of transconjugants for low-efficiency recipients. Conjugation efficiencies may increase for some recipients after growth at elevated temperatures or heat shock, possibly owing to inactivation of restriction enzymes or other protein-based inhibitors (Irani & Rowe, 1997; Zeng, Ardeshna, & Lin, 2015).
Transfer, integration, or both can be limiting for obtaining transconjugants from Tn 7 - and ICE Bs1 -based Mobile-CRISPRi systems. For instance, Tn 7 systems fail to produce transconjugants when B. subtilis 168 is the recipient strain; however, the B. subtilis att Tn7 site is functional for integration when cloned into E. coli , suggesting that transfer of the two-plasmid Tn 7 system is limiting in this case (Peters et al., 2019). In contrast, Tn 7 integration into wild-type Agrobacterium tumefaciens occurs at low frequency because insertion into the native att Tn7 site disrupts a gene immediately downstream of glmS ; introducing a second att Tn7 site at a neutral locus alleviated this issue (Figueroa-Cuilan, Daniel, Howell, Sulaiman, & Brown, 2016). If mating fails for Tn 7 Mobile-CRISPRi, we recommend adding a second copy of the att Tn7 site on a plasmid to test for integration into that site either in E. coli or in the relevant recipient. Further, we have observed that inclusion of a third donor strain containing a self-transferrable RP4 helper plasmid—either pRK2013 (Ditta, Stanfield, Corbin, & Helinski, 1980) or pEVS104 (Stabb & Ruby, 2002)—is required to obtain transconjugants when A. baumannii is the recipient (Peters et al., 2019); although the mechanism is unknown, inclusion of the RP4 helper may also increase recovery of transconjugants for other recipients as well. We strongly prefer use of pEVS104 as the RP4 helper plasmid due to its pir -dependent origin that does not replicate in recipient strains (pRK2013 has a ColE1 origin which replicates in species related to E. coli). For strains in which restriction endonucleases are a major barrier to conjugation, such as Z. mobilis , we have used mutants deleted for genes encoding restriction systems to increase recovery of Tn 7 Mobile-CRISPRi transconjugants by several orders of magnitude (Banta et al., 2020). Tn 7 systems can also be electroporated into cells if mating is inefficient or inconvenient (Choi & Schweizer, 2006). Although the Tn 7 recognition site in the 3′ end of glmS is generally conserved across bacteria, the ICE Bs1 recognition site in the leu2 tRNA is present only in bacteria related to B. subtilis (e.g., various Bacillus species, L. monocytogenes , and S. aureus). However, work from the Voigt lab has shown that a smaller, engineered ICE element (mini-ICE) can insert at noncanonical sites (Brophy et al., 2018), raising the possibility that the ICE Bs1 host range is broader than previously appreciated. Therefore, we recommend testing ICE Bs1 Mobile-CRISPRi in Gram-positives that lack an obvious leu2 gene and mapping the insertion sites using arbitrary PCR (Saavedra, Schwartzman, & Gilmore, 2017).
Cloning into Mobile-CRISPRi modules
Mobile-CRISPRi vectors are composed of functional “modules” flanked by restriction sites to enable swapping or cloning of new components by restriction digestion and Gibson assembly (Gibson et al., 2009) or ligation. The modules are as follows: (1) antibiotic-resistance genes (e.g., conferring resistance to kanamycin, chloramphenicol, gentamicin, and spectinomycin) flanked by XhoI sites; (2) reporter genes (e.g., mRFP , sfGFP) flanked by PmeI sites; (3) an sgRNA gene with an existing spacer or BsaI sites to clone new spacers (see Cloning new sgRNA spacers into Mobile-CRISPRi, above) flanked by EcoRI sites; (4) promoter-regulation genes (e.g., lacI and tetR) flanked by SmaI sites; (5) the dcas9 promoter and ribosome-binding site (RBS) flanked by SpeI sites, and (6) dcas9 flanked by SpeI and AscI sites (cutting with SpeI also removes the dcas9 promoter and RBS module).
Optimizing Mobile-CRISPRi knockdown
CRISPRi knockdown from published Mobile-CRISPRi vectors varies by strain and is not currently predictable (Peters et al., 2019). If knockdown is insufficient for desired experiments, we recommend testing whether sgRNA or dcas9 expression is limiting by cloning these genes individually onto multicopy plasmids and testing knockdown alongside Mobile-CRISPRi integrated into the chromosome. We used this strategy to determine that sgRNA expression was limiting during optimization of Mobile-CRISPRi for Z. mobilis (Banta et al., 2020). Optimizing Mobile-CRISPRi knockdown involves cloning new components into the function modules listed above (specifically, the sgRNA and dcas9 modules 3, 5, and 6). If expression of the sgRNA is limiting, we clone synthetic DNA with a strong promoter and sgRNA gene into the EcoRI sites. If a stronger promoter has not been characterized for the relevant recipient bacterium, we empirically determine the activity of strong promoters from model bacteria (i.e., E. coli and B. subtilis) in the context of CRISPRi in the recipient. If dcas9 expression is limiting, we clone synthetic DNA containing a strong promoter and an RBS optimized for the relevant recipient strain into the SpeI sites. We use the RBS Calculator (https://salislab.net/software/) from the Salis lab to estimate RBS efficacy and design sites with higher translation rates (Espah Borujeni et al., 2017). Alternatively, codon-optimized variants of dcas9 (along with a new promoter and RBS) can be cloned into the SpeI-AscI sites of Mobile-CRISPRi for bacteria with high or low GC content. Overexpression of dCas9 is toxic in some bacteria (Cho et al., 2018; Lee, Hoynes-O'Connor, Leong, & Moon, 2016; Qu et al., 2019; Rock et al., 2017), even when expressed in single copy from the chromosome (e.g., in P. aeruginosa). In these cases, dcas9 expression must be reduced to avoid pleiotropic effects; we previously used low-level constitutive promoters from the Anderson promoter series (Qu et al., 2019) to drive dcas9 expression in P. aeruginosa , avoiding toxic effects while maintaining knockdown. The Bikard lab has developed a screening strategy to avoid toxicity from certain sgRNA seed sequences (known as “bad seeds”) by randomizing bases within the dcas9 RBS to identify constructs that minimize toxicity but still offer robust knockdown (Depardieu & Bikard, 2020).
Understanding Results
Basic Protocol 3: Tn7 Transfer of Mobile-CRISPRi to Gram-negative bacteria
Tn 7 Mobile-CRISPRi transfer and integration efficiency varies by strain. In our experience, efficiency ranges from ∼1% (E. coli) to ∼10–7% (P. mirabilis) with a median efficiency of ∼10–2-10–3% (Peters et al., 2019).
Basic Protocol 4: ICEBs1 transfer of Mobile-CRISPRi to Bacillales
ICE Bs1 Mobile-CRISPRi transfer and integration efficiency varies by strain. In our experience, efficiency ranges from ∼1% (B. subtilis) to ∼10-7% (E. faecalis) with a median efficiency of ∼10-3%-10-4% (Peters et al., 2019).
Support Protocol 1: Quantifying CRISPRi repression using fluorescent reporters
Mobile-CRISPRi knockdown efficacy varies by strain and ranges from 150-fold knockdown (S. aureus) to 8-fold knockdown (P. aeruginosa) with a median efficacy of ∼40-fold (Peters et al., 2019).
Time Considerations
The mating process can be performed in a single day, but many other aspects of the protocols listed here are dependent on the growth rate of the recipient strain. For instance, transconjugants can be evaluated the next day when using E. cloacae as a recipient, but Z. mobilis transconjugants take 3-4 days to form robust colonies.
Acknowledgments
This work was supported by a Career Transition Award from the NIH National Institute of Allergy and Infectious Diseases (K22AI137122) to Jason M. Peters and by the DOE Office of Biological and Environmental Research, Great Lakes Bioenergy Research Center (DE-SC0018409). Jennifer S. Tran was supported by the Biotechnology Training Program (NIH 5T32GM135066) and a GRFP from the NSF. Ryan D. Ward was supported by the Predoctoral Training Program in Genetics (NIH 5T32GM007133-46).
Author Contributions
Amy B. Banta : Conceptualization; methodology; writing-original draft; writing-review & editing. Ryan D. Ward : Software; writing-original draft; writing-review & editing. Jennifer S. Tran : Visualization; writing-original draft; writing-review & editing. Emily E. Bacon : Writing-original draft; writing-review & editing. Jason M. Peters : Conceptualization; funding acquisition; methodology; writing-original draft; writing-review & editing.
Literature Cited
- Banta, A. B., Enright, A. L., Siletti, C., & Peters, J. M. (2020). A high-efficacy CRISPRi system for gene function discovery in Zymomonas mobilis. Applied and Environmental Microbiology , 86, e01621-20. doi: 10.1128/AEM.01621-20.
- Bikard, D., Jiang, W., Samai, P., Hochschild, A., Zhang, F., & Marraffini, L. A. (2013). Programmable repression and activation of bacterial gene expression using an engineered CRISPR-Cas system. Nucleic Acids Research , 41, 7429–7437. doi: 10.1093/nar/gkt520.
- Blodgett, J. A. V., Thomas, P. M., Li, G., Velasquez, J. E., van der Donk, W. A., Kelleher, N. L., & Metcalf, W. W. (2007). Unusual transformations in the biosynthesis of the antibiotic phosphinothricin tripeptide. Nature Chemical Biology , 3, 480–485. doi: 10.1038/nchembio.2007.9.
- Brophy, J. A. N., Triassi, A. J., Adams, B. L., Renberg, R. L., Stratis-Cullum, D. N., Grossman, A. D., & Voigt, C. A. (2018). Engineered integrative and conjugative elements for efficient and inducible DNA transfer to undomesticated bacteria. Nature Microbiology , 3, 1043–1053. doi: 10.1038/s41564-018-0216-5.
- Burnett, L. C., Lunn, G., & Coico, R. (2009.) Biosafety: Guidelines for working with pathogenic and infectious microorganisms. Current Protocols in Microbiology , 13, 1A.1.1–1A.1.14. doi: 10.1002/9780471729259.mc01a01s13.
- Calvo-Villamañán, A., Ng, J. W., Planel, R., Ménager, H., Chen, A., Cui, L., & Bikard, D. (2020). On-target activity predictions enable improved CRISPR-dCas9 screens in bacteria. Nucleic Acids Research , 48, e64. doi: 10.1093/nar/gkaa294.
- Cho, S., Choe, D., Lee, E., Kim, S. C., Palsson, B., & Cho, B.-K. (2018). High-level dCas9 expression induces abnormal cell morphology in Escherichia coli. ACS Synthetic Biology , 7, 1085–1094. doi: 10.1021/acssynbio.7b00462.
- Choi, K.-H., & Schweizer, H. P. (2006). mini-Tn 7 insertion in bacteria with single attTn 7 sites: Example Pseudomonas aeruginosa. Nature Protocols , 1, 153–161. doi: 10.1038/nprot.2006.24.
- Cui, L., Vigouroux, A., Rousset, F., Varet, H., Khanna, V., & Bikard, D. (2018). A CRISPRi screen in E. coli reveals sequence-specific toxicity of dCas9. Nature Communications , 9, 1912. doi: 10.1038/s41467-018-04209-5.
- Datsenko, K. A., & Wanner, B. L. (2000). One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proceedings of the National Academy of Sciences , 97, 6640–6645. doi: 10.1073/pnas.120163297.
- Depardieu, F., & Bikard, D. (2020). Gene silencing with CRISPRi in bacteria and optimization of dCas9 expression levels. Methods , 172, 61–75. doi: 10.1016/j.ymeth.2019.07.024.
- Ditta, G., Stanfield, S., Corbin, D., & Helinski, D. R. (1980). Broad host range DNA cloning system for gram-negative bacteria: Construction of a gene bank of Rhizobium meliloti. Proceedings of the National Academy of Sciences of the United States of America , 77, 7347–7351. doi: 10.1073/pnas.77.12.7347.
- Ellis, N. A., Kim, B., & Machner, M. P. (2020). A multiplex CRISPR interference tool for virulence gene interrogation in an intracellular pathogen [Preprint]. bioRxiv , 2020.06.17.157628. Retrieved from https://www.biorxiv.org/content/10.1101/2020.06.17.157628v1.
- Espah Borujeni, A., Cetnar, D., Farasat, I., Smith, A., Lundgren, N., & Salis, H. M. (2017). Precise quantification of translation inhibition by mRNA structures that overlap with the ribosomal footprint in N-terminal coding sequences. Nucleic Acids Research , 45, 5437–5448. doi: 10.1093/nar/gkx061.
- Figueroa-Cuilan, W., Daniel, J. J., Howell, M., Sulaiman, A., & Brown, P. J. B. (2016). Mini-Tn7 insertion in an artificial attTn7 site enables depletion of the essential master regulator CtrA in the phytopathogen Agrobacterium tumefaciens. Applied and Environmental Microbiology , 82, 5015–5025. doi: 10.1128/AEM.01392-16.
- Gibson, D. G., Young, L., Chuang, R.-Y., Venter, J. C., Hutchison, C. A., & Smith, H. O. (2009). Enzymatic assembly of DNA molecules up to several hundred kilobases. Nature Methods , 6, 343–345. doi: 10.1038/nmeth.1318.
- Gilbert, L. A., Horlbeck, M. A., Adamson, B., Villalta, J. E., Chen, Y., Whitehead, E. H., … Weissman, J. S. (2014). Genome-scale CRISPR-mediated control of gene repression and activation. Cell , 159, 647–661. doi: 10.1016/j.cell.2014.09.029.
- Hawkins, J. S., Silvis, M. R., Koo, B.-M., Peters, J. M., Jost, M., Hearne, C. C., … Gross, C. A. (2020). Mismatch-CRISPRi reveals the co-varying expression-fitness relationships of essential genes in Escherichia coli and Bacillus subtilis. Cell Systems , 11(5), 523–535.e9. doi: 10.1016/j.cels.2020.09.009.
- Irani, V. R., & Rowe, J. J. (1997). Enhancement of transformation in Pseudomonas aeruginosa PAO1 by Mg2+ and heat. BioTechniques , 22, 54–56. doi: 10.2144/97221bm09.
- Jinek, M., Chylinski, K., Fonfara, I., Hauer, M., Doudna, J. A., & Charpentier, E. (2012). A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science , 337, 816–821. doi: 10.1126/science.1225829.
- Lee, H. H., Ostrov, N., Wong, B. G., Gold, M. A., Khalil, A. S., & Church, G. M. (2019). Functional genomics of the rapidly replicating bacterium Vibrio natriegens by CRISPRi. Nature Microbiology , 4, 1105–1113. doi: 10.1038/s41564-019-0423-8.
- Lee, Y. J., Hoynes-O'Connor, A., Leong, M. C., & Moon, T. S. (2016). Programmable control of bacterial gene expression with the combined CRISPR and antisense RNA system. Nucleic Acids Research , 44, 2462–2473. doi: 10.1093/nar/gkw056.
- Li, X., Jun, Y., Erickstad, M. J., Brown, S. D., Parks, A., Court, D. L., & Jun, S. (2016). tCRISPRi: Tunable and reversible, one-step control of gene expression. Scientific Reports , 6, 39076. doi: 10.1038/srep39076.
- Liu, X., Gallay, C., Kjos, M., Domenech, A., Slager, J., van Kessel, S. P., … Veening, J. (2017). High-throughput CRISPRi phenotyping identifies new essential genes in Streptococcus pneumoniae. Molecular Systems Biology , 13, 931. doi: 10.15252/msb.20167449.
- Peters, J. M., Colavin, A., Shi, H., Czarny, T. L., Larson, M. H., Wong, S., … Gross, C. A. (2016). A comprehensive, CRISPR-based functional analysis of essential genes in bacteria. Cell , 165, 1493–1506. doi: 10.1016/j.cell.2016.05.003.
- Peters, J. M., Koo, B.-M., Patino, R., Heussler, G. E., Hearne, C. C., Qu, J., … Rosenberg, O. S. (2019). Enabling genetic analysis of diverse bacteria with Mobile-CRISPRi. Nature Microbiology , 4, 244–250. doi: 10.1038/s41564-018-0327-z.
- Qi, L. S., Larson, M. H., Gilbert, L. A., Doudna, J. A., Weissman, J. S., Arkin, A. P., & Lim, W. A. (2013). Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell , 152, 1173–1183. doi: 10.1016/j.cell.2013.02.022.
- Qu, J., Prasad, N. K., Yu, M. A., Chen, S., Lyden, A., Herrera, N., … Rosenberg, O. S. (2019). Modulating pathogenesis with Mobile-CRISPRi. Journal of Bacteriology , 201(22), e00304–19. doi: 10.1128/JB.00304-19.
- Reis, A. C., Halper, S. M., Vezeau, G. E., Cetnar, D. P., Hossain, A., Clauer, P. R., & Salis, H. M. (2019). Simultaneous repression of multiple bacterial genes using nonrepetitive extra-long sgRNA arrays. Nature Biotechnology , 37, 1294–1301. doi: 10.1038/s41587-019-0286-9.
- Rock, J. M., Hopkins, F. F., Chavez, A., Diallo, M., Chase, M. R., Gerrick, E. R., … Fortune, S. M. (2017). Programmable transcriptional repression in mycobacteria using an orthogonal CRISPR interference platform. Nature Microbiology , 2, 16274. doi: 10.1038/nmicrobiol.2016.274.
- Rousset, F., Cui, L., Siouve, E., Becavin, C., Depardieu, F., & Bikard, D. (2018). Genome-wide CRISPR-dCas9 screens in E. coli identify essential genes and phage host factors. PLoS Genetics , 14, e1007749. doi: 10.1371/journal.pgen.1007749.
- Saavedra, J. T., Schwartzman, J. A., & Gilmore, M. S. (2017). Mapping transposon insertions in bacterial genomes by arbitrarily primed PCR. Current Protocols in Molecular Biology , 118, 15.15.1–15.15.15. doi: 10.1002/cpmb.38.
- Specht, D. A., Xu, Y., & Lambert, G. (2020). Massively parallel CRISPRi assays reveal concealed thermodynamic determinants of dCas12a binding. Proceedings of the National Academy of Sciences , 117, 11274–11282. doi: 10.1073/pnas.1918685117.
- Stabb, E. V., & Ruby, E. G. (2002). RP4-based plasmids for conjugation between Escherichia coli and members of the vibrionaceae. Methods in Enzymology , 358, 413–426. doi: 10.1016/S0076-6879(02)58106-4.
- Sternberg, S. H., Redding, S., Jinek, M., Greene, E. C., & Doudna, J. A. (2014). DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature , 507, 62–67. doi: 10.1038/nature13011.
- Thomas, C. M., & Nielsen, K. M. (2005). Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nature Reviews Microbiology , 3, 711–721. doi: 10.1038/nrmicro1234.
- Vigouroux, A., & Bikard, D. (2020). CRISPR tools to control gene expression in bacteria. Microbiology and Molecular Biology Reviews , 84, e00077-19. doi: 10.1128/MMBR.00077-19.
- Vigouroux, A., Oldewurtel, E., Cui, L., Bikard, D., & van Teeffelen, S. (2018). Tuning dCas9's ability to block transcription enables robust, noiseless knockdown of bacterial genes. Molecular Systems Biology , 14, e7899. doi: 10.15252/msb.20177899.
- Walton, R. T., Christie, K. A., Whittaker, M. N., & Kleinstiver, B. P. (2020). Unconstrained genome targeting with near-PAMless engineered CRISPR-Cas9 variants. Science , 368, 290–296. doi: 10.1126/science.aba8853.
- Wang, T., Guan, C., Guo, J., Liu, B., Wu, Y., Xie, Z., … Xing, X.-H. (2018). Pooled CRISPR interference screening enables genome-scale functional genomics study in bacteria with superior performance. Nature Communications , 9, 2475. doi: 10.1038/s41467-018-04899-x.
- Wright, A. V., Nuñez, J. K., & Doudna, J. A. (2016). Biology and applications of CRISPR systems: Harnessing nature's toolbox for genome engineering. Cell , 164, 29–44. doi: 10.1016/j.cell.2015.12.035.
- Yao, L., Shabestary, K., Björk, S. M., Asplund-Samuelsson, J., Joensson, H. N., Jahn, M., & Hudson, E. P. (2020). Pooled CRISPRi screening of the cyanobacterium Synechocystis sp PCC 6803 for enhanced industrial phenotypes. Nature Communications , 11, 1666. doi: 10.1038/s41467-020-15491-7.
- Zeng, X., Ardeshna, D., & Lin, J. (2015). Heat shock-enhanced conjugation efficiency in standard Campylobacter jejuni Strains. Applied and Environmental Microbiology , 81, 4546–4552. doi: 10.1128/AEM.00346-15.
- Zheng, Y., Han, J., Wang, B., Hu, X., Li, R., Shen, W., … Peng, W. (2019). Characterization and repurposing of the endogenous Type I-F CRISPR–Cas system of Zymomonas mobilis for genome engineering. Nucleic Acids Research , 47, 11461–11475. doi: 10.1093/nar/gkz940.
Internet Resources
Conda package manager and environment management system.
GitHub development platform.
sgRNA design scripts for CRISPRi in bacteria.
Original sgRNA design code by John Hawkins that was modified for these protocols.
NCBI Nucleotide Database.
Citing Literature
Number of times cited according to CrossRef: 13
- Steven J. Russell, Amanda K. Garcia, Betül Kaçar, A CRISPR interference system for engineering biological nitrogen fixation, mSystems, 10.1128/msystems.00155-24, 9 , 3, (2024).
- Logan J. Geyman, Madeline P. Tanner, Natalia Rosario-Meléndez, Jason M. Peters, Mark J. Mandel, Julia C. van Kessel, Mobile-CRISPRi protocol optimization for Vibrionaceae , Microbiology Resource Announcements, 10.1128/mra.00040-24, 13 , 8, (2024).
- Amy L. Enright, William J. Heelan, Ryan D. Ward, Jason M. Peters, CRISPRi functional genomics in bacteria and its application to medical and industrial research, Microbiology and Molecular Biology Reviews, 10.1128/mmbr.00170-22, 88 , 2, (2024).
- Ryan D. Ward, Jennifer S. Tran, Amy B. Banta, Emily E. Bacon, Warren E. Rose, Jason M. Peters, Essential gene knockdowns reveal genetic vulnerabilities and antibiotic sensitivities in Acinetobacter baumannii , mBio, 10.1128/mbio.02051-23, 15 , 2, (2024).
- Ashley N. Hall, Benjamin W. Hall, Kyle J. Kinney, Gabby G. Olsen, Amy B. Banta, Daniel R. Noguera, Timothy J. Donohue, Jason M. Peters, Tools for genetic engineering and gene expression control in Novosphingobium aromaticivorans and Rhodobacter sphaeroides , Applied and Environmental Microbiology, 10.1128/aem.00348-24, 90 , 10, (2024).
- Logan J. Geyman, Madeline P. Tanner, Natalia Rosario-Meléndez, Jason M. Peters, Mark J. Mandel, Julia C. van Kessel, Mobile-CRISPRi as a powerful tool for modulating Vibrio gene expression , Applied and Environmental Microbiology, 10.1128/aem.00065-24, 90 , 6, (2024).
- Christine M. Hustmyer, Robert Landick, Bacterial chromatin proteins, transcription, and DNA topology: Inseparable partners in the control of gene expression, Molecular Microbiology, 10.1111/mmi.15283, 122 , 1, (81-112), (2024).
- Alexa FS Gomberg, Alan D Grossman, It's complicated: relationships between integrative and conjugative elements and their bacterial hosts, Current Opinion in Microbiology, 10.1016/j.mib.2024.102556, 82 , (102556), (2024).
- Yeganeh Hajizadeh, Farzad Badmasti, Mana Oloomi, Inhibition of the blaOXA-48 gene expression in Klebsiella pneumoniae by a plasmid carrying CRISPRi-Cas9 system, Gene, 10.1016/j.gene.2024.148332, 910 , (148332), (2024).
- Chunmei Chen, Pu Zheng, Effects of down-regulation of ackA expression by CRISPR-dCpf1 on succinic acid production in Actinobacillus succinogenes, AMB Express, 10.1186/s13568-023-01518-x, 13 , 1, (2023).
- Amy L. Enright, Amy B. Banta, Ryan D. Ward, Julio Rivera Vazquez, Magdalena M. Felczak, Michael B. Wolfe, Michaela A. TerAvest, Daniel Amador-Noguez, Jason M. Peters, The genetics of aerotolerant growth in an alphaproteobacterium with a naturally reduced genome, mBio, 10.1128/mbio.01487-23, 14 , 6, (2023).
- Michelle A. Yu, Amy B. Banta, Ryan D. Ward, Neha K. Prasad, Michael S. Kwon, Oren S. Rosenberg, Jason M. Peters, Investigating Pseudomonas aeruginosa Gene Function During Pathogenesis Using Mobile-CRISPRi, Pseudomonas aeruginosa, 10.1007/978-1-0716-3473-8_2, (13-32), (2023).
- N. I. Nadolinskaia, A. V. Goncharenko, CRISPR Interference in Regulation of Bacterial Gene Expression, Molecular Biology, 10.1134/S0026893322060139, 56 , 6, (823-829), (2022).

