Production of CRISPR-Cas9 Transgenic Cell Lines for Knocksideways Studies
Michael Wagenbach, Michael Wagenbach, Juan Jesus Vicente, Juan Jesus Vicente, Wren Wagenbach, Wren Wagenbach, Linda Wordeman, Linda Wordeman
Abstract
Protein activity is generally functionally integrated and spatially restricted to key locations within the cell. Knocksideways experiments allow researchers to rapidly move proteins to alternate or ectopic regions of the cell and assess the resultant cellular response. Briefly, individual proteins to be tested using this approach must be modified with moieties that dimerize under treatment with rapamycin to promote the experimental spatial relocalizations. CRISPR technology enables researchers to engineer modified protein directly in cells while preserving proper protein levels because the engineered protein will be expressed from endogenous promoters. Here we provide straightforward instructions to engineer tagged, rapamycin-relocalizable proteins in cells. The protocol is described in the context of our work with the microtubule depolymerizer MCAK/Kif2C, but it is easily adaptable to other genes and alternate tags such as degrons, optogenetic constructs, and other experimentally useful modifications. Off-target effects are minimized by testing for the most efficient target site using a split-GFP construct. This protocol involves no proprietary kits, only plasmids available from repositories (such as addgene.org). Validation, relocalization, and some example novel discoveries obtained working with endogenous protein levels are described. A graduate student with access to a fluorescence microscope should be able to prepare engineered cells with spatially controllable endogenous protein using this protocol. © 2023 The Authors. Current Protocols published by Wiley Periodicals LLC.
Basic Protocol 1 : Choosing a target site for gene modification
Basic Protocol 2 : Design of gRNA(s) for targeted gene modification
Basic Protocol 3 : Split-GFP test for target efficiency
Basic Protocol 4 : Design of the recombination template and analytical primers
Support Protocol 1 : Design of primers for analytical PCR
Basic Protocol 5 : Transfection, isolation, and validation of engineered cells
Support Protocol 2 : Stable transfection of engineered cells with binding partners
INTRODUCTION
Knocksideways is a technique to experimentally move an intracellular protein of interest and anchor it to other places in the cell. Briefly, this involves fusing the FKBP domain of FKBP12 to a protein of interest (often called a “bait” protein). Upon addition of rapamycin to your bait-protein-expressing cells, the bait protein will dimerize with an FRB moiety (the FKBP rapamycin-binding domain of mTOR) that you have anchored, or otherwise labeled, elsewhere in the cell. Both the FKBP and FRB moieties are relatively small and have a good record of not interfering with protein function in the absence of rapamycin. The usual regime is to use rapamycin to remove the bait protein from a subcellular location, where it is presumably performing a function, to another region of the cell and assay the effect of this loss on cellular processes. Rapamycin-dependent knocksideways experiments (Robinson et al., 2010) have the advantage of removing key proteins (and their interactors) from cellular structures on a timescale of minutes and in a manner that is compatible with live imaging (Cherry et al., 2019; Wagenbach et al., 2020; Wordeman et al., 2016). This can provide an instantaneous picture of the functional contribution of the bait protein to dynamic processes within the cell. Knocksideways experiments have been successfully used with microtubule (MT) modulators to define their roles in dynamic cellular processes such as cytoskeletal turnover (Cheeseman et al., 2013). When knocksideways experiments are performed with transiently transfected or stably expressed proteins, the expression levels of the target protein to be relocalized are invariably superphysiological and may suffer from lack of complete relocalization. Furthermore, existing endogenous levels of unmodified (and therefore unrelocalized) protein may impact or dilute the experimental measurements. These problems can be avoided by engineering homozygous CRISPR-Cas9 cell lines in which a chosen endogenous bait protein of interest is dually modified with both an FKBP moiety and a fluorescent protein. This allows researchers to visually monitor the extent and timing of translocation of the bait protein from its region of activity to the FRB moiety anchored elsewhere. The impact of this translocation on the cellular function and spatial organization of other proteins can be assayed and the results compared with those for the pre-rapamycin-treated cell.
The CRISPR protocol described here can be adapted to engineer a variety of transgenic cell lines with different bait proteins of interest. The workflow for the engineering and isolation of homozygous cell lines is described in Figure 1. We have used this protocol primarily to prepare adherent human cell lines possessing engineered microtubule (MT) regulatory genes (such as MCAK/Kif2C, ch-TOG, EB-1, and CENP-F, among others). In our previously published studies (Wordeman et al., 2016), we have employed induced dimerization between GFP-FKBP-modified MT regulators (such as MCAK/Kif2C and Kif18A) and the SH4-rapamycin-binding domain of mTOR (FRB). FRB is a 100-amino-acid (aa) domain (E2015 to Q2114) of the mammalian target of rapamycin (mTOR) known as the FKBP−rapamycin binding domain (Suh et al., 2006). The FRB construct is, in turn, modified with both SBFP2, a blue fluorescent protein used to assess the localization of the dimerization partner, and an SH4 (Lyn11) domain to anchor FRB in the membrane (Wordeman et al., 2016). If desired, our CRISPR-engineered cell lines can be further modified by stably transfecting them with constructs expressing SH4-FRB-BFP2 and using drug selection for transformants. As CRISPR-engineered cells are not under drug selection once isolated, stable transfectants of various types can be produced with CRISPR lines using selectable markers. Upon addition of rapamycin, GFP-FKBP-MCAK/Kif2C will translocate from MT tips, the MT lattice, or other cellular structures to the plasma membrane, leaving the dynamic MT structures free of this MT regulator. Using live microscopy, one can assess whether MT growth rates or turnover are altered in the moment that a regulator is lost. Additionally, the FKBP-modified protein could conceivably be engineered to relocate anywhere that the FRB moiety is anchored, such as centrosomes or kinetochores. In these cases the SH4 domain would be replaced by another protein or domain that maintains a stable association with another cellular structure.

An advantage of this CRISPR protocol over others is that it is designed to minimize screens of replicate cell clones that involve isolating large numbers of cell lines whose recombinant status is unknown. Additionally, this protocol includes a preliminary screen of three or four predicted single guide RNAs (sgRNAs) using a split-GFP reporter developed by the Ikawa lab (Mashiko et al., 2013) to identify the sgRNA that produces the highest number of recombinants. These are detectable in this assay as producing the highest proportion of GFP-fluorescent cells and represents an easily quantifiable means to identify the most efficient sgRNA to use to prepare recombinants. After the best sgRNA is identified, the most efficient Cas9 sgRNA plasmid is co-transfected with linearized donor DNA synthesized from bait protein genomic sequence. Enrichment for recombinant cells is facilitated by a two-step process: a preliminary puromycin-based drug selection to remove cells that are not transfected with the CRISPR plasmids followed by a single-cell sorting to identify and purify fluorescent cells (we use a BD Aria III cell sorter). Sparse plating of the sorted cells enables the identification by fluorescence microscopy of suitable clones of uniform cells exhibiting proper localization of the engineered gene product. Suitable-appearing clones are isolated as colonies directly from the plate, and subjected to further purification if necessary. This imaging-centric approach usually provides clones within 6 to 8 weeks. Should the procedure fail, there is less time investment up front. Replicates of good-looking clones can then be rapidly confirmed through western blotting, genomic DNA digestion, and sequencing. Finally, it is worthwhile to compare the newly engineered clones to the parent cell line for parameters such as cell cycle length, cell motility, appearance, and size.
There are several considerations to address before attempting to engineer cells for manipulation of endogenous protein. These are described in detail in the Strategic Planning section and will help increase success in obtaining a homozygous cell line expressing a fluorescently tagged, relocalizable endogenous protein for future studies. The complete process is described in order of execution as five Basic Protocols. Basic Protocol 1 consists of choosing your genomic target site in your gene of interest (we use MCAK/Kif2C as an example) using the NCBI bioinformatics gene database (https://www.ncbi.nlm.nih.gov/gene) and CRISPRdirect (crispr.dbcls.jp). Basic Protocol 2 covers the design and synthesis of the guide RNA you will use to target your gene of interest. Basic Protocol 3 identifies the most efficient target site (of which there are often several predicted sites in close apposition) by synthesizing and testing several (usually two to four) protospacers predicted as potential target sites for cutting efficiency using a split-GFP reporter developed by the Ikawa lab (Mashiko et al., 2013). Once you have chosen a targeting site, Basic Protocol 4 details the design and modification of your recombination template to target the homologous region of your gene of interest. And finally, in Basic Protocol 5, the engineered cell clones are isolated and validated by fluorescence microscopy, genomic DNA digestion, and western blotting. In further studies, the target gene modified with an FKBP (FK506 binding protein) moiety should relocalize to any cellular region decorated with the FRB (FKBP–rapamycin binding) domain of the mammalian target of rapamycin (mTOR) with high affinity (K D ∼ 1-2 nM). This localization is irreversible and can occur under the microscope on a timescale of seconds. This technique can be used to evaluate the immediate functional contribution of your engineered bait protein to dynamic processes within the cell.
STRATEGIC PLANNING
Experimental potential
Knocksideways experiments involve the addition of an FKBP moiety to a fluorescent, or otherwise detectable, protein that is to be moved elsewhere in the cell (Cheeseman et al., 2013; DeRose et al., 2012; Ding et al., 2014; Robinson et al., 2010). Upon addition of rapamycin, the protein (bait) to be functionally tested will dimerize with an FRB moiety that is anchored to another structure in the cell, the easiest and most common being the plasma membrane. This effectively removes a protein from its key site of activity, allowing researchers to evaluate the cellular effects of the loss of this activity. It is advisable to test your protein of interest using a transient transfection format to determine whether it can be easily relocalized and whether it localizes properly while tagged with a fluorescent protein (FP) and FKPB fusion. It is best to begin with proteins you are familiar with, using tags and tag positioning that you know will work. For example, Figure 2A and B show diagrams of two genes (MCAK/Kif2C and ch-TOG) in which we have engineered the endogenous protein to be visible (GFP) and relocalizable (FKBP). These are positions (N- vs. C-terminal tags) that have been determined empirically not to interfere with protein function (Brouhard et al., 2008; Wordeman et al., 1999). Thus, the tagged elements are positioned accordingly to maintain functionality. The FRB moiety has been fused to BFP and SH so that it can be anchored in the plasma membrane and evaluated for expression level by preparing transient stable lines expressing blue plasma membranes under drug selection. Live cell lines stably expressing FRB-BFP (Fig. 2C, blue) and endogenously engineered MCAK/Kif2C (Fig. 2C-E, green) and ch-TOG (Fig. 2F-H, green) before and after rapamycin administration are shown.
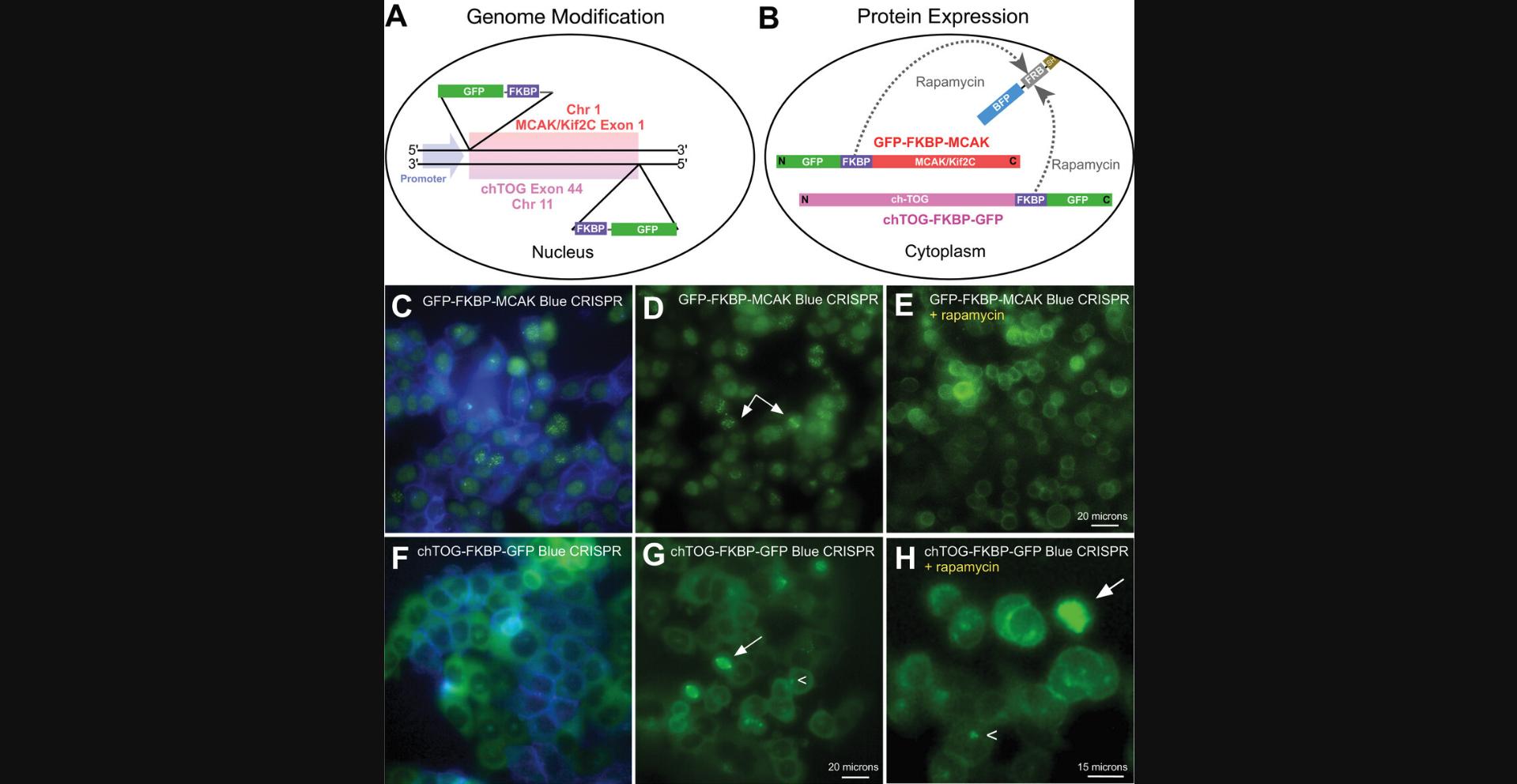
This CRISPR protocol can be easily adapted to prepare cell lines that express proteins modified with moieties suitable for other applications, such as optogenetics (in which a module is inserted into a protein rendering its activity sensitive to light; Dema et al., 2022) or auxin-based degrons (in which an AID tagged protein is degraded in the presence of auxin and TIR1). For our studies, we chose the rapamycin knocksideways approach because it is easy to perform on an entire culture dish, it is easy to visually assess for completion, and we liked the irreversible nature of the approach because it allowed us to evaluate greater numbers of cells after treatment as compared to optogenetics approaches, where cell recovery can be quite rapid. Furthermore, our experience with auxin-based degrons is that degradation is difficult to assess visually and that it requires more time (2 hr in our hands) to take effect (Wordeman et al., 2016). The degradation time of proteins in these systems is very protein specific and can range from a few minutes to many hours. However, if the time scale of your experimental assay is longer than minutes, auxin-based degrons can be a good option for relatively rapid protein removal.
Choice of parent cell line
This protocol is designed for adherent human cell lines that are amenable to imaging and for which there is a great deal of DNA sequence information available. However, because many human cell lines are derived from cancer cells, which possess many abnormalities in both chromosomal number and structure, it is worth investigating the karyotype of your preferred cell line. We have found that we have the most success using diploid or pseudo-diploid cell lines rather than more chromosomally unstable cell lines that are likely to exhibit high levels of aneuploidy and chromosomal rearrangements. Modal numbers of chromosomes per cell are generally 46 in stable human cell lines, whereas they can be quite variable in cells exhibiting chromosome instability (CIN). Some commonly studied but chromosomally unstable human cells are HeLa (modal number 70 to 164 depending on the strain), U2OS (hypertriploid), Hek293 (modal number 64), and HT29 (modal number = 71), to name a few. In addition to the possibility of encountering structural chromosomal rearrangements that may cause your homologous recombination to fail, the ploidy must be considered. When the desire is to engineer a homozygous cell line, there may be several alleles of the same gene within one cell. Although a number of endogenously engineered genes have been produced in chromosomally unstable cell lines (Neumann et al., 2010), you may be able to increase your chance of success by choosing a chromosomally stable cell line with minimal chromosomal rearrangements. Human cell lines that tend to maintain a close to diploid cell number include Hct116 (modal number 45) and hTERT-RPE-1 (modal number 46). We have successfully engineered both of these cell lines. CRISPR-engineered cells can be prepared from chromosomally unstable parent cell lines, but your success will be more unpredictable. We advise commencing with stable lines as unstable lines may require preliminary sequencing of homology domains and more extensive analysis and monitoring after engineering. See also the Critical Parameters section of the Commentary for more details.
Limitations
There are five important limitations to consider when employing CRISPR knocksideways approaches to visualize dynamic cellular processes. First, proteins expressed at endogenous levels are sometimes poorly detectable under the microscope. Fluorescent cells are often easily isolated by fluorescence-activated cell sorting (FACs), which can be highly sensitive. Such cells, once validated by PCR, can be directly usable in experiments. However, it is best to further cross-correlate them with other controls such as fixation and signal amplification (immunofluorescence) to ensure the protein target is properly distributed within the cell and properly relocalized after rapamycin treatment. Second, every protein behaves a little differently during relocalization. Some are more difficult to relocalize or consistently exhibit only partial relocalization (Cheeseman et al., 2013). Third, knocksideways experiments using rapamycin-dependent localization are irreversible and have to be robustly controlled. They cannot be rescued by rapamycin withdrawal because rapamycin has a high affinity (K D < 1 nM) for FRB when bound to FKBP12. Reversible rapamycin analogs have been researched, but they are not widely available and may compromise the speed and efficiency of the reaction (Bayle et al., 2006). Fourth, rapamycin alone has some reported effects on cultured cells due to its ability to inhibit the mTOR pathway. Thus, we advise controlling knocksideways experiments with the CRISPR parent cell line lacking BFP-FRB, which will not relocalize your FKBP fusion protein in the presence of rapamycin. Such cells will control for the effects of rapamycin without relocalizing your FKBP-modified protein. They will also control for any likelihood that your protein will pair with endogenous mTOR, which is present in cells at a concentration of ∼110 nM (Cho et al., 2022). We routinely cross correlate our experimental data with two controls: (i) a CRISPR cell line expressing BFP-FRB plus added vehicle and (ii) a CRISPR cell line without BFP-FRB plus added rapamycin. Finally, when using chromosomally unstable cells, in addition to whole-chromosome losses and gains, chromosomes may sustain structural rearrangements that may alter a particular target or homology region, requiring genomic re-sequencing. Unfortunately, this will only manifest itself if/when homologous recombination fails; it will not be evident in the split-GFP target sequence test.
Basic Protocol 1: CHOOSING A TARGET SITE FOR GENE MODIFICATION
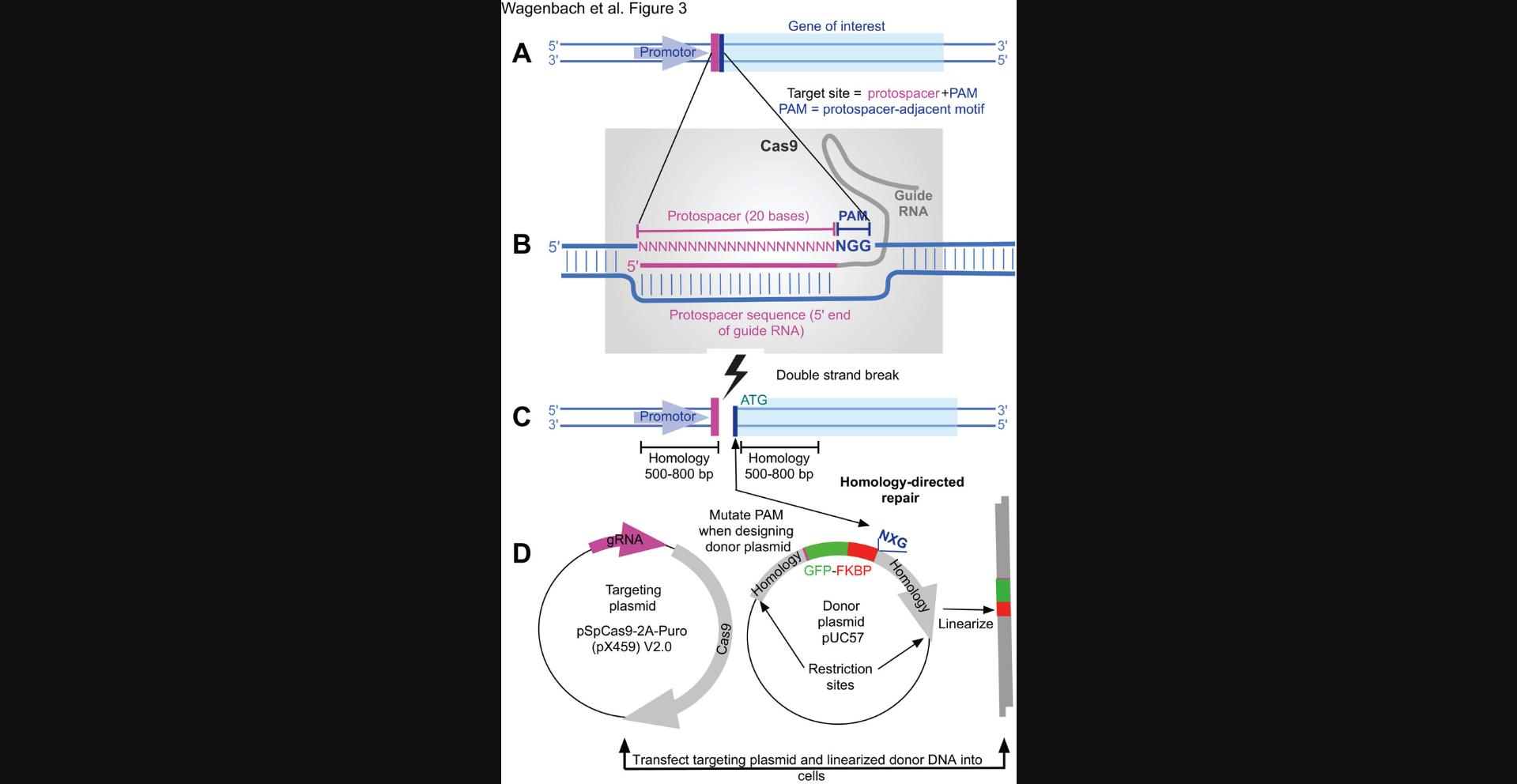
CRISPR engineering of endogenous cellular proteins requires the construction of a targeting vector that encodes an RNA (the guide RNA) that is complementary to a 20-base-pair (bp) section of the chromosome. A homologous recombination region should span the sequence (target sequence) that the user wishes to target for cleavage and gene modification (Fig. 3A). A CRISPR targeting plasmid (we used pX459 v2.0) also includes a gene for an endonuclease, Cas9, that forms a complex with the transcribed guide RNA and acts at the target site to cut the chromosome (Fig. 3B). The wild-type Cas9 enzyme catalyzes a double-stranded break, so only one targeting construct is needed to completely cleave the chromosome at the target location (Fig. 3C). This will serve as the site for the introduction of your linearized donor DNA by your chosen guard RNA (gRNA; Fig. 3D). If greater specificity is needed, a mutant enzyme can be chosen (in conjunction with a related targeting vector, pX462 v2.0) that creates only a single-strand nick in the chromosome (often called a Cas9 nickase), which requires using two closely spaced targets on opposite strands of the chromosome. We used the wild-type Cas9 enzyme for all of our lines because it, predictably, gave us better efficiency than the nickase method. To date, we have not used the mutant endonuclease to prepare our lines and have not noticed second-site problems. although these could be admittedly difficult to detect.
Necessary Resources
Hardware
- Computer (Mac or PC)
Software
- Modern browser (Chrome, Firefox, Safari)
- DNA sequence analysis program: SnapGene (Dotmatics, ),MacVector(MacVector,Inc,,),Geneious(Dotmatics,),DNASTAR(DNASTAR,Inc.,$), BioEdit (Free), ApE plasmid editor (Free), Serial Cloner (Free), or SeaView (PRABI-Doua, Free), matched to your computer type
Files
- Appropriate FASTA files
1.Use the “Gene” search at NCBI to find the genomic view of your gene of interest. For human MCAK/Kif2C, we find https://www.ncbi.nlm.nih.gov/gene/11004.
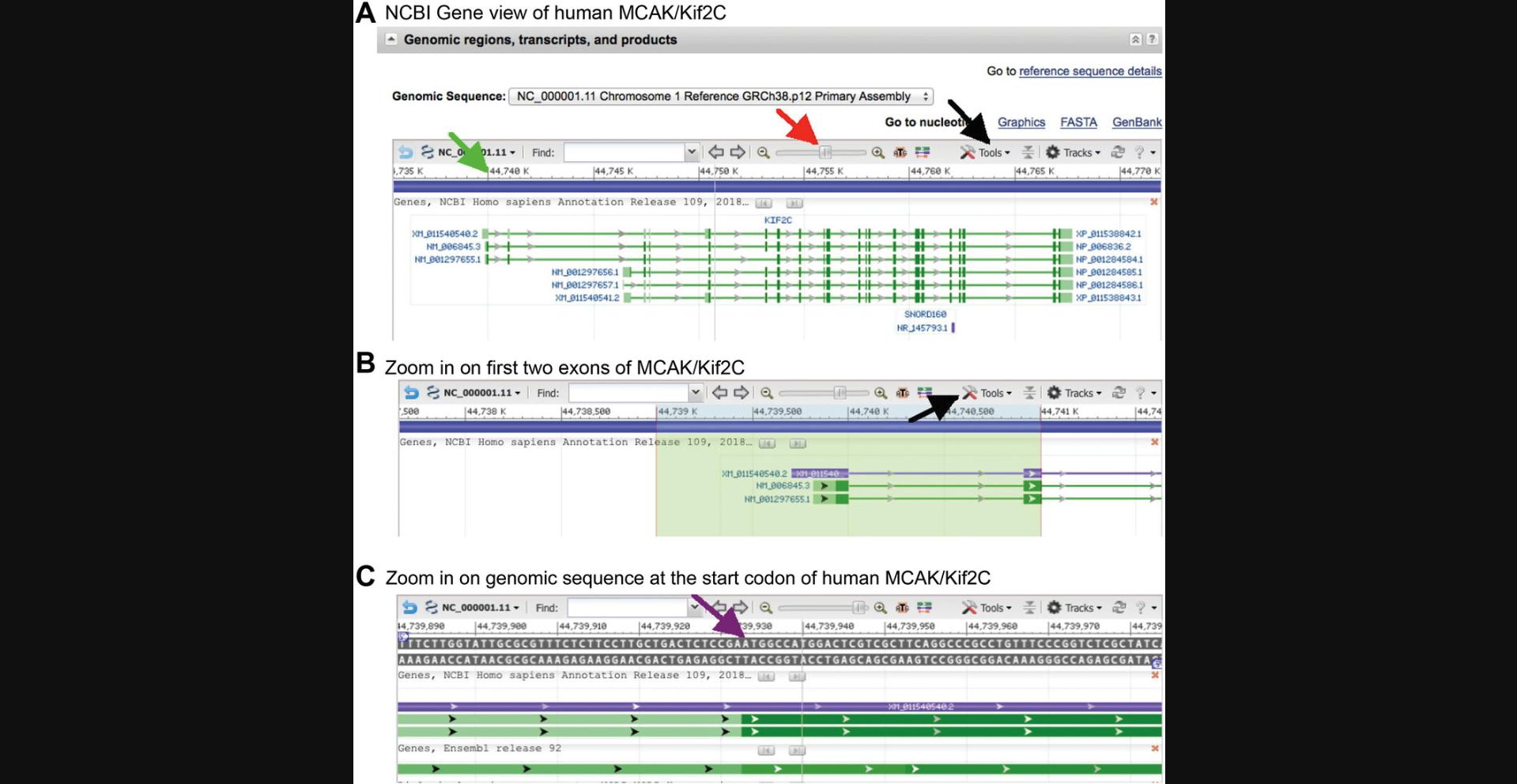
2.Center the 5′ end of the gene and zoom in until the window is about ∼5-10 kb wide (that is, the 65%-70% zoom level; the zoom level is revealed by clicking and holding on the slider button; red arrow in Fig. 4A). Starting ∼1 kb to the left of the 5′ end of the gene, click and drag across the ruler to ∼1 kb to the right of the 5′ end to highlight a region of a size that will be convenient to download.
3.Copy the DNA sequence text from the file in step 2 to your preferred DNA sequence analysis program, so that you now have ∼2000 bp of genomic sequence in a file you can manipulate. Return to the NCBI window, center the start of the mRNA, and zoom up to the 95% or 100% level so that you can see the actual nucleotide sequence around the start codon. Make a note of ∼10 bp that include the start site (purple arrow in Fig. 4C), and then search the genomic sequence file for this and mark the start codon.
Basic Protocol 2: DESIGNING AND ENGINEERING THE CRISPR TARGETING PLASMID(S)
These plasmids will cleave the chromosome at your chosen target site (Fig. 5A). The broad goal is to use a predictive algorithm (such as CRISPRdirect) to locate target sequences and predict the number of off-target sites found in the genome (Fig. 5B). Different optimal-appearing target sites will operate at different efficiencies. For this reason it is advisable to choose three or four putative target sites and test their efficiencies in Basic Protocol 3.To do this you will need to synthesize the oligos for each site, and clone pairs of them into the Bbs1 sites (positions 252 and 260) of your cloning vector (pX459; Fig. 5C). These three or four vectors will code for the three or four guide RNAs you would like to test. Each vector will direct to a slightly different target site (with likely different cutting efficiencies) as defined by your chosen oligos. We typically make three or four different targeting plasmids, in case some are more effective than others. An oligonucleotide with the target + sticky ends is purchased for each sgRNA desired and of course a complementary strand to that oligo to form a small double-stranded piece of DNA for ligation (Fig. 5D). We have not found it necessary to have these oligos purified because they are so short; the crude oligo will contain enough full-length product to perform successfully.

Materials
- pX459 (pSpCas9(BB)-2A-Puro V2.0 (Addgene plasmid #62988)
- Oligos for gRNA construction (Table 1)
- pXforward oligo (Table 1)
- BbsI-HF (New England Biolabs R3539S)
- Phenol (Sigma P4557)
- Chloroform
- 3 M sodium acetate, pH 5.2 (see recipe)
- Isopropanol
- 70% (v/v) ethanol (prepared from 200-proof ethanol, Fisher 04-355-222)
- TE buffer (see recipe)
- T4 DNA ligase (New England Biolabs M0202S)
- NEB 5-alpha competent Escherichia coli (high efficiency; New England Biolabs C2987H)
- LB agar ± 50 µg/ml ampicillin plates (see recipe)
- NotI-HF (New England Biolabs R3189S)
- TB medium ± 50 µg/ml ampicillin
- 10,000× SYBR Safe DNA stain (Invitrogen. S33102)
- Agarose (VWR 0710-25G)
- 50× TAE (see recipe)
- Lambda DNA, BstEII digest (New England Biolabs N3014L)
- 6× gel loading dye (New England Biolabs B7024S)
- QIAquick gel extraction kit (Qiagen. 28704)
- QIAprep spin miniprep kit (Qiagen. 27106)
- Qiagen HiSpeed Midi column kit (Qiagen 12643)
- BigDye 3.1 sequencing reagent (Fisher 43-374-54)
| Protocol(s) | Oligo name | Sequence (5′→3′) | Purpose |
|---|---|---|---|
| 3 | Test target oligo sense | 5′-gatccNNNNNNN…NNNNNNNNNN-3′ | Clone multiple target sequence into pCAG-EGxxFP |
| 3 | Test target oligo antisense | 5′-aattNNNNNN…NNNNNNNNNNNg-3′ | Clone multiple target sequence into pCAG-EGxxFP |
| 2 | gRNA sense | 5′-caccgNNNNNNNNNNNNNNNNNNNN-3′ | Clone protospacer into pX459 (sense) |
| 2 | gRNA antisense | 5′-aaacNNNNNNNNNNNNNNNNNNNNc-3′ | Clone protospacer into pX459 (antisense) |
| 2, 4 | gRNA sense (MCAK/Kif2C) | 5′-caccgGCTGACTCTCCGAATGGCCA-3′ | Successful protospacer for cloning a representative MCAK/Kif2C target site into pX459 (sense) |
| 2, 4 | gRNA antisense (MCAK/Kif2C) | 5′-aaacTGGCCATTCGGAGAGTCAGCc-3′ | Successful protospacer for cloning a representative MCAK/Kif2C target site into pX459 (antisense) |
| 2, 3; Support Protocol 1 | pXforward | 5′-GAGGGCCTATTTCCCATGATTCCTT-3′ | Primer for sequencing the pX459 vector |
| 3 | pCAGforward | 5′-CTCCTGGGCAACGTGCTGGTT-3′ | Primer for sequencing the pCAG-EGxxFP vector |
- 15-ml conical tube (Fisher 12-565-269)
- 10-ml sterile syringe (Fisher 14-955-459)
- 0.22-µm sterile filter (Fisher 09-720-004)
- 100-mm Falcon petri dish (Fisher 08-757-100D)
- 500-ml 0.2-µm-pore-size filter unit (Fisher FB12566504)
- Bacteriological incubator (VWR 10055-006 or equivalent), 37°C
- Shaker (VWR 76407-108 or equivalent), 37°C
- Horizontal electrophoresis apparatus (VWR 10001-836 or equivalent)
- Electrophoresis DC power supply (VWR 76196-454 or equivalent)
- Microcentrifuge for 1.5-ml tubes, >10,000 × g (VWR 76533-876 or equivalent)
- Thermal cycler for 0.2-ml tubes (VWR 89217-558 or equivalent)
Design and order the oligos to prepare several targeting plasmids to test for sgRNA efficiency
1.Duplicate the genomic sequence file (created in Basic Protocol 1, step 3) so that you have the original for reference, as well as copies to alter when designing your various CRISPR sequences.
2.Copy a small region of 40-50 bp on either side of the start codon and paste into the “or Paste a nucleotide sequence” window at http://crispr.dbcls.jp/ after deleting the sample text. Leave the PAM setting on the NGG default. Change the default genome setting only if you are not using human cells. Analyze by clicking the “design” button. The output should look like Figure 5B.
3.Choose one or more target sequences to create in CRISPR targeting vectors. Try to select those with fewer 12-mer and 8-mer target sites. If you plan to use the wild-type Cas9 (in pX459), only one target sequence will be needed (although trying several different targets is one of the easiest parts of the process, and may increase your chances of success). Putative targets can be located on either the plus or minus strand of your chromosomal sequence.
4.Design and order oligos for targeting vectors as shown in Figure 5D (refer to Fig. 5E for examples from MCAK/Kif2C). Do not include the PAM sequence (shown in the box in Fig. 5B, third column) as part of the oligo.
5.Have the oligos synthesized (we use IDTdna.com) and also order primer pXforward (Table 1): GAGGGCCTATTTCCCATGATTCCTT for sequencing the vector. Gel purification of full-length oligos should not be necessary.
Ligate the oligos into pX459 to create several targeting plasmids to test for efficiency
6.Digest 5 µg pX459 with 20 U BbsI-HF in a 50-µl reaction with NEB-supplied 1× Cutsmart buffer for 3 hr at 37°C. Purify vector DNA from reaction using QIAquick gel extraction kit.
7.Add 25 µl phenol, pH 8, and vortex for 5 s.
8.Add 170 µl additional 1× Cutsmart buffer and then 50 µl chloroform, vortex 5 sec, and centrifuge 5 min in a microcentrifuge at >10,000 × g , room temperature.
9.Transfer 200 µl of aqueous (upper) phase to a clean microcentrifuge tube.
10.Add 20 µl 3 M sodium acetate, pH 5.2, and 220 µl isopropanol, vortex 5 sec, let stand at room temperature for 15 min, and centrifuge 5 min in microcentrifuge at >10,000 × g , room temperature.
11.Carefully remove supernatant with a micropipettor, avoiding pellet (pellet will be very small).
12.Wash pellet gently with 50 µl of 70% (v/v) ethanol and remove wash with micropipettor. Dissolve pellet in 50 µl TE buffer.
13.Dissolve the oligos at 100 µM in sterile, distilled water.
14.Dilute 20 pM (2 µl) of each top-strand and bottom-strand oligo pair into a 20-µl reaction containing 1× NEB-supplied ligase buffer, heat briefly to 90°C, let cool to <45°C, and put on ice.
15.Ligate 2 µl (∼200 ng) of linear vector from step 6 to 2 µl (2 pM) of annealed oligo duplex from step 7 using 0.5 µl T4 DNA ligase in 10 µl of 1× ligase buffer for >12 hr at 15°C (one ligation reaction for each pair of targeting oligos designed in step 4).
16.Transform 50 µl NEB 5-alpha competent bacteria with 1 µl of ligation reaction according to NEB-supplied instructions.
17.Incubate 1 bacteria for 1 hr at 37° and then plate on LB agar + 50 µg/ml ampicillin.
18.Grow a few colonies for miniprep DNA preps in 2 ml TB medium + 50 µg/ml ampicillin.
19.Prepare DNA with QIAprep spin mini columns per the manufacturer's instructions.
20.Digest clones with BbsI + NotI for 1 hr.
21.Electrophorese clones on a 1% agarose gel in 1× TAE as described in Sambrook et al. (1989) or Voytas et al. (2000). Stain gel with 1× SYBR Safe DNA stain.
22.Identify clones with inserted oligos: these clones should have lost the BbsI site and appear as a single band on the gel, whereas incorrect clones will form two bands.
23.Sequence one or two “correct” clones (those lacking BbsI sites) with primer pXforward to verify that the insertion is correct.
24.Grow a large-scale (100 µg or more) prep of each completed targeting vector using 50 ml of TB + 50 µg/ml ampicillin medium and Qiagen High Speed midi columns, for transfection into cultured cells in Basic Protocol 3, step 5, and Basic Protocol 5, step 2.
Basic Protocol 3: SPLIT-GFP TEST FOR TARGET EFFICIENCY
When choosing a precise location to which to target your recombination event using predictive algorithms such as CRISPRdirect, you are likely to have several choices of reasonable locations to choose from. Certain sites will be more efficient at producing recombinants than others, often for unknown reasons. Thus, it is useful to empirically test likely sites before settling on a final recombination site. This protocol describes how to choose a target site within your target region by empirically testing them for efficiency using a simple visual microscope assay.
This protocol requires the purchase of two additional oligos (Table 1) that contain the genomic sequence of only the region sufficient to contain all of the sgRNAs, plus sticky ends for different enzymes found in a vector called pCAG-EgxxFP (https://www.addgene.org/50716/). This vector contains a direct repeat of two copies of GFP, one lacking the 3′ end of the gene and the other lacking the 5′ end of the gene, with several cloning sites in between them. The test oligos (Table 1) corresponding to the entire targeted genomic region (Fig. 6A) are cloned between the two copies of GFP (Fig. 6B). Because these oligos are long, typically 50 to 70 bp, we have ordered them gel-purified because the crude oligo would probably contain a fairly low fraction of full-length product and therefore perform poorly in a ligation reaction. Alternately, they could be assembled from four or more shorter, overlapping oligos.
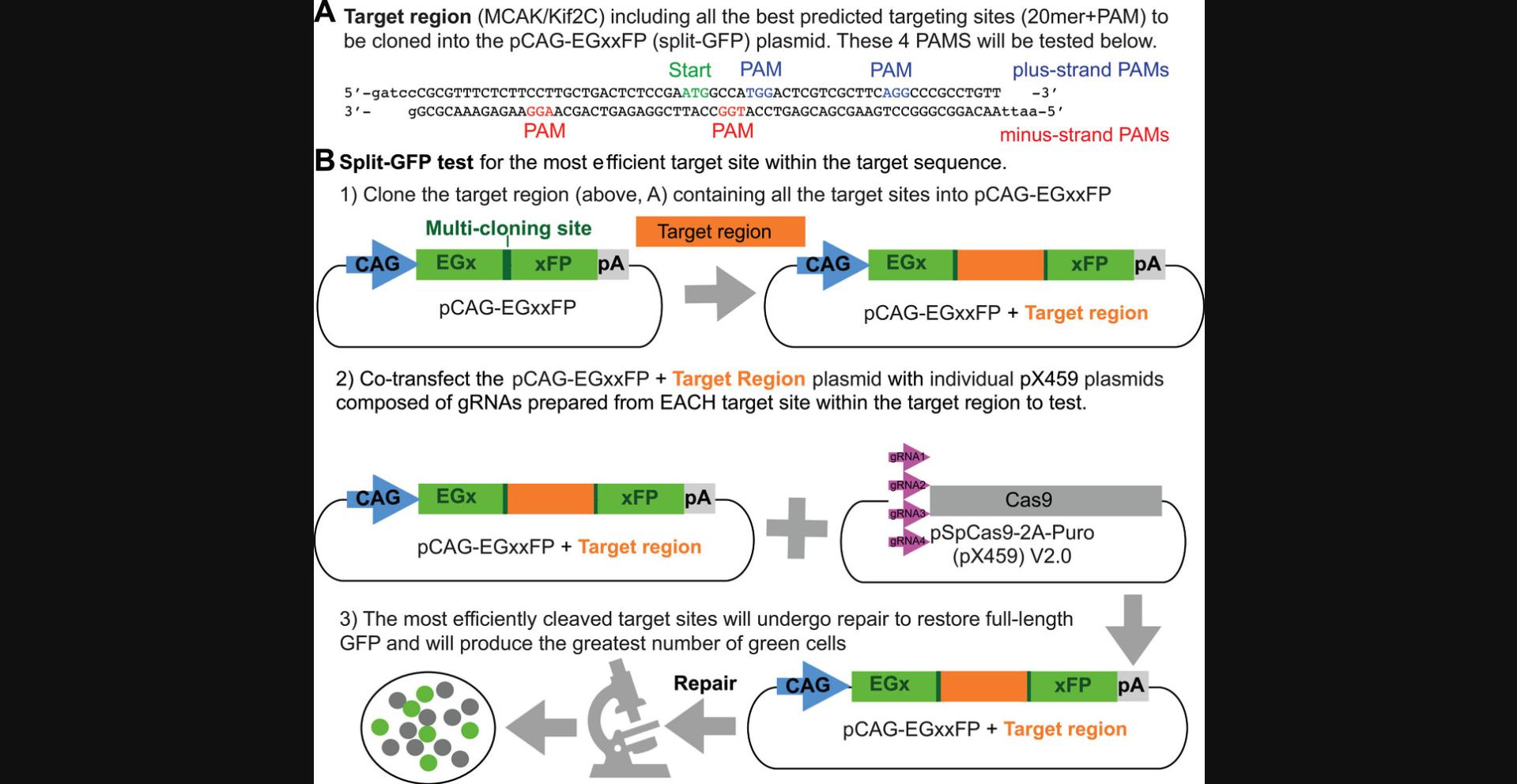
When the test plasmid is transiently transfected into cultured cells without a CRISPR targeting plasmid (Fig. 3D), a very low rate of spontaneous recombination will occur between the two defective copies of each GFP to generate one functional EGFP open reading frame (ORF) and thus a few green cells. In the presence of a functional CRISPR targeting plasmid (Fig. 3D), however, the cleavage of the targeted genomic sequence will generate an increased rate of recombination and an increased number of green cells, which can validate which of several targeting vectors is likely to be the most successful in generating an actual CRISPR cell line (Fig. 6B). Use of this EGFP recombination test (which takes only a couple of days) is not required, but we found it to be reassuring before investing the time (2 to 3 months) required to perform a CRISPR cell line construction. Our successful MCAK CRISPR cell line was derived from a transfection with the targeting plasmid (Fig. 3D) engineered from the successful protospacers (gRNA sense and antisense for MCAK/Kif2C, Table 1) that scored the best in this assay.
Note that you do not need to consider the reading frame of this sequence relative to the EGFP genes in pCAG-EgxxFP and it does not matter if your genomic sequence contains a stop codon in any reading frame. The in vivo recombination that generates the functional EGFP gene in this test assay works regardless of the sequence of the test insert.
Materials
-
pCAG-EgxxFP vector (Addgene plasmid #50716)
-
BamHI-HF (New England Biolabs R3136S)
-
EcoRI-HF (New England Biolabs R3101S)
-
T4 DNA ligase (New England Biolabs M0202S)
-
NEB 5-alpha c ompetent E. coli (New England Biolabs C2987H)
-
LB agar + ampicillin plates (see recipe)
-
TB medium + 50 µg/ml ampicillin (see recipe)
-
QIAprep spin miniprep kit (Qiagen 27106)
-
Qiagen HiSpeed Midi column kit (Qiagen 12643)
-
Appropriate mammalian cell line
-
Cell culture medium appropriate for your cell line (consult American Type Culture Collection [ATCC] for guidance)
-
10,000× SYBR Safe DNA stain (Invitrogen S33102)
-
50× TAE (see recipe)
-
Agarose (VWR 0710-25G)
-
Lambda DNA, BstEII Digest (New England Biolabs N3014L)
-
6× gel loading dye (New England Biolabs B7024S)
-
QIAquick gel extraction kit (Qiagen 28704)
-
BigDye 3.1 sequencing reagent (Fisher 43-374-54)
-
15-ml conical tube (Fisher 12-565-269)
-
10-ml sterile syringe (Fisher 14-955-459)
-
0.22-µm-pore-size sterile filter (Fisher 09-720-004)
-
100-mm Falcon petri dish (Fisher 08-757-100D)
-
6-well plate (Fisher 12-556-004)
-
500-ml 0.2-µm-pore-size filter unit (Fisher FB12566504)
-
Bacteriological incubator (VWR 10055-006 or equivalent), 37°C
-
Shaker (VWR 76407-108 or equivalent), 37°C
-
Horizontal electrophoresis apparatus (VWR 10001-836 or equivalent)
-
Electrophoresis DC power supply (VWR 76196-454 or equivalent)
-
Microcentrifuge for 1.5-ml tubes, >10,000 × g (VWR 76533-876 or equivalent)
-
Thermal cycler for 0.2-ml tubes (VWR 89217-558 or equivalent)
-
CO2 cell culture incubator (VWR 10810-744 or equivalent), 37°C
1.From your genomic sequence file (Basic Protocol 1, step 3), copy the minimum region needed to contain all of the targeting sites for which you made vectors in Basic Protocol 2, including the sequence of their PAMs.
2.To this sequence and its reverse complement, add sticky ends as shown in Figure 6A to give ends compatible with BamHI and EcoRI. Order oligos from IDT, specifying gel purification of full-length oligos. Dissolve received oligos at 100 µM concentration in sterile, distilled water.
3.Digest 5 µg pCAG-EgxxFP with 40 U BamHI-HF + 40 U EcoRI-HF as described in Basic Protocol 2, step 6; purify vector DNA from reaction using QIAquick gel extraction kit; anneal pair of test oligos as described in Basic Protocol 2, step 13 and 14; and ligate and transform competent bacteria as described in Basic Protocol 2, step 15 and 16.Plate on LB agar + ampicillin and grow up several minipreps in 2 ml TB medium + ampicillin, as described in Basic Protocol 2, steps 17 and 18.Prepare miniprep DNA with QIAspin mini columns, stain gel with 1× SYBR Safe DNA stain, screen for loss of the EcoRI site, and then sequence one or two “correct” clones with primer CAGforward to verify the insertion is correct, as described in Basic Protocol 2, steps 19 to 23, except using EcoRI.
4.Grow a large-scale (≥100 µg, as in Basic Protocol 2, step 24) prep of completed test vector with sequenced insert for transfection into cultured cells.
5.Set up cell transfections: mix equal amounts (amount as appropriate for the transfection method you prefer) of the EGxxFP test vector with your target region insert from step 4 and one of the targeting plasmids from Basic Protocol 2, step 24.Also set up a control transfection using your test vector and “empty” pX459. Transfect 106 mammalian cells and plate at a density of 200 cells/mm2 in cell culture medium appropriate for your cell line (consult ATCC for guidance). Seed cells on sterile round coverslips in multiwell TC plates if you don't have an inverted fluorescence scope that can directly view your TC plate.
6.Check dishes every 24 hr and replace with puromycin-free medium when significant cell death (floating cells) is observed.
7.Count green cells in a few fields of a fluorescence microscope. The control with the “empty” vector will have some green cells. These are spontaneous recombinants. Compare the number of bright cells in each targeted recombinant with each other and the control cells.
8.Make a note of the targeting plasmid(s) that produce the greatest number of green cells. These plasmids are likely to produce recombinants with the greatest efficiency.
Basic Protocol 4: DESIGN OF THE RECOMBINATION TEMPLATE AND ANALYTICAL PRIMERS
The recombination template DNA should possess regions that correspond closely to the genomic sequence outside any experimental insertions or mutations being made. Linearly speaking, from 5′ to 3′, it will consist of some homologous sequence 5′ to the start codon of your gene of interest (∼700 bp) and then DNA coding for a marker (such as GFP) that will contribute the new start codon for your protein of interest if you are modifying the N-terminus, followed by an in-frame linker, DNA coding for FKBP, a longer in-frame linker, and finally the DNA sequence coding for (in the case of MCAK/Kif2C) the expressed N-terminus (first exon) of your gene of interest and the following intron (∼700 bp). An example of a successful homologous recombination donor template used to engineer endogenous GFP-FKBP-MCAK/Kif2C is shown in Figure 7A. It is important to alter the sequence of the template to disrupt remaining CRISPR targeting sites or else the template DNA itself will be cleaved by the CRISPR targeting construct. If this occurs, the potential for generating the desired recombinant chromosome will be destroyed. CRISPR targeting sites are shown in Figure 7B and their positions indicated in the donor template sequence (Fig. 7A). One simple way to disrupt the remaining CRISPR targeting sites is to alter the PAM so that it no longer contains the GG dinucleotide, by replacing one or both Gs with any other base through silent mutation. Base-pairing of the sgRNA targeting sequence within the 20-mer chosen can also be altered by silent mutations. Thus, the recombination construct (other than inserted regions) will almost completely match the genomic DNA sequence. However, in the region corresponding to the targeting locations, it must contain at least one nucleotide that does not match the genomic sequence. For example, if you are constructing a fusion protein and the sgRNA targeting sequence contains either the start codon or the stop codon (a “stop” of course will be deleted to create a fusion), the addition of the fusion protein sequence will disrupt the sgRNA targeting sequence and may make other alterations unnecessary. An example of this is shown in Figure 7B, wherein three PAM sequences whose sgRNAs do not significantly overlap the start codon are altered, while a fourth PAM very close to the start codon is left unchanged because the homology in that region will already be disrupted by the newly introduced linker sequences. If you are in doubt, you may still want to disrupt the PAM sequence or the target near the PAM if possible, just to be sure that it will not be used for cleavage. Compare the sequence of the first few base pairs of the MCAK recombination template (the target region) from Figure 6A, ATGGCaA, with the corresponding sequence from successful MCAK CRISPR targeting vector 1+ from Table 1, row 5: ATGGCCA. The PAM immediately follows this sequence (Fig. 7B, red nucleotides) but could not be altered by silent mutation, so one C-to-A silent mutation in the target was made (Fig. 7A, lower red box). The rest of the 20-mer target for this construct is on the other side of the EGFP-FKBP insert, leaving only 7 bp of target homology before the PAM, so it may be that this site would have been essentially safe from degradation without this mutation, but it was easy to include it.
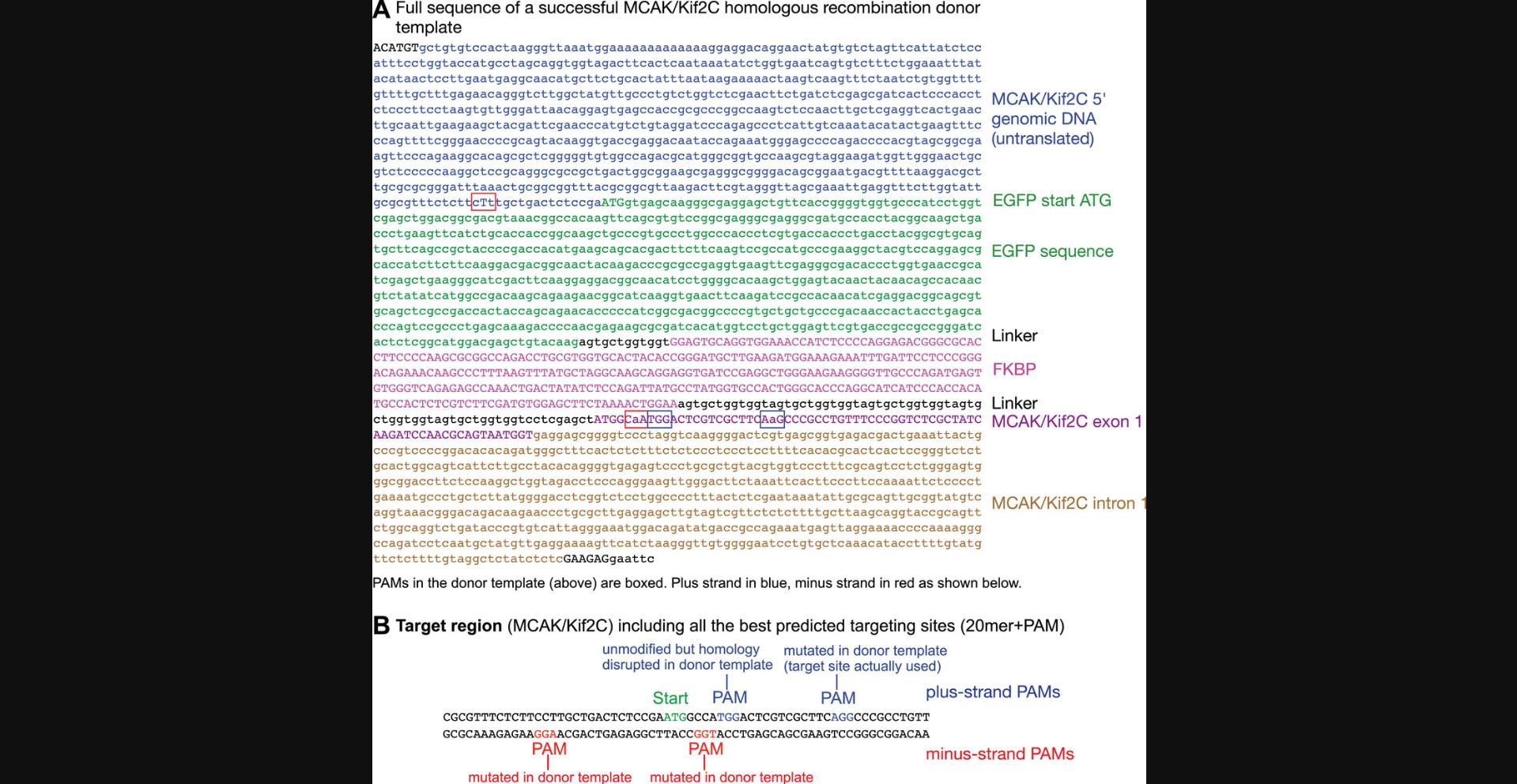
This protocol requires no sequence differences in the 5′ UTR before the start codon, except as described in the preceding paragraph for protecting the template from CRISPR degradation (if the target site is in the 5′ UTR). This would be the situation most likely to result in normal endogenous levels of protein production, because the 5′ UTR in your CRISPR cell will have no changes relative to the original wild-type sequence.
Materials
- Computer and DNA analysis software as in Basic Protocol 1
1.From the sequence file created in Basic Protocol 1, step 3, copy everything from 700 bp before the start codon through 700 bp after the start codon to a new file. This is your recombination homology region.
2.Simply insert the additional sequences desired (e.g., EGFP and FKBP) into the sequence from Basic Protocol 4, step 1, immediately after the start codon of your gene, so that they form a continuous ORF with the first exon.
3.Locate the targeting sites selected in Basic Protocol 2, step 3, in the sequence(s) designed in Basic Protocol 4, step 2.If the insertions you have made in the sequence have not completely severed the PAMs from the targeting sites, edit the sequence file from Basic Protocol 4, step 2, to make silent changes in the PAM or target (as described in the introduction to Basic Protocol 4) to prevent CRISPR attack on the template.
4.To the extreme ends of the sequence(s) from step 3, add restriction sites that are not found anywhere in the template sequence, to be used to cut the template from the vector prior to co-transfection of cells with the linearized template and the CRISPR targeting vector. These two sites can be for a single enzyme, because there is no need for directional ligation of the digestion product.
5.Order the synthetic DNA for the sequence(s) from step 4, and design and order analytical primers as described below in Support Protocol 1.
6.When synthetic DNA is received, make a large-scale plasmid preparation (as in Basic Protocol 2, step 24) and proceed to Basic Protocol 5.
Support Protocol 1: DESIGN OF PRIMERS FOR ANALYTICAL PCR
Verification of CRISPR-directed recombination into the chromosomal locus can be conveniently performed by PCR of genomic DNA made from a clonal population of CRISPR-treated cells. This will give you a band whose length corresponds to the predicted size of a recombinant gene. Non-recombined clones will produce shorter PCR products. The primers designed in steps 1 and 2 are both capable of analyzing whether a recombination has occurred at your expected CRISPR targeting site at the locus of interest. However, it is formally possible (although not very likely) that your entire recombination template could integrate in an unexpected site elsewhere in the genome (an “off-target recombination”). If that were to occur, the primers in step 2 would still amplify a product from that random integration of the template that looked like the desired recombinant, while also amplifying the wild-type-sized product from the unmodified locus of interest. In other words, the PCR from this off-target recombination event would look like that for a hemizygous clone (one with one modified allele and one wild-type allele at the locus of interest). Meanwhile, the primers from step 1 would amplify only the wild-type-sized product and would reveal that an off-target recombination had occurred, not a recombination at the locus of interest, because they would amplify only a wild-type-sized product. In contrast, the oligos designed in step 2 produces a smaller product, which is likely to be technically easier to amplify and which will show a greater relative difference in length for recombinant vs. wild-type cell clones. As primers are easy to design and cheap, and PCR is fairly quick, it is worth having both pairs to use and generate redundant evidence of the recombinant product.
Materials
- Computer and DNA analysis software as in Basic Protocol 1
1.Choose a pair of forward and reverse primers that bind to the chromosome on either side of the CRISPR targeting site, near but outside the region included in the recombination template plasmid as defined in Basic Protocol 4, step 1.These will typically be ∼1.5 kb apart on the wild-type chromosome.
2.Choose a pair of forward and reverse primers that bind 200-300 bp on either side of your target site. From the wild-type chromosome, these will give a product of ∼0.5 kb, which is about the smallest we find convenient to look at on a gel. The EGFP-FKBP insert that we used created a recombinant chromosome that produced a PCR product ∼1.1 kb larger.
3.Choose at least one primer within your insert that is not expected to be found in the genome (or at least not nearby on the relevant chromosome) of your cells. This can be used for PCR with the opposite-strand primer from either primer pair in step 1 or 2 to give a product only in the case of chromosomes that have your specific insert.
Basic Protocol 5: TRANSFECTION, ISOLATION, AND VALIDATION OF ENGINEERED CELLS
If you have completed Basic Protocols 1 to 4 correctly and are using a diploid human cell line with minimal structural rearrangements to the chromosomes, it is likely that you will be able to successfully engineer cell lines possessing the modifications you desire. The remaining challenge, and this can be significant, is to find the engineered cells within the vast majority of rapidly growing non-recombinant cells. Your GFP tag, which will prove useful for future imaging and performing experiments, will be helpful for this process as well. You can use a microscope and select clones by eye (we have done this successfully). But often the endogenous protein is expressed at modest levels, making this method challenging. More routinely, we use FACs to isolate the cells expressing GFP at the highest levels and then plate them sparsely, after which we search for clones by eye using a fluorescence microscope. This method assumes that you are using a cell line that is tolerant of being grown highly dispersed on plates in cloning steps 9 and 10.An advantage of this method is that you can prioritize clones that are uniform, normal looking, and show appropriate spatial localization of your protein of interest up front, rather than isolating clones blind and using PCR to detect recombinants. It is also possible, depending on the microscope, to measure the expressed protein fluorescently “on the fly” and parse lower- versus higher-expressing cells to enrich for homozygosity.
Materials
-
Recombination plasmid (Basic Protocol 4)
-
Appropriate restriction enzymes
-
CRISPR targeting plasmid (Basic Protocol 2)
-
Phenol (Sigma P4557)
-
Chloroform
-
3 M sodium acetate, pH 5.2 (see recipe)
-
Isopropanol
-
70% (v/v) ethanol (prepared from 200-proof ethanol, Fisher 04-355-222)
-
TE buffer (see recipe)
-
pX459
-
GFP reporter plasmid, such as pmaxGFP
-
Appropriate cultured cells
-
Cell Line Nucleofector Kit V (for Hct116 or hTERT-RPE; Lonza VCA-1003)
-
Medium appropriate for the cell line: e.g., we use RPMI 1640 (Fisher 11-875-093) + 10% FBS (Hyclone SH30070.03) and 1× Pen-Strep (Fisher 30-002-CI)
-
Puromycin (Fisher BP 2956-100)
-
Trypsin/EDTA (Fisher 25-200-056)
-
Dimethyl sulfoxide (DMSO)
-
PBS (Fisher 10-010-023)
-
Proteinase K (Fisher FEREO0491)
-
Pfusion High-Fidelity PCR kit (Fisher F553S)
-
Lonza nucleofector II
-
BD Aria III cell sorter
-
Inverted fluorescence microscope (such as a Nikon Ti2)
-
MatTek 35-mm glass-bottom dish, no. 1.5, poly-D-lysine coated. (cat. no. P35GC-1.5-14-C)
-
Optional: MatTek 50-mm dish, no. 1.5, poly-D-lysine coated (cat. no. P50GC-1.5-14-F)
-
6-well plate (Fisher 12-556-004)
-
10-cm plates (Fisher FB012924)
1.Digest the recombination template plasmid at the restriction sites added in Basic Protocol 4, step 4, and then inactivate the restriction enzyme(s), typically by phenol extraction and isopropanol precipitation, as described in Basic Protocol 2, steps 7-12.
2.Mix 4-µg aliquots of digested recombination template + 1 µg of each CRISPR targeting plasmid from Basic Protocol 2, step 24, or preferably use one or two of the targeting vectors that were most successful in the EGxxFP test in Basic Protocol 3.
3.Set up a control transfection with 1 µg of empty pX459 and an appropriate amount of a GFP reporter plasmid such as pmaxGFP.
4.Transfect 2 × 106 cultured cells with DNA mixes from steps 2 and 3.Plate as in Basic Protocol 3, step 5, preferably in 10-cm dishes to maximize the isolation of clones of cells. The greater the number of cells you plate, the higher the likelihood of finding recombinant colonies of cells, which are often present at low numbers.
5.After ∼24 hr, view the reporter plate from step 3 and verify that the number and proportion of transfected cells is as expected for a typical, successful transfection with your method and cell type. If the transfection looks good, add puromycin to all plates at a concentration appropriate for your cell line.
6.Check dishes every day to look for cell death (floating cells). Puromycin kills quickly, so 1-3 days of treatment should be enough.
7.Recombinant cells must be located among the surviving cells. Let cells grow after puromycin selection until the survivors appear to have formed small colonies (a few dozen to a couple of hundred cells per colony, depending on how crowded the plates are). View these in an inverted fluorescence microscope each day or so. If you are lucky, your gene will be expressed at a high enough level that you will be able to see GFP-expressing colonies. If green colonies are visible, you may be able to remove them directly from the plate; to do this, proceed to step 9.Alternatively, you can considerably enrich your cells for recombinants by subjecting them to a cell sorting step to collect GFP-labeled cells.
8.Allow the cells to grow until a 10-cm plate is near confluence, and then use single-cell FACS to try to sort out the brightest 1% of the cells. Generally this will be performed in a university FACS facility on a cost-per-run basis and under the guidance of a skilled technician. At the University of Washington, a typical FACS sort will cost ∼$300, but the procedure greatly increases the likelihood that we will find recombinant cells. After collecting several thousand cells, plate a proportion (about one-third) with sufficient dilution to ensure the formation of isolated colonies in step 9i. Culture the remainder in a 6- or 10-cm dish for a few days. Trypsinize cells, dilute into two or three aliquots, and freeze in cell culture medium + 10% DMSO. These cells will serve as a back-up reserve in case it becomes necessary to isolate more colonies at a future date.
9.Clone recombinant cells by limiting dilution (meaning from single cells). If you prefer, you can do this using a 96-well plate. However, we prefer the utility and speed of selecting individual colonies on a larger, sparse plate by eye, as described below.
1.Wash the plate with PBS or serum-free medium and leave a thin layer of the same solution on the plate. 2.Locate patches of green cells under the inverted fluorescence microscope and leave plate on stage. 3. Gently apply several drops of trypsin/EDTA solution mixed 1:1 with PBS to each uniform and brightly labeled colony. 4.Wait until cells appear to be coming loose from the plate. Then, aspirate each colony with a pipettor (a 200-µl [P200] pipettor set to 30-50 µl usually works well) and dilute cells from each colony into 1-ml aliquots of complete medium with serum. 5.Estimate how many cells are in the aliquots. Replate these cells such that you expect them to be disperse enough to grow up into well-isolated colonies of ∼100 cells. We usually use 6-well plates for this step.
Ideally, you will work up clonal cells from five to ten colonies from the initial transfection (probably not all will be homozygous; see step 20). If you do not plate all the cells, you may want to save the remainder growing in another plate, at least until you are fairly confident that the cells plated for colonies are growing properly. This excess cell population could also be frozen to clone out more colonies later, just to hedge your bets.
10.If you are confident that the colonies in step 9 are truly clonal, proceed to step 12.If you want to be extra cautious, pick one colony from each “clone” and repeat step 9 once more, plating clones into individual wells of 6-well plates.
11.At this time, you should also start growing some wild-type cells to be used for a control genomic DNA prep in steps 13-18.
12.Trypsinize cells gently (as described in step 9i) while on the microscope stage and pull a colony of ∼100 cells from each clone. Replate each clone in one well of a 6-well plate. Let grow until near confluence.
13.Trypsinize each clone in each well of the 6-well plate from step 12 and suspend in 3 ml RPMI + 10% FBS, as follows:
- Remove growth medium from each well and add PBS to the cells.
- Remove PBS and add 0.2 µl trypsin/EDTA to each well.
- Triturate (pipet up and down repeatedly) cells in each well with a fresh pipet tip.
14.Roughly count the number of cells collected from each well.
15.Replate 10% of the cells in a 10-cm dish, and process these cells as described in step 21.These will be your recombinant CRISPR cells, assuming that they pass the genomic DNA analysis.
16.Gently pellet the remainder of the cells and resuspend in 100 µl PBS. Collect a similar sample of wild-type cells. If not all cells are ready on the same day, cells in PBS can be frozen until all are ready.
17.Perform genomic DNA preps with proteinase K digestion and phenol extraction as described in Koh (2013) or Moore and Dowhan (2002) or using a similar protocol, scaled as appropriate for the number of cells collected.
18.Dissolve the final DNA pellet in 200 µl TE buffer. Genomic DNA can be hard to redissolve, so incubate overnight at room temperature. Mix thoroughly but gently.
19.Do PCRs with analytical primers from Support Protocol 1 and 1 µl of each genomic DNA prep, as directed in your PCR kit instructions. Analyze products on 1% agarose gel as described in Sambrook et al. (1989) or Voytas (2000).
20.Score band patterns for expected lengths from wild-type or recombinant chromosomes. For primer pairs from Basic Protocol 5, step 1 or 2, homozygous clones (which have had both alleles modified by recombination) will give only a larger band relative to the wild type, whereas hemizygous clones (only one allele modified) will give both the wild-type and the expected larger band.
21.When the 10-cm dishes from step 12 are near confluence, passage to a new dish, collect cells that are not replated, aliquot, and freeze in 10% DMSO. Store frozen cell aliquots in liquid N2. Cells that scored correctly for PCR band patterns in step 16 are now ready to use experimentally or can be assayed by western blotting (according to Harlow, & Lane 1988, pp. 480–507, or Gallagher et al., 2008) for the correct size gene product, as an optional confirmation that the expected fusion protein is being expressed. Once you are satisfied with the status of the isolated cell lines, you can prepare stable cell lines from your isolates that express binding partners (Support Protocol 2).
Support Protocol 2: STABLE TRANSFECTION OF ENGINEERED CELLS WITH BINDING PARTNERS
Obtaining a CRISPR-engineered cell line expressing a GFP-FKBP-modified gene can be useful without further modification because such cell lines enable researchers to track endogenous proteins live. However, once modified with an FKBP tag, the proteins can be relocalized to any region of the cell in which FRB stably resides. We have engineered a plasmid, pMX1463 (available from Addgene.com), in which FRB has been modified with an 18-aa Src homology 4 domain (SH4) to localize FRB to the membrane (Suh et al., 2006) and SBFP2 (Strongly-enhanced Blue Fluorescent Protein; Kremers et al., 2007) to visualize and assess this localization. The labeling enables cell selection of stable transformants using G418 that can be visually assayed. One disadvantage of labeling with SBFP2 is that it is antigenically identical to GFP and will be labeled with anti-GFP antibodies should you want to amplify your signal using immunofluorescence. Thus, BFP labeling will interfere with immunodetection of GFP should you want to do that. A solution is to use another fluorescent protein or labeling tag that does not share antigenicity with GFP to evaluate the localization of your FRB binding partner.
Materials
-
CRISPR cell line (Basic Protocol 5)
-
pMX1463, LDR-FRB-SBFP2 plasmid (AddGene 208805)
-
Cell Line Nucleofector Kit V (for Hct116 or hTERT-RPE; Lonza VCA-1003)
-
RPMI 1640 (Fisher 11-875-093), with and without 10% FBS (Hyclone SH30070.03) and 1× Pen-Strep (Fisher 30-002-CI)
-
Puromycin (Fisher BP 2956-100)
-
G418 (geneticin)
-
200 mM L-glutamine (Fisher 25-030-081)
-
Rapamycin
-
Lonza nucleofector II
-
BD Aria III cell sorter
-
Inverted fluorescence microscope (such as a Nikon Ti2)
-
MatTek 35-mm glass-bottom dish, no. 1.5, poly-D-lysine coated (cat. no. P35GC-1.5-14-C)
-
Optional: MatTek 50-mm dish, no. 1.5, poly-D-lysine coated (cat. no. P50GC-1.5-14-F)
-
6-well plate (Fisher 12-556-004)
-
10-cm plates (Fisher FB012924)
1.Transfect your CRISPR cell line with 1-2 µg plasmid pMX1463 into 2 × 106 cells according to the Nucleofector II instructions.
2.Once the cells have recovered from transfection, plate the entire 500-µl tube of transfected cells in RPMI (no additives) into a 10-ml dish containing standard cell culture medium (with 10% FBS, 1× Pen/Strep, and L-glutamine). Let the cells grow and express overnight.
3.The next day, replace the medium with medium containing G418.We have used 1000 µg/ml for Hct116 cells.
4.After 6-8 days, you should begin to see massive cell death (in the form of floating cells) and small colonies of surviving cells.
5.Observe the colonies in a fluorescence inverted microscope. Not all of the colonies will be fluorescent even if you transfect your cells a fluorescent-protein-expressing construct.
6.Systematically look over all of the dish and identify colonies with bright, nicely even-looking cells that appear healthy. Mark the best-looking colonies with a Sharpie pen by either circling them or putting a dot beside them on the bottom of the dish. Hint: the red Sharpies can be seen under the fluorescence microscope!
7.Isolate colonies as described in Basic Protocol 4, steps 9-11.Once the cells are pure, you can lower the G418 concentration to a maintenance level of ∼200 µg/ml.
8.Plate the purified cells on a MatTek 35-mm glass-bottom dish. Let them grow for 24-48 hr, add 0.5 µM rapamycin, and evaluate the cells under the fluorescence microscope live to score for relocalization of your GFP-engineered protein.
REAGENTS AND SOLUTIONS
Ampicillin, 100 mg/ml
Place 9.5 ml sterile (previously autoclaved and cooled) distilled water in a sterile 15-ml conical tube. Add 1 g ampicillin sodium salt (Sigma A0166), cap, and mix by inversion until dissolved. Attach a 0.22-µm-pore-size sterile filter (Fisher 09-720-004) to a 10-ml sterile syringe(Fisher 14-955-459). Decant ampicillin solution into syringe and filter into a new sterile 15-ml conical tube. Store at −20° up to 1 year. Thaw by standing tube in a beaker of water at room temperature for 20 min (do not heat solution).
EDTA-Na, 0.4 M, pH 8.0
Place 400 ml distilled water in a 0.5-liter beaker. Add 74.4 g EDTA disodium salt dihydrate (Sigma ED2SS) and stir, slowly adding NaOH (Fisher BP358-212) until all EDTA dissolves (wear appropriate personal protective equipment [PPE] for working with NaOH). Adjust pH to 8.0 with additional NaOH, and then adjust volume to 0.5 liter. Filter sterilize through 500-ml 0.2-µm-pore-size filter unit. Store up to 5 years at room temperature.
LB agar ± 50 µg/ml ampicillin plates
- 1 L distilled water
- 20 g LB Broth Base (ThermoFisher 12780052; 2% w/v)
- 17 g agar (Sigma A1296; 1.7% w/v)
Place all reagents in 2-liter glass Erlenmeyer flask, capped loosely with aluminum foil. Autoclave 40 min at 121°C. Let cool to 45-50°C. Add 0.5 ml of 100 mg/ml ampicillin solution (see recipe). Swirl well to mix, and immediately pour into 100-mm petri dishes, filling dishes half to two-thirds full. Let stand at room temperature until completely solidified (2-16 hr) and then dishes place in plastic bag and store up to 10 weeks at 4°C.
Sodium acetate, 3 M, pH 5.2
Place 120 ml distilled water in a 250-ml beaker. Add 81.6 g sodium acetate trihydrate (J.T. Baker 3460-01) and stir to dissolve. Adjust pH to 5.2 with concentrated acetic acid (Fisher A38C-212; wear appropriate PPE and work in a fume hood). Adjust volume to 200 ml with additional distilled water. Place in a 250-ml glass bottle and autoclave 15 min at 121°C. Store up to 1 year at room temperature.
TAE, 50×
Place 700 ml distilled water in a 1-liter graduated cylinder. Add 242 g Tris base (Fisher BP152-1) and stir to dissolve. Add 57 ml concentrated acetic acid (Fisher A38C-212; wear appropriate PPE and work in a fume hood) and then 125 ml of 0.4 M EDTA-Na, pH 8.0 (see recipe). Adjust final volume to 1 liter. Store up to 5 years at room temperature.
TB medium ± 50 µg/ml ampicillin
Solution A : Place 900 ml distilled water in a 1-liter bottle. Add 12 g N-Z Case Plus (Sigma N4642), 24 g yeast extract (Sigma Y-0875), and 5 g glycerol (VWR BDH172-1LP); swirl until reagents dissolve. Autoclave 40 min at 121°C. Let cool to room temperature. Store up to 1 year at room temperature.
Solution B : Place 450 ml distilled water in a 0.5-liter glass bottle. Add 62.6 g dibasic potassium phosphate (Fisher P288-500) and 11.5 g monobasic potassium phosphate (Fisher P285-500); swirl until reagents dissolve. Autoclave 40 min at 121°C. Let cool to room temperature. Store up to 5 years at room temperature.
To prepare TB medium : Add 100 ml solution B to 900 ml solution A (both must be at room temperature or a precipitate will form). Add 0.5 ml of 100 mg/ml ampicillin (see recipe) to completed (A + B) TB medium. Store up to 10 weeks at 4°C.
TE buffer
- 10 mM Tris base (Fisher BP152-1), pH adjusted to 8.0 with HCl
- 1 mM EDTA (Sigma ED2SS), pH adjusted to 8.0 with NaOH
- Store up to 1 year at room temperature
COMMENTARY
Background Information
Most protocols relying on gene product depletion (such as siRNA or CRISPR knockout) are dependent on protein and mRNA turnover before their depletion phenotypes can be identified and analyzed. This has been a disadvantage of mammalian cell culture systems for many years because it allows cells to activate compensatory mechanisms that can obscure the phenotype. Both rapamycin-dependent “knocksideways” and optogenetic approaches can provide almost instantaneous (<1 to 10 min) removal of select proteins from their cellular locations to allow evaluation of their functional contribution to cellular processes. This surpasses even degron activation, which, in our hands, requires at least 30 min to 2 hr to noticeably deplete MCAK/Kif2C protein (Wordeman et al., 2016), although this timing may be somewhat protein specific. Ultimately, by combining knocksideways with CRISPR engineering, it is possible to perform experiments with endogenous protein levels in a cellular background. This increases the sensitivity of the protein depletion assay overall.
Critical Parameters
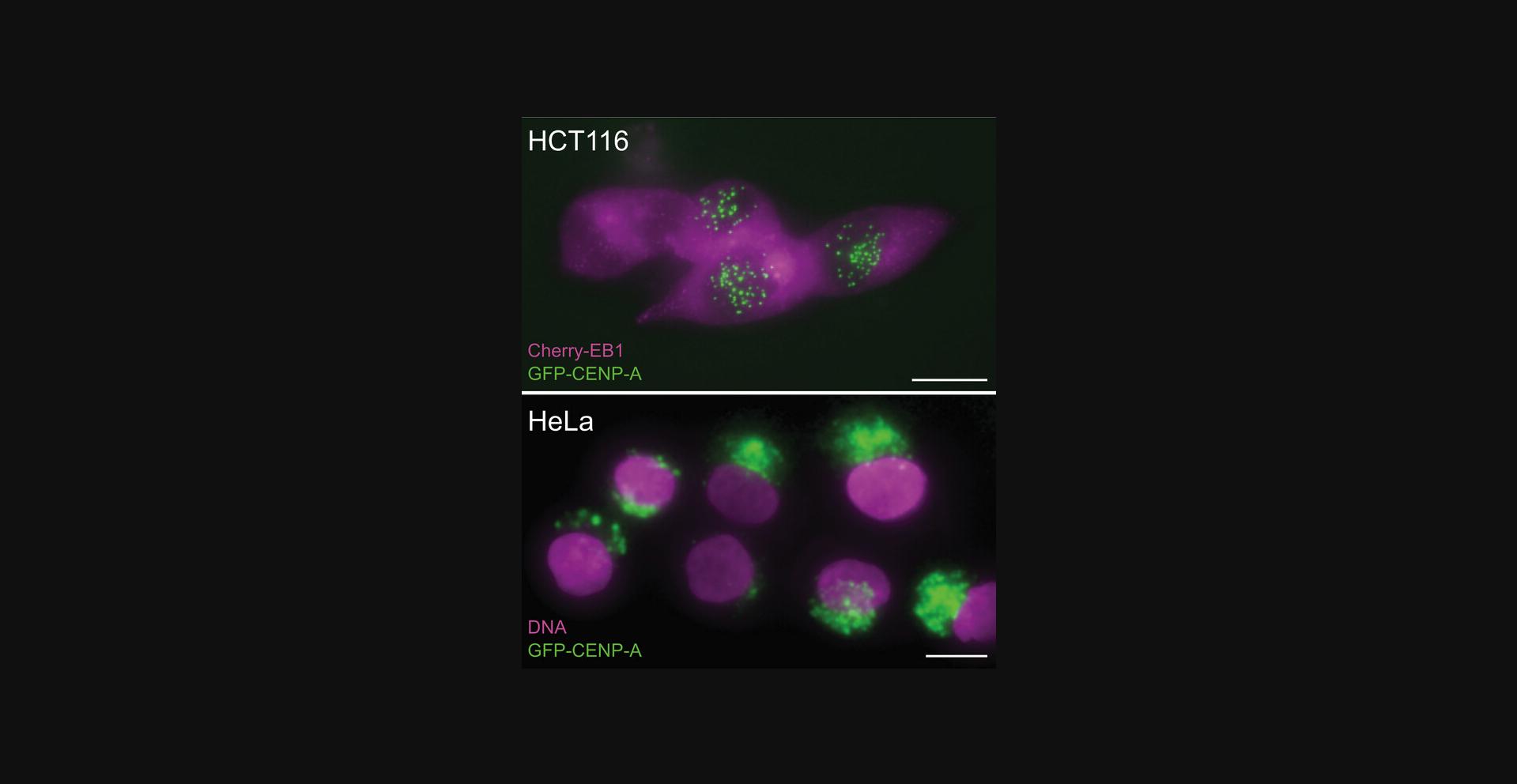
The most critical parameter we have encountered related to the success in engineering the cell lines is the use of parent cell lines with minimal chromosomal structural rearrangements surrounding the gene of interest. We have been able to engineer our GFP-FKBP onto every gene that we have so far attempted (MCAK/Kif2C, ch-TOG, CENP-F, Mad1) and also fluorescent tracers (CENP-A, EB1) plus combinations thereof. The only consistent lack of success we have had is in attempts to engineer HeLa cells—a cell line with notoriously rearranged chromosomal structure. Our attempt to prepare endogenously modified GFP-CENP-A is illustrated in Figure 8. The top panel shows live Hct-116 cells endogenously labeled with GFP-CENP-A and EB1-Cherry. Green centromeres are visible within the nucleus. In contrast, live HeLa cells engineered using the same constructs are shown in the lower panel. It is interesting to note that while Neumann et al. (2010) were able to successfully tag a large number of proteins using CRISPR in HeLa-Kyoto cells, they did not, to our knowledge, succeed in CRISPR engineering GFP-CENP-A in that cell line. Instead, stable GFP-CENP-A HeLa-Kyoto cell lines were prepared (Cai et al., 2018; Mahen et al., 2014). In this case the green label is found in autophagosomes, which is indicative of increased turnover of GFP-linked protein, likely due to misfolding (Metcalf et al., 2012). Thus, based on our experience, parent cell line choice is the most critical determinant of success.
Troubleshooting
A troubleshooting guide for some common problems is provided in Table 2.
| Problem | Possible cause | Solution |
|---|---|---|
| No green cells | Recombination is rare | Try recombination again and use FACs to isolate the brightest 1% of cells. |
| Recombination failed | Check for nearby active PAM sites. | |
| Green signal in lysosomes or autophagosomes | Off-target recombination due to chromosome structural rearrangements | Try a chromosomally stable cell line. |
| Anomalous genomic PCR | Off-target recombination | Pick a less promiscuous PAM; switch to a nickase-based protocol. |
| No homozygous recombinants | Recombination is rare | Try rescreening potential recombinants from FACs for the very brightest cells relative to positive cells. |
Understanding Results
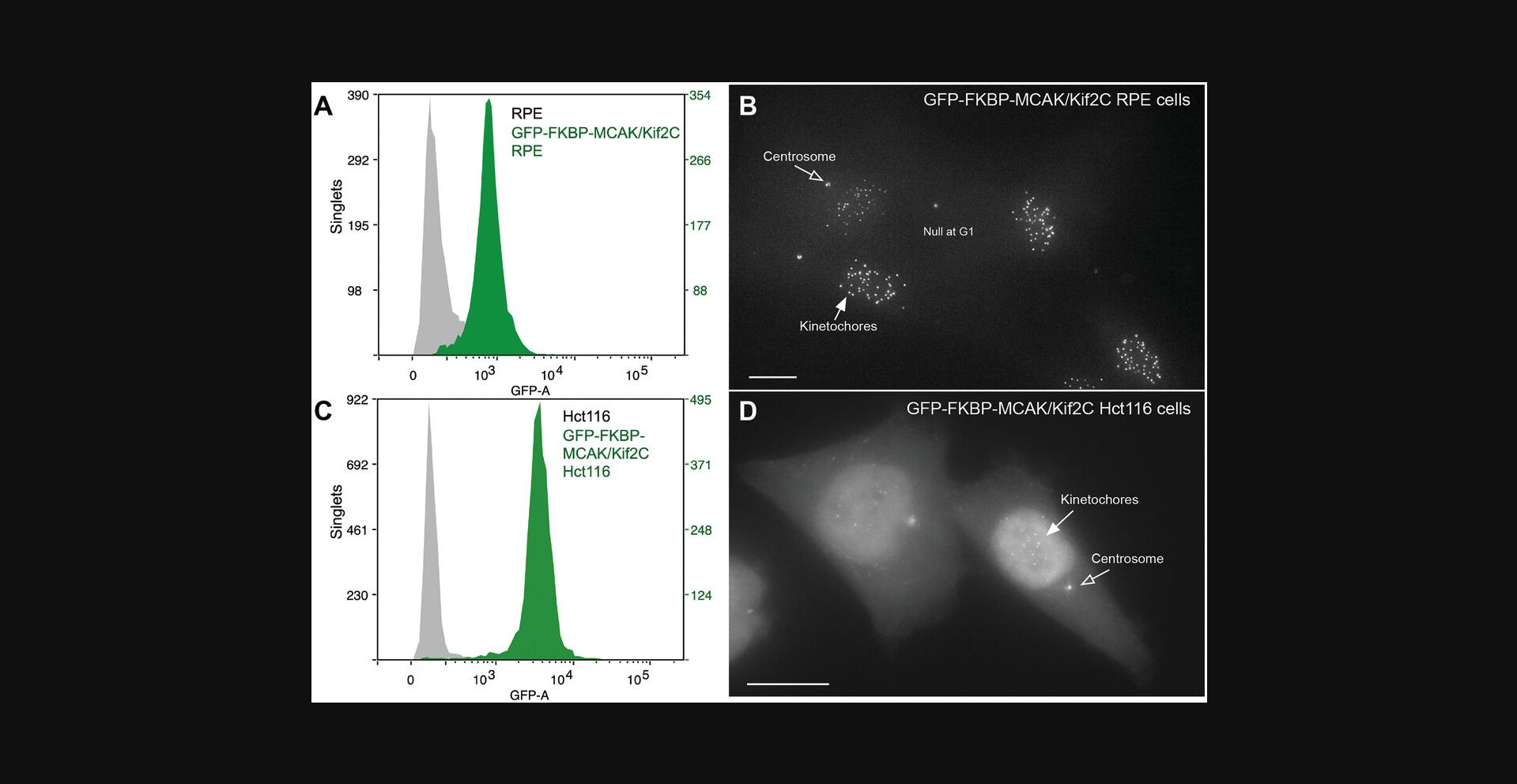
Two successful live cell lines in which either MCAK/Kif2C or ch-TOG can be relocalized to the cell membrane once the lines are stably transfected with BFP-FRB-SH are shown in Figure 2. When two different human cell lines are used, the endogenous protein is often expressed at different levels. For example, Figure 9A and B show a purified TERT-RPE cell line that has been engineered to express GFP-FKBP-MCAK at endogenous levels. This cell line often serves as a model for normal, untransformed cells. Consistent with the published literature, a cancer cell line (Hct-116) expressing GFP-FKBP-MCAK does so at measurably higher endogenous levels (Fig. 9C and D).
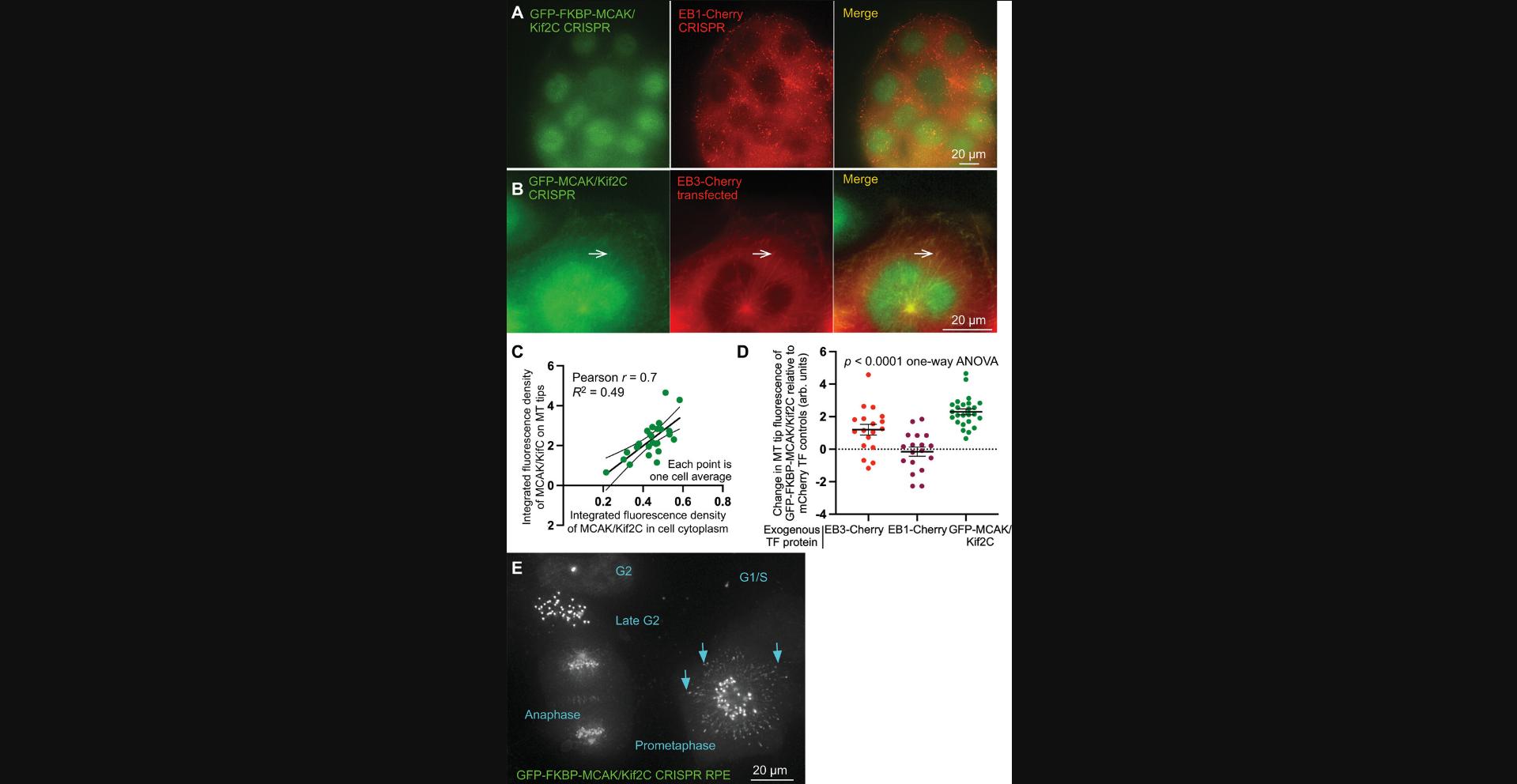
Additionally, careful analysis of the CRISPR-engineered MCAK/Kif2C cell lines reveals some deviations from results in the published literature simply by virtue of the fact that the engineered protein is expressed at endogenous levels (Fig. 10). MCAK/Kif2C possesses a SxIP domain that allows it to be recruited to microtubule plus-ends via end-binding (EB) proteins (Honnappa et al., 2009; Montenegro Gouveia et al., 2010). In human cells, there are three EB variants: EB1 (which is ubiquitously expressed), EB2 (ubiquitously expressed but at low levels), and EB3 (specific to brain and skeletal muscle) (Straube & Merdes, 2007). The tissue-specific variant, EB3, is used most often to measure microtubule dynamics because it associates robustly with microtubules with high visibility (Stepanova et al., 2003). When transfected into cells, GFP-MCAK/Kif2C appears robustly on microtubule ends as predicted. However, GFP-MCAK/Kif2C-FKBP CRISPR-engineered cells do not show an association of MCAK/Kif2C with interphase microtubule plus-ends (Fig. 10A). In contrast, when EB3-Cherry is transfected, MCAK/Kif2C becomes visibly associated with microtubule plus ends (Fig. 10B, arrows). Interestingly, when excess GFP-MCAK/Kif2C is transfected into cells, it associates with microtubule plus-ends in a dose-dependent manner (Fig. 10C). Furthermore, Figure 10D shows that the quantity of GFP-MCAK measured that was associated with MT plus-ends (y -axis) was dependent on the exogenous expression of excess GFP-MCAK/Kif2C (green points) or excess EB3-Cherry (red points) but was not influenced by excess EB1-Cherry expression (dark red points). It was surprising that endogenous MCAK/Kif2C was not associated with interphase MT plus-ends because it possesses an SxIP domain to recruit the protein to that position. The solution to this conundrum is found in Figure 10E, which shows that MCAK/Kif2C associates with MT tips only transiently at prometaphase (arrows). This is interesting as it pinpoints the time at which MT-plus-end-associated MCAK/Kif2C is likely to have its greatest influence. Importantly, these new data were discovered using live cells engineered with tagged endogenous protein.
Time Considerations
Basic Protocol 1, choosing a target site for gene modification: 1 day
Basic Protocol 2, design and construction of gRNA(s) for targeted gene modification: 1 week
Basic Protocol 3, split-GFP test for target efficiency: 1 week (may partially overlap with time spent on Basic Protocol 2)
Basic Protocol 4, design of the recombination template and analytical primers: 2 days
Basic Protocol 5, transfection, isolation, and validation of engineered cells: 6-10 weeks
Acknowledgments
This work was supported by awards GM069429 and GM145567 from the US National Institutes of Health and award 2319918 from the National Science Foundation. J.J.V. is also supported by the Royalty Research Fund of the University of Washington.
Author Contributions
Michael Wagenbach : Conceptualization; formal analysis; methodology; validation; writing—original draft. Juan Jesus Vicente : Data curation; methodology; software; visualization. Wren Wagenbach : Formal analysis; investigation; methodology; visualization. Linda Wordeman : Conceptualization; funding acquisition; project administration; validation; visualization; writing—review and editing.
Conflict of Interest
We have no conflicts to report.
Open Research
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request. Materials used in this study are openly available at Addgene.org.
Literature Cited
- Bayle, J. H., Grimley, J. S., Stankunas, K., Gestwicki, J. E., Wandless, T. J., & Crabtree, G. R. (2006). Rapamycin analogs with differential binding specificity permit orthogonal control of protein activity. Chemistry & Biology, 13(1), 99–107. https://doi.org/10.1016/j.chembiol.2005.10.017
- Brouhard, G. J., Stear, J. H., Noetzel, T. L., Al-Bassam, J., Kinoshita, K., Harrison, S. C., Howard, J., & Hyman, A. A. (2008). XMAP215 is a processive microtubule polymerase. Cell , 132(1), 79–88. https://doi.org/10.1016/j.cell.2007.11.043
- Cai, Y., Hossain, M. J., Heriche, J. K., Politi, A. Z., Walther, N., Koch, B., Wachsmuth, M., Nijmeijer, B., Kueblbeck, M., Martinic-Kavur, M., Ladurner, R., Alexander, S., Peters, J. M., & Ellenberg, J. (2018). Experimental and computational framework for a dynamic protein atlas of human cell division. Nature , 561(7723), 411–415. https://doi.org/10.1038/s41586-018-0518-z
- Cheeseman, L. P., Harry, E. F., McAinsh, A. D., Prior, I. A., & Royle, S. J. (2013). Specific removal of TACC3-ch-TOG-clathrin at metaphase deregulates kinetochore fiber tension. Journal of Cell Science , 126(Pt 9), 2102–2113. https://doi.org/10.1242/jcs.124834
- Cherry, A. E., Vicente, J. J., Xu, C., Morrison, R. S., Ong, S. E., Wordeman, L., & Stella, N. (2019). GPR124 regulates microtubule assembly, mitotic progression, and glioblastoma cell proliferation. Glia , 67(8), 1558–1570. https://doi.org/10.1002/glia.23628
- Cho, N. H., Cheveralls, K. C., Brunner, A. D., Kim, K., Michaelis, A. C., Raghavan, P., Kobayashi, H., Savy, L., Li, J. Y., Canaj, H., Kim, J. Y. S., Stewart, E. M., Gnann, C., McCarthy, F., Cabrera, J. P., Brunetti, R. M., Chhun, B. B., Dingle, G., Hein, M. Y., … Leonetti, M. D. (2022). OpenCell: Endogenous tagging for the cartography of human cellular organization. Science , 375(6585), eabi6983. https://doi.org/10.1126/science.abi6983
- Dema, A., van Haren, J., & Wittmann, T. (2022). Optogenetic EB1 inactivation shortens metaphase spindles by disrupting cortical force-producing interactions with astral microtubules. Current Biology , 32(5), 1197–1205.e1194. https://doi.org/10.1016/j.cub.2022.01.017
- DeRose, R., Pohlmeyer, C., Umeda, N., Ueno, T., Nagano, T., Kuo, S., & Inoue, T. (2012). Spatio-temporal manipulation of small GTPase activity at subcellular level and on timescale of seconds in living cells. Journal of Visualized Experiments: JoVE , (61). https://doi.org/10.3791/3794
- Ding, L., Laor, D., Weisman, R., & Forsburg, S. L. (2014). Rapid regulation of nuclear proteins by rapamycin-induced translocation in fission yeast. Yeast , 31(7), 253–264. https://doi.org/10.1002/yea.3014
- Gallagher, S., Winston, S. E., Fuller, S. A., & Hurrell, J. G. R. (2008). Immunoblotting and immunodetection. Current Protocols in Molecular Biology , 83, 10.8.1–10.8.28. https://doi.org/10.1002/0471142727.mb1008s83
- Harlow, E., & Lane, D. (1988). Antibodies: A laboratory manual. Cold Spring Harbor Laboratory.
- Honnappa, S., Gouveia, S. M., Weisbrich, A., Damberger, F. F., Bhavesh, N. S., Jawhari, H., Grigoriev, I., van Rijssel, F. J., Buey, R. M., Lawera, A., Jelesarov, I., Winkler, F. K., Wuthrich, K., Akhmanova, A., & Steinmetz, M. O. (2009). An EB1-binding motif acts as a microtubule tip localization signal. Cell , 138(2), 366–376. https://doi.org/10.1016/j.cell.2009.04.065
- Koh, C. M. (2013). Isolation of genomic DNA from mammalian cells. Methods in Enzymology , 529, 161–169. https://doi.org/10.1016/B978-0-12-418687-3.00013-6
- Kremers, G. J., Goedhart, J., van den Heuvel, D. J., Gerritsen, H. C., & Gadella, T. W. Jr. (2007). Improved green and blue fluorescent proteins for expression in bacteria and mammalian cells. Biochemistry , 46(12), 3775–3783. https://doi.org/10.1021/bi0622874
- Mahen, R., Koch, B., Wachsmuth, M., Politi, A. Z., Perez-Gonzalez, A., Mergenthaler, J., Cai, Y., & Ellenberg, J. (2014). Comparative assessment of fluorescent transgene methods for quantitative imaging in human cells. Molecular Biology of the Cell , 25(22), 3610–3618. https://doi.org/10.1091/mbc.E14-06-1091
- Mashiko, D., Fujihara, Y., Satouh, Y., Miyata, H., Isotani, A., & Ikawa, M. (2013). Generation of mutant mice by pronuclear injection of circular plasmid expressing Cas9 and single guided RNA. Scientific Reports , 3, 3355. https://doi.org/10.1038/srep03355
- Metcalf, D. J., Garcia-Arencibia, M., Hochfeld, W. E., & Rubinsztein, D. C. (2012). Autophagy and misfolded proteins in neurodegeneration. Experimental Neurology , 238(1), 22–28. https://doi.org/10.1016/j.expneurol.2010.11.003
- Montenegro Gouveia, S., Leslie, K., Kapitein, L. C., Buey, R. M., Grigoriev, I., Wagenbach, M., Smal, I., Meijering, E., Hoogenraad, C. C., Wordeman, L., Steinmetz, M. O., & Akhmanova, A. (2010). In vitro reconstitution of the functional interplay between MCAK and EB3 at microtubule plus ends. Current Biology , 20(19), 1717–1722. https://doi.org/10.1016/j.cub.2010.08.020
- Moore, D., & Dowhan, D. (2002). Manipulation of DNA. Current Protocols in Molecular Biology , 59, 2.1.1–2.1.10. https://doi.org/10.1002/0471142727.mb0201as59
- Moore, A. T., Rankin, K. E., von Dassow, G., Peris, L., Wagenbach, M., Ovechkina, Y., Andrieux, A., Job, D., & Wordeman, L. (2005). MCAK associates with the tips of polymerizing microtubules. Journal of Cell Biology , 169(3), 391–397. https://doi.org/10.1083/jcb.200411089
- Neumann, B., Walter, T., Heriche, J. K., Bulkescher, J., Erfle, H., Conrad, C., Rogers, P., Poser, I., Held, M., Liebel, U., Cetin, C., Sieckmann, F., Pau, G., Kabbe, R., Wunsche, A., Satagopam, V., Schmitz, M. H., Chapuis, C., Gerlich, D. W., … Ellenberg, J. (2010). Phenotypic profiling of the human genome by time-lapse microscopy reveals cell division genes. Nature , 464(7289), 721–727. https://doi.org/10.1038/nature08869
- Robinson, M. S., Sahlender, D. A., & Foster, S. D. (2010). Rapid inactivation of proteins by rapamycin-induced rerouting to mitochondria. Developmental Cell , 18(2), 324–331. https://doi.org/10.1016/j.devcel.2009.12.015
- Sambrook, J., Fritsch, E. R., & Maniatis, T. (1989). Molecular cloning: A laboratory manual ( 2nd ed.). Cold Spring Harbor Laboratory Press.
- Stepanova, T., Slemmer, J., Hoogenraad, C. C., Lansbergen, G., Dortland, B., De Zeeuw, C. I., Grosveld, F., van Cappellen, G., Akhmanova, A., & Galjart, N. (2003). Visualization of microtubule growth in cultured neurons via the use of EB3-GFP (end-binding protein 3-green fluorescent protein). Journal of Neuroscience , 23(7), 2655–2664. https://doi.org/10.1523/JNEUROSCI.23-07-02655.2003
- Straube, A., & Merdes, A. (2007). EB3 regulates microtubule dynamics at the cell cortex and is required for myoblast elongation and fusion. Current Biology , 17(15), 1318–1325. https://doi.org/10.1016/j.cub.2007.06.058
- Suh, B. C., Inoue, T., Meyer, T., & Hille, B. (2006). Rapid chemically induced changes of PtdIns(4,5)P2 gate KCNQ ion channels. Science , 314(5804), 1454–1457. https://doi.org/10.1126/science.1131163
- Voytas, D. (2000). Agarose gel electrophoresis. Current Protocols in Molecular Biology , 51, 2.5A.1–2.5A.9. https://doi.org/10.1002/0471142727.mb0205as51
- Wagenbach, M., Vicente, J. J., Ovechkina, Y., Domnitz, S., & Wordeman, L. (2020). Functional characterization of MCAK/Kif2C cancer mutations using high-throughput microscopic analysis. Molecular Biology of the Cell , 31(7), 580–588. https://doi.org/10.1091/mbc.E19-09-0503
- Wordeman, L., Decarreau, J., Vicente, J. J., & Wagenbach, M. (2016). Divergent microtubule assembly rates after short- versus long-term loss of end-modulating kinesins. Molecular Biology of the Cell , 27(8), 1300–1309. https://doi.org/10.1091/mbc.E15-11-0803
- Wordeman, L., Wagenbach, M., & Maney, T. (1999). Mutations in the ATP-binding domain affect the subcellular distribution of mitotic centromere-associated kinesin (MCAK). Cell Biology International , 23(4), 275–286. https://doi.org/10.1006/cbir.1999.0359
Internet Resources
This site is used for a “Gene” search of the National Center for Biotechnology Information. Such a Gene search will show the sequence, genomic regions, transcripts, and gene products for your gene of interest.
This site (CRISPRdirect) is used to predict targeting regions within your gene of interest based on nucleotide sequence around the region of interest supplied by the user.

