Processing frozen cells for population-scale Oxford Nanopore long-read DNA sequencing SOP
Kimberley J Billingsley, Pilar Alvarez Jerez, Cornelis Blauwendraat, on behalf of the CARD Long-read Team
Disclaimer
In development
We are still developing and optimizing this protocol.
Abstract
Processing frozen cells for population-scale Oxford Nanopore long-read DNA sequencing SOP At the NIH's Center for Alzheimer's and Related Dementias (CARD) https://card.nih.gov/research-programs/long-read-sequencing we will generate long-read sequencing data from roughly 4000 patients with Alzheimer's disease, frontotemporal dementia, Lewy body dementia, and healthy subjects. With this research, we will build a public resource consisting of long-read genome sequencing data from a large number of confirmed people with Alzheimer's disease and related dementias and healthy individuals.For this project the long-read sequencing data will be derived from brain tissue, but for benchmarking purposes we also developed a protocol for processing frozen cells.The frozen cell protocol was developed to generate long-read sequencing data for the Coriell human-genome cell-lines that is comparable to the data output of the brain based protocol described herehttps://www.protocols.io/view/processing-human-frontal-cortex-brain-tissue-for-p-kxygxzmmov8j/v2) i.e. targeting an N50 of ~30kb and ~30X coverage.
†Correspondence to: Kimberley Billingsley billingsleykj@nih.gov and Cornelis Blauwendraat cornelis.blauwendraat@nih.gov
Acknowledgements: We would like to thank the Nanopore team (Androo Markham &Hannah Lucio), Circulomics Inc team (Jeffrey Burke, Michelle Kim, Duncan Kilburn & Kelvin Liu) and the whole CARD long-read team listed below => UCSC : Benedict Paten, Mikhail Kolmogorov, Miten Jain, Kishwar Shafin, Trevor Pesout; NHGRI : Adam Phillippy, Arang Rhie; Baylor: Fritz Sedlazeck; JHU : Winston Timp; NINDS: Sonja Scholz; NIA : Cornelis Blauwendraat, Kimberley Billingsley, Pilar Alvarez Jerez, Frank Grenn, Bryan Traynor, Shannon Ballard, Caroline Pantazis. Laksh Malik, Edmund Roberts; CZI : Paolo Carnevali.
Steps
Part 1: Extracting Cell DNA (~2 hours per 8 samples)
From: Circulomics HMW DNA Extraction Cultured Cells Protocol
- Kit required: Nanobind CBB Big DNA Kit or Nanobind Tissue Big DNA Kit
- Cell Input Requirements: 1x106 – 5x106 diploid human cells or equivalent
- Cell counts should be accurately determined using a hemocytometer or cell counter.
- Cell pellets should be frozen dry with as much liquid removed as possible.
- For non-diploid or non-human cells, the cell input should be scaled appropriately to contain 5–30 µg of DNA.
Thaw frozen cell pellets on ice (about 5-10 minutes)
Add Nanobind disk to cell lysate and add 225µL of isopropanol. Inversion mix 5X.
- The Nanobind disk must be added before isopropanol.
Mix on tube rotator at 9 rpm at RT for 0h 10m 0s.
- Some white precipitate may be seen attached to the DNA as it binds to the disk. This is normal.
Place tubes on the magnetic tube rack.
- Use the method described in the Kit Handbook Magnetic Rack Handling Procedure section.
#尊敬的用户,由于网络监管政策的限制,部分内容暂时无法在本网站直接浏览。我们已经为您准备了相关原始数据和链接,感谢您的理解与支持。
https://lh4.googleusercontent.com/qTeNmUP6awubHESg0cjD31S3jhcZjC8W_0aPAVrSIZnX52F_cgncaKNWLBDWz_TeWr4HKcpB1POTy7P7Jy0Stp6DXQLF7FrjuFAW_nPj7Wy8KYGZk5UCispvohaY7i2ySgKHzc_a
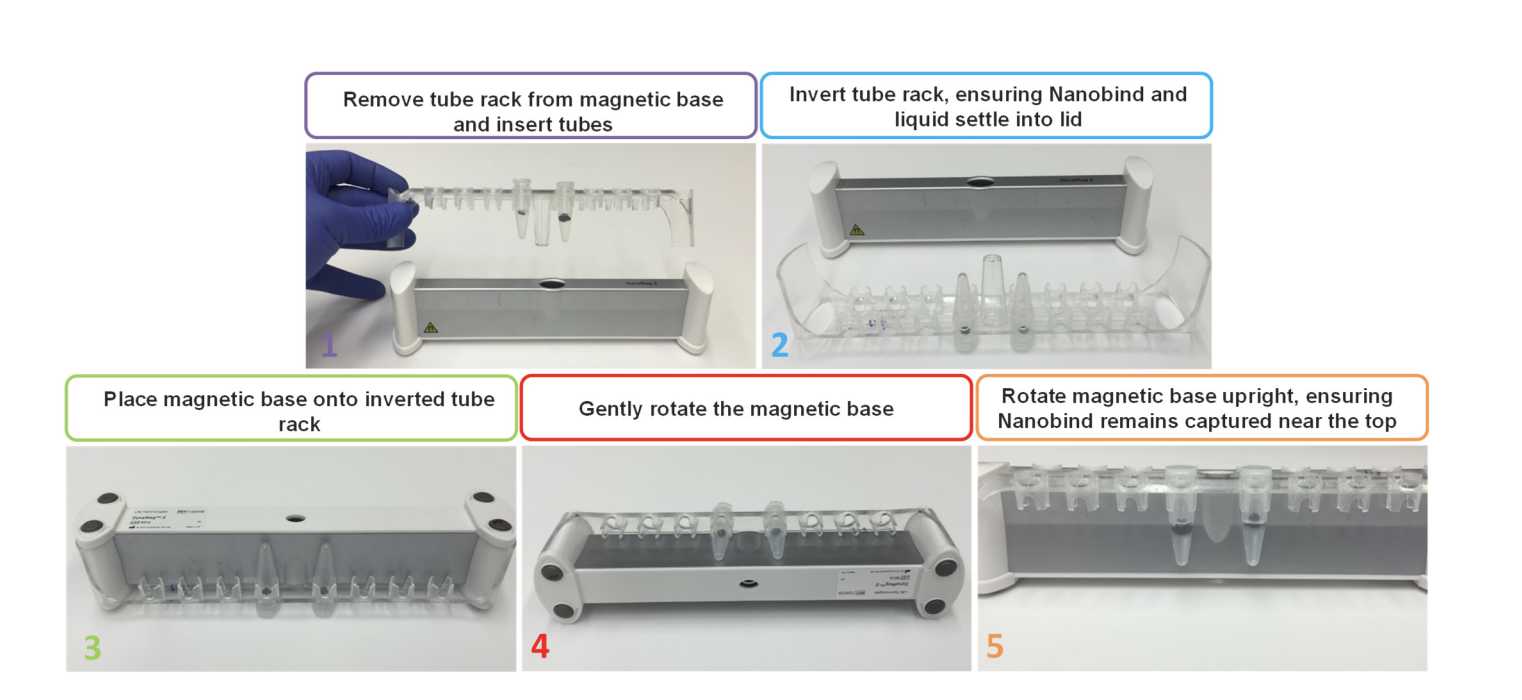
Discard the supernatant with a pipette, taking care to avoid pipetting the DNA or contacting the Nanobind disk.
Add 700µL of Buffer CW1, remove tube rack from magnetic base, inversion mix 4X, replace tube rack on the magnetic base, and discard the supernatant.
- Remove excess liquid from the tube cap to minimize carryover contamination.
Add 500µL of Buffer CW2, remove tube rack from magnetic base, inversion mix 4X, replace tube rack on the magnetic base, and discard the supernatant.
- Remove excess liquid from the tube cap to minimize carryover contamination.
Repeat step 1.15.
Spin the tube on a mini-centrifuge for 0h 0m 2s. With the tube rack already on the magnetic base and right-side-up, place the tube on tube rack and remove residual liquid.
- If the Nanobind disk is blocking the bottom of the tube, gently push it aside with the tip of the pipette. At this stage, DNA is tightly bound to the disk and gently manipulating the disk with a pipette tip should not cause any damage.
Repeat step 1.17
Add 100µL of Buffer EB and incubate at RT for 0h 15m 0s.
Add 70µL μL of 1x PBS to the pellet and pipette mix 10X with a standard P200 pipette to re-suspend cells.
-
Mix until the cell pellet is fully resuspended without visible lumps. Sticky cell types may require additional pipette mixing or vortexing.
-
Aggressive mixing at this step will not affect DNA size. However, incomplete resuspension will result in inefficient lysis and digestion which will lead to low yield, low purity, and high heterogeneity.
-
Vortexing is your friend.
Collect DNA by transferring eluate to a new 1.5mL microcentrifuge tube with a standard P200 pipette. Repeat until all eluate is transferred.
Spin the tube containing the Nanobind disk on a micro-centrifuge at 10,000 x g for 0h 0m 15s and combine any additional liquid that comes off the disk with the previous eluate. Repeat if visible DNA remains on the disk.
- This 15 s spin is CRITICAL for recovering the DNA. We do not recommend a second elution.
- This should not require any more than 1–2 spins.
Pipette mix the sample 10X with a standard P200 pipette to homogenize and disrupt any unsolubilized “jellies” that may be present.
- Take care to disrupt any regions that feel more viscous than other regions.
- Limited pipette mixing will not noticeably reduce DNA size or sequencing read lengths but is important for accurate quantitation and consistent sequencing performance.
Let sample rest at RT for overnight to allow DNA to solubilize.
- Visible “jellies” should disperse after resting.
Following overnight rest, pipette mix 10X with a standard P200 pipette and analyze the recovery and purity as described in QC Procedure (Step 2).
After overnight rest at RT, DNA can be stored at 4°C for up to four weeks, or -80°C indefinitely.
Add 20µL of Proteinase K. Pulse vortex for 0h 0m 1sx 10 times (max setting).
Add 10µL of Buffer CLE3 and pulse vortex for 0h 0m 1s x 10 times (max setting).
- Do not skip vortexing steps, mix aggressively. Even with aggressive vortexing, the DNA will be hundreds of kilobases in length.
Incubate on a ThermoMixer at 55°C and 900 rpm for 0h 10m 0s.
- If a ThermoMixer is not available, a heat block or water bath can instead be used with periodic agitation to ensure lysis.
Optional for removal of RNA: Add 20µL of RNase A, pulse vortex for 0h 0m 1s x 5 times (max setting), and incubate at RT (18-25°C) for 0h 3m 0s.
Add 100µL of Buffer BL3 and pulse vortex for 0h 0m 1s x 10 times (max setting).
- A white precipitate may form after addition of BL3. This is completely normal and usually disappears during the step 8 incubation.
- Insufficient mixing in step 1.2, step 1.4, and step 1.7 will result in very large DNA but also low purity, low yield, high heterogeneity, and difficult elution.
Incubate on a ThermoMixer at 55°C and 900 rpm for 0h 10m 0s.
- Cell inputs greater than 5x10^6 may require longer incubation times to ensure complete lysis. If cellular aggregates are visible, extend lysis time by 10 min (up to 30 min).
Pulse vortex for 0h 0m 1s x 10 times (max setting).
Pre-size selection DNA quantification (~30 minutes for 8 samples)
Hand-shear each sample 20X with 1mL Luer-Lock syringes and 1.5” needles (bringing sample up into needle and depressing plunger counts as 1 cycle).
Quantify by taking triplicate measurements (top, middle, bottom) on the Qubit.
If CV is <10% for the three measurements, move on to the size selection.
If the CV is greater than 10%, further hand shearing is needed. Shear another 10x and take triplicate measurements on the Qubit. Repeat this until the CV is less than 10%. Once achieved, measure on the Nanodrop and size on the Agilent Tapestation 4200.
Size selection (~1:15 hours for 8 samples).
Ensure your sample is between 50-150 ng/uL. If it is > 150 ng/uL then it will need dilution. The optimal range for the sample going into size selection is between 100-150ng/uL but protocol will work as long as it's above 50 ng/uL.
Remove as much EtOH as possible without disturbing the pellet.
Repeat steps 3.8 - 3.10.
Add 50-100µL of Buffer EB from the Short Read Eliminator Kit.
Let sample rest at RT for 0h 20m 0s.
After 0h 20m 0s, flick the tube to ensure proper mixing.
Start a microcentrifuge at 10,000 rpm and 25°C for 0h 5m 0s.
In the meantime, once the sample is at the correct concentration, aliquot the desired volume (enough to have at least 8µg of DNA, but more is ok) into a 1.5mL DNA LoBind tube.
To the DNA LoBind tube with the sample, add equal volume of Buffer SRE from the Circulomics Short Read Eliminator Kit.
Flick the tube to mix.
Place tube in microcentrifuge at 10,000 rpm and 25°C for 0h 30m 0s with hinge facing out as shown.
- Inserting tube with the hinge out is crucial to avoid aspirating the pellet if not visible.
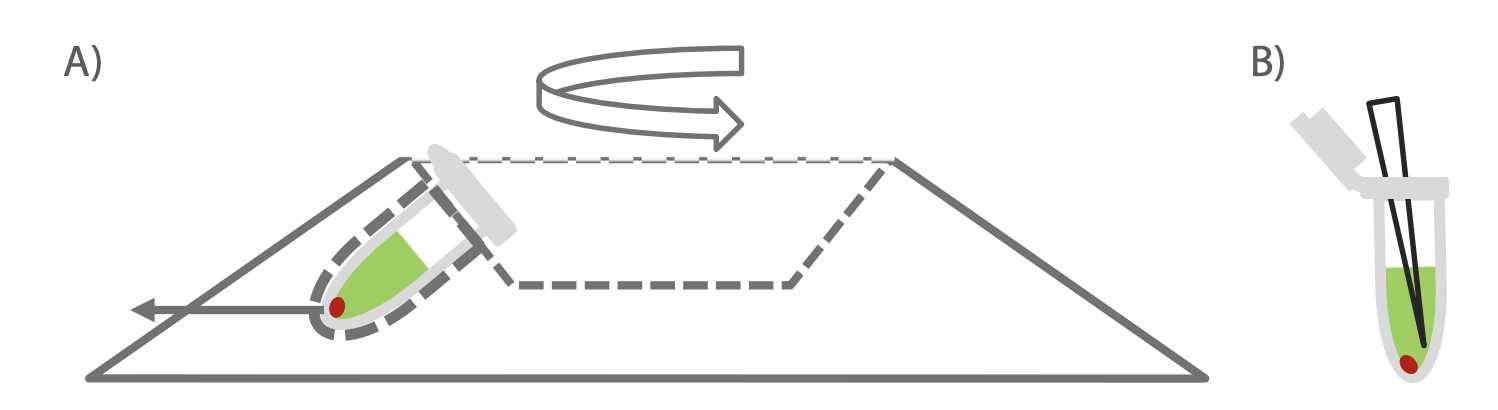
Figure 4. Taken from Circulomics Short Read Eliminator Kit Family Handbook v2.0 (07/2019) A) Note orientation of tube in centrifuge. Pellet will form on the side of the tube facing outwards, in the case underneath the hinge region. B) Pipette from opposite side of tube on the thumb lip side to avoid disturbing pellet (pellet may not be visible).
Remove tube from centrifuge, and remove supernatant from opposite wall of pellet formation.
Note: Do not remove all of the supernatant if close to aspirating pellet or if pellet has not properly formed (can happen with very viscous DNA). Pellet will form a tighter knot after the EtOH washes.
Wash pellet with 70% fresh EtOH, pipette on opposite wall so as to not disturb the pellet.
Centrifuge at 10,000 rpm and 25°C for 0h 2m 0s with hinge facing out.
Post-size selection DNA quantification (~10 minutes for 8 samples)
Analyze size selection recoveries via Qubit.
Note: Quantification post size-selection can be difficult. If necessary, gently pipette mix the sample x5 before quantification to homogenize. The most accurate quantification will come after Megaruptor shearing, but an estimate of DNA concentration is important in this step to standardize the samples going into shearing.
Shearing (~2.5 hours for 8 samples)
In a Diagenode DNA Fluid + tube, make up the sample to 150µL and 40-50ng/uL with H2O.
Note: It is ok if target concentration cannot be reached for all samples. If concentration differs from this range, make sure the Megaruptor 3 shearing settings are updated to reflect your sample concentration. If shearing more than one sample try to get all the concentrations to be as close as possible to this target range.
Attach the DNA Fluid+ needle onto the tube and push the entire item into the Megaruptor 3 slot until it fits snugly.
Note: If running fewer than 8 samples, put the tubes in the 1st and/or 8th slots, working your way in. Samples should always be balanced. If running an odd number of samples, samples can be balanced with an empty corresponding tube.
Shear at speed 45 for two rounds.
Once done, remove item from Megaruptor 3 and carefully remove needle from tube. Make sure the plunger is fully depressed in order to avoid losing sample.
Avoid any vortexing of DNA from this point on to avoid any unnecessary further shearing, instead mix by gently flicking the tube and spin down.
Post-shearing DNA Quantification (~30 minutes for 8 samples)
Upload the TapeStation reports and quantifications to tracking google sheet.
At this point, at least 4µg of DNA is necessary to move on to library prep.
DNA can be stored at 4°C for up to four weeks, or -80°C indefinitely.
Library Prep (~6 hours, including reloads but not including flushing and returning cells)
Note: Library prep is the same as the brain library prep with two exceptions. First, in step 7.1A we mix beads into the sample by flicking and not pipetting up and down. Second, the loading amount is 350ng/load for cells.
A. DNA Repair and End-Prep
-
Place all the necessary reagents on ice to thaw and the Agencourt AMPure XP beads out at room temperature.
-
Prepare the following in a
0.2mLthin-walled PCR tube:
48µLDNA (input 3ug, this might be over 48 μL but that is fine. Adjust the amount of beads to match the total volume of this mixture (sample + buffer + enzyme)3.5µLNEBNext FFPE DNA Repair Buffer (vortex and spin down)3.5µLUltra II End-prep reaction buffer (vortex and spin down)3µLUltra II End-prep enzyme mix (do not vortex, spin down)2µLNEBNext FFPE DNA Repair Mix (do not vortex, spin down)
-
Mix thoroughly by gently flicking the tube or very gently pipetting up and down x10, and then spin down.
-
Using a Thermocycler, incubate samples at
20°Cfor0h 5m 0sand65°Cfor0h 5m 0s.
- Start and pause Thermocycler to allow lid to come to
85°Cbefore putting samples in.
-
Allow Thermocycler to cool to
4°Cand then remove your samples. -
Re-suspend the AMPure XP beads by vortexing.
-
Transfer DNA samples to clean
1.5mLEppendorf DNA LoBind tube. -
Add
60µL(or equivalent volume, see step 2) of resuspended beads to the reaction and mix by flicking the tube x10. Do not pipette mix here as beads may clump around pipette tip. -
Incubate for
0h 5m 0sat room temperature. -
Prepare
500µLper sample of fresh 75% ethanol in nuclease-free water. -
Spin down and pellet sample on magnet until eluate is clear and colorless, about
0h 2m 0s. -
Pipette off the supernatant and retain, just in case the following quant is uncharacteristically low.
-
With the samples remaining on the magnet, wash the beads with 200uL of 75% ethanol, pipetting on the opposite wall making sure not to disturb the pellet. Count to 3 and remove and discard ethanol.
Note: The goal here is to make sure the beads are fully covered, if initial volume of beads was a lot higher than 60uL, more ethanol may be used.
-
Repeat previous step
-
Spin down and place the tube back on magnet, pipetting off any residual ethanol.
-
Allow to dry for ~
0h 0m 30sbut do not over-dry. -
Remove the tube from the magnetic rack and re-suspend the pellet in
61µLnuclease-free water, incubate for0h 3m 0sat RT gently flicking every so often. -
Spin down and pellet the samples on a magnet until eluate is clear and colorless.
-
Remove and retain
61µLof eluate into a clean1.5mLEppendorf DNA LoBind tube. -
The sample concentration must be > 40ng/ul . If the sample does not reach this requirement restart from Part 5.
-
It is possible to store samples at
4°Covernight at this step if needed
B. Adapter Ligation and Clean-Up
- Spin down the AMX-F, Quick T4 ligase, and LNB, then return to ice.
Note: Do not allow AMX-F to remain at room temperature for too long.
-
Thaw LNB at RT and mix by pipetting up and down (vortexing is ineffective due to viscosity).
-
Thaw EB at RT, mix by vortexing, spin down, and place on ice.
-
Thaw SFB at RT, mix by vortexing, spin down, and keep at RT.
-
In a
1.5mLEppendorf DNA LoBind tube, mix the following in order:
60µLDNA sample (if not 60uL, make up with water)25µLLNB10µLQuick T45µLAMX-F
-
Mix by gently flicking the tube and spin down.
-
Incubate the reaction for
0h 10m 0sat RT. -
During this time, put flow cells out at RT.
-
Re-suspend beads by vortexing.
-
Add
45µLof resuspended beads to the reaction and mix by flicking. -
Incubate for
0h 5m 0sat RT. -
Spin down sample and pellet on magnet.
-
Pipette off the supernatant and retain, just in case the final elution quantification is uncharacteristically low.
-
Wash the beads with
250µLSFB, remove from magnet and flick to re-suspend, spin down and re-pellet on magnet and then remove and discard supernatant. -
Repeat previous step.
-
Spin down and place the tube back on magnet, pipetting off any residual SFB.
-
Allow to dry for ~
0h 0m 30s, but do not over-dry. -
Remove the tube from magnet and re-suspend pellet in
25µLEB, spin down, and incubate for0h 20m 0sat37°C. -
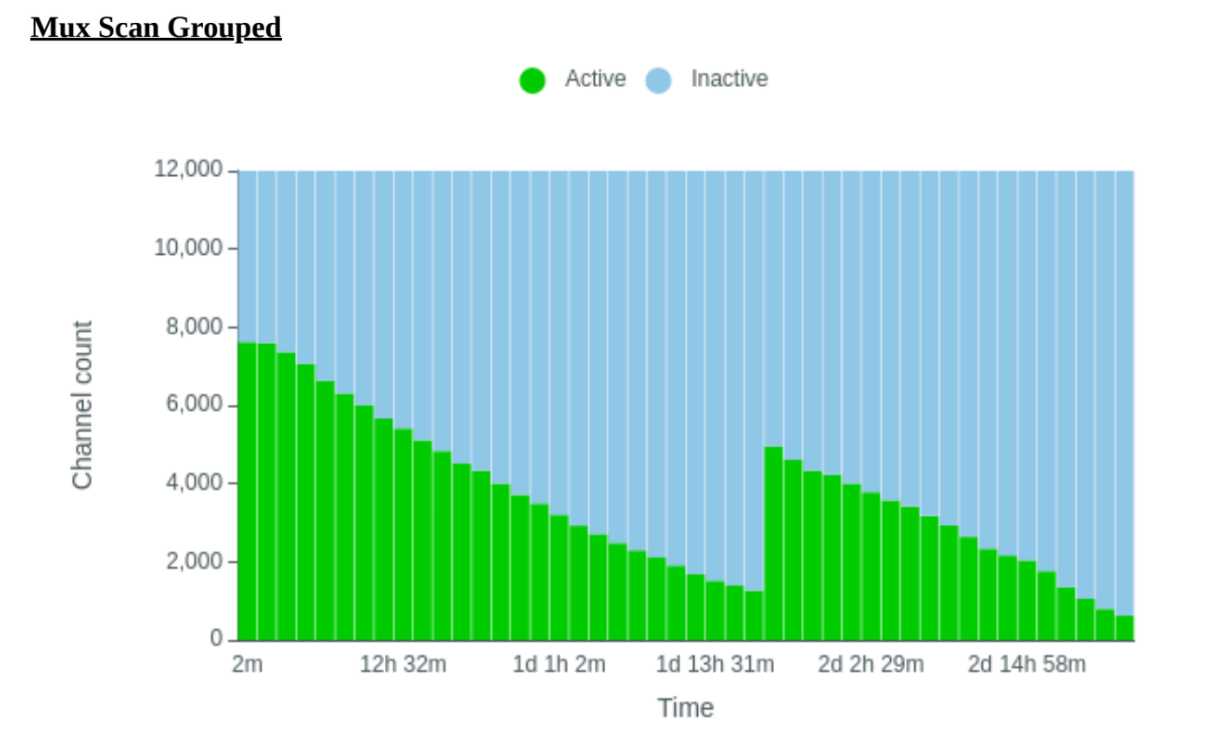
During this time, QC the flow cells (only use flow cells with >6000 pores).
-
Pellet the beads on magnet until eluate is clear and colorless.
-
Remove and retain
25µLof eluate into a clean1.5mLEppendorf DNA LoBind tube (this is the DNA library). -
Quantify samples on Qubit.
-
Re-prep library from Part 6 if < 1,050ng.
-
Keep libraries on ice until ready to load on flow cell.
C. Priming and Loading Flow Cell
- Thaw SBII, FLT, and FB, vortex, and spin down.
- Thaw LBII.
- Add
30µLof thawed and mixed FLT directly to tube of FB and vortex. - Expose inlet port on flow cell, set P1000 pipette to
200µLand draw back a small amount of volume to remove any air bubbles (usually about20-30µL, just until a small volume of buffer enters the pipette tip). - Flush
500µLof Priming Mix into the inlet port of the flow cell, being extremely careful to avoid the introduction of air bubbles at the end. - Wait
0h 5m 0s. - During this time, separate the DNA library into three equal aliquots of 350 ng each. Bring each aliquot up to
24µLwith EB. (i.e, if your final elution is exactly 1050 ng in 24 uL, move 8 µL to three separate tubes and add 16 µl of EB to each.) - Prepare the first library mix for loading:
75µLSBII (vortex and spin down)51µLLB (pipette up and down immediately before use)24µLDNA library in EB (350 ng)
-
Flush
500µLof Priming Mix a second time into the inlet port of the flow cell, being extremely careful to avoid the introduction of air bubbles at the end. -
Load all
150µLof the library mix. -
Close valve to seal inlet port and close PromethION lid.
-
Wait
0h 10m 0sand then initiate sequencing. -
Ideally, the library quants yielded at least 1050 ng to allow for 3X 350 ng loads, the latter 2 loaded approximately after 24 and 48 hours. However this will vary slightly depending on pore usage, data generated, as well as other factors i.e. if after 24 hours there are still +3000 pores then the sample does not need to be reloaded until the pores are between 2000-3000.
-
To wash and reload a flow cell:
-
Begin by thawing Wash Mix (WMX) on ice and Wash Diluent (DIL) at RT. Note: DIL should be vortexed. WMX should NOT be vortexed, only spun.
-
In a new tube, add
2µLWMX into398µLDIL and pipette mix. -
Pause the PromethION runs and export .pdf reports.
-
With inlet port 1 closed, remove waste from port 2 or 3.
-
Rotate the inlet port cover to reveal inlet port 1.
-
Using a P1000, insert tip into inlet port and draw back a small volume using the wheel to remove any air (usually around
20-30µL). -
Load
400µLFlow Cell Wash Mix into the inlet port, avoiding any introduction of air. -
Wait
1h 0m 0s. -
Repeat priming steps and reload samples (steps 1 - 12).
D. Flushing and Recycling Flow Cells (~15 minutes per set of 4 flow cells)
- Following the completion of the sequencing, flow cells may be removed from the sequencer.
- Place enough absorbent material to take up approximately
4mLof flush waste. - Rotate valve to reveal inlet port 1.
- Place flow cell at a 45° angle on the absorbent material and, using a P1000, flush
1mLof DI water into the inlet port. - Repeat 3 more times for a total of
4mL. - Once complete, close the input port cover and remove all liquid from the waste port.
- Dispose of absorbent material as local biological waste guidelines dictate.
- Return flow cells to clear plastic tray in which it was shipped, making sure to record the flow cell IDs.
- Seal the tray with the sticker provided in the packaging.
- Put the clear plastic lid back on the tray.
- Place the tray back in the packaging.
- Place packaged cells in the returns box (large box can hold 80).
- Once returns box is filled, follow the instructions here here and follow the prompts to request the box to be sent back to Nanopore.