Preparing Gene of interest for GateWay cloning (2 step PCR process)
Johannes Wolfram JWD Debler
Abstract
GateWay recombination cloning is achieved by flanking your gene of interest with GateWay attachment sites. In our case attB1 and attB2. Those sites are added to the PCR product via primers with 5' extensions. Since those primes create 31 bp and 30 bp 5' primer extensions respectively, plus about 20 bp of actual binding primer sequence it becomes expensive fast if you need 2 x ~50 bp primers for every GOI. We therefore use a 2 step PCR process to attach GateWay attB1 and attB2 sites. We first run a gene specific PCR with primers carrying short 5' extesions, and then a second PCR utilizing universal GateWay primers which bind to the short extension of the first PCR product to create the full attB1 and attB2 sites.
This protocol has been adapted from: 2-STEP GATEWAY PCR EXPERIMENTS
Steps
Primer design
There are a few things we need to keep in mind when designing the primers in the wider context of Gateway cloning. The idea is that you create an "entry clone" containing your gene of interest. Ususally INCLUDING the start codon, but EXCLUDING the stop codon. This is because one of the points of Gateway recombination is that you use the same entry clone to shuttle your gene of interest into different destination vectors with different properties (different N- or C- terminal tags). If you included the stop codon you would prevent yourself from being able to attach C-terminal tags.
Gene specific forward primer (PCR 1): 5'-AA AAA GCA GGC T NN -(15 to 20 bp template specific sequence)-3'
The 'NN' here can be any base. Don't use AA, AG or GA though, as that would introduce an in-frame stop codon! They are inserted to keep the reading frame if a destination vector with an N-terminal tag is used. I use 'CC' in all of my primer design.
Gene specific reverse primer (PCR 1): 5'-A GAA AGC TGG GT N -(15 to 20 bp template specific sequence)-3'
The 'N' here can be any base, just be careful to not accidentally introduce a stop codon. I use 'A' in all of my primer design.
attB1 adapter primer (PCR 2): 5'-GGG GAC AAG TTT GTA CAA AAA AGC AGG CT-3'
attB2 adapter primer (PCR 2): 5'-GGG GAC CAC TTT GTA CAA GAA AGC TGG GT-3'
This is what we are trying to achieve:
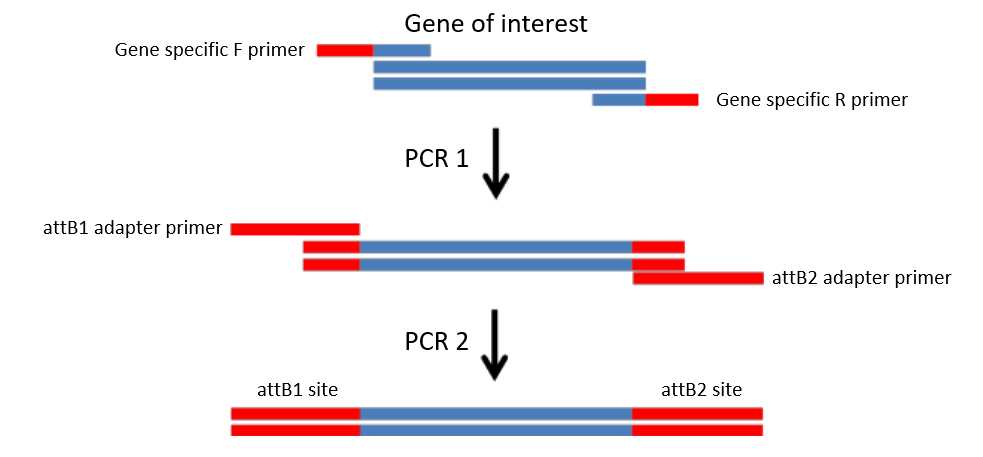
PCR 1 - gene specific PCR
Use your favourite High Fidelity Polymerase. I have used NEB Q5 and Thermo Fisher SuperFi Mastermix as listed below with great success.
| A | B | C | D | E | F |
|---|---|---|---|---|---|
| 10 ul total | |||||
| 5x Q5 Buffer | 2 ul | 98°C | 1 min | ||
| 2 mM dNTPs | 1 ul | 98°C | 10 s | ||
| 10 uM F primer | 0.5 ul | 50°C | 20 s | } 40 x | |
| 10 uM R primer | 0.5 ul | 72°C | 30 s (per kb) | ||
| Q5 polymerase | 0.1 ul | 72°C | 2 min | ||
| H2O | 5.4 ul | 10°C | hold | ||
| template | 0.5 ul |
| A | B | C | D | E | F |
|---|---|---|---|---|---|
| 10 ul total | |||||
| 2x SuperFi Mastermix | 5 | 98°C | 30 s | ||
| 10 uM F primer | 0.5 ul | 98°C | 10 s | ||
| 10 uM R primer | 0.5 ul | 50°C | 20 s | } 40 x | |
| H2O | 3.5 ul | 72°C | 30 s (per kb) | ||
| template | 0.5 ul | 72°C | 2 min | ||
| 10°C | hold |
PCR 2 - GateWay attB1 and attB2 adapter primers
Use PCR 1 product as input for PCR 2.
| A | B | C | D | E | F |
|---|---|---|---|---|---|
| 10 ul total | 98°C | 1 min | |||
| 5x Q5 Buffer | 2 ul | 98°C | 10 s | ||
| 2 mM dNTPs | 1 ul | 45°C | 30 s | } 40 x | |
| Q5 polymerase | 0.1 ul | 72°C | 30 s (per kb) | ||
| 10 uM attB1 primer | 0.5 ul | 72°C | 2 min | ||
| 10 uM attB2 primer | 0.5 ul | 10°C | hold | ||
| PCR 1 product | 1 ul | ||||
| Cresol red | 1.7 ul | ||||
| H2O | 3.2 ul |
| A | B | C | D | E | F |
|---|---|---|---|---|---|
| 10 ul total | 98°C | 1 min | |||
| 2x SuperFi Mastermix | 5 ul | 98°C | 10 s | ||
| 10 uM attB1 primer | 0.5 ul | 45°C | 20 s | } 40 x | |
| 10 uM attB2 primer | 0.5 ul | 72°C | 30 s (per kb) | ||
| PCR 1 product | 1 ul | 72°C | 2 min | ||
| H2O | 3 ul | 10°C | hold |
Run a gel
Run PCR 2 product on a gel to make sure you have a crisp band (and to know if the PCR has worked).
Gel extraction
Extract the PCR 2 band with your gel extraction kit of choice.

