Prediction and Characterization of miRNA/Target Pairs in Non-Model Plants Using RNA-seq
Kira C. M. Neller, Kira C. M. Neller, Alexander Klenov, Alexander Klenov, Katalin A. Hudak, Katalin A. Hudak
de novo transcriptome assembly
differential expression
miRNA prediction
miRNA target
non-model plant
RNA-seq
small RNA
Abstract
Plant microRNAs (miRNAs) are ∼20- to 24-nucleotide small RNAs that post-transcriptionally regulate gene expression of mRNA targets. Here, we present a workflow to characterize the miRNA transcriptome of a non-model plant, focusing on miRNAs and targets that are differentially expressed under one experimental treatment. We cover RNA-seq experimental design to create paired small RNA and mRNA libraries and perform quality control of raw data, de novo mRNA transcriptome assembly and annotation, miRNA prediction, differential expression, target identification, and functional enrichment analysis. Additionally, we include validation of differential expression and miRNA-induced target cleavage using qRT-PCR and modified RNA ligase–mediated 5′ rapid amplification of cDNA ends, respectively. Our procedure relies on freely available software and web resources. It is intended for users that lack programming skills but can navigate a command-line interface. To enable an understanding of formatting requirements and anticipated results, we provide sample RNA-seq data and key input/output files for each stage. © 2019 The Authors. This is an open access article under the terms of the Creative Commons Attribution-NonCommercial License, which permits use, distribution and reproduction in any medium, provided the original work is properly cited and is not used for commercial purposes.
INTRODUCTION
Plant small RNAs are ∼20 to 24 nucleotides (nt) in length and are classified as either microRNAs (miRNAs) or small interfering RNAs (siRNAs), depending on their biogenesis. miRNAs are derived from single-stranded mRNA precursors (pre-miRNAs) that adopt a characteristic hairpin structure. The biological relevance of an miRNA is defined by the functional role of its mRNA target. Plant miRNAs tend to have high sequence complementarity with targets and act by inducing transcript cleavage, resulting in mRNA decay. This differs from animal miRNAs, which tend to share a smaller ‘seed’ region of complementarity with targets and act through translational inhibition. By modulating the level of transcript abundance or protein accumulation, miRNAs provide an important layer of gene expression regulation.
This article outlines the steps involved in large-scale miRNA prediction and characterization, with a focus on non-model plants that lack genomic references. The workflow of the protocols in this article is provided in Figure 1. We cover strategic planning, including RNA-seq experimental design and considerations for library preparation and sequencing; quality control of raw small RNA and mRNA reads (Basic Protocol 1); de novo mRNA transcriptome assembly and annotation of protein-coding genes through homology and protein domain identification (Basic Protocol 2); miRNA prediction and identification of conserved versus novel sequences (Basic Protocol 3); miRNA and mRNA differential expression analysis (Basic Protocol 4); prediction of miRNA/target pairs (Basic Protocol 5); and gene ontology (GO) term functional enrichment analysis (Basic Protocol 6). We also include wet-lab methods to validate bioinformatic predictions derived from the above analyses: qRT-PCR and stem-loop qRT-PCR to confirm mRNA and miRNA differential expression, respectively (Basic Protocol 8), and modified RNA ligase–mediated 5′ rapid amplification of cDNA ends (RLM-RACE) to detect miRNA-induced cleavage of target transcripts (Basic Protocol 7). This protocol enables the detection of miRNAs that act through transcript cleavage rather than translational inhibition of targets. Although both mechanisms of miRNA activity exist in plants, the former is more prevalent.

This protocol is based on our previous work to identify defense-associated miRNA/target pairs in the non-model plant Phytolacca americana (Neller, Klenov, & Hudak, 2016, 2018). To provide greater context for the reader, we carry out the bioinformatic workflow using an example RNA-seq dataset that was generated by random sampling of our published data. We aim to be user-friendly, introducing graphical interface options when available. However, we assume that the reader can navigate a UNIX-like operating system from a command-line interface, since this is commonly required to conduct large-scale computational analysis.
STRATEGIC PLANNING
Experimental design
The method described in this article is based on small RNA and mRNA samples that are both derived from one control and one experimental condition. The sample data that we provide were used to characterize miRNAs associated with defense in P. americana. In this case, plants were sprayed with either ethanol (control) or jasmonic acid (JA, experimental treatment). JA is a plant hormone that mediates plant defense against insect herbivores and necrotrophic pathogens. It is possible to follow this procedure without incorporating a conditional variable; however, we advocate the use of differential expression as a filter to identify the most biologically meaningful miRNA/target pairs.
To reduce the impact of experimental variability on miRNA and target expression, we suggest the use of paired small RNA and mRNA samples. This means that for each sample, both the small RNA and mRNA fractions are extracted. Additionally, the use of biological replicates (i.e., different plants that undergo the same treatment) is recommended for robust differential expression analysis. Due to the expense of RNA-seq, technical replicates (i.e., identical samples) are not usually included in the experimental design. Furthermore, it is common to pool equal volumes of RNA from independent plants undergoing the same treatment. Such ‘pooled biological replicates’ deviate from the definition of true biological replicates, but ease downstream analysis. Specifically, this strategy increases the number of plants tested (leading to greater biological representation) while reducing technical variability across replicates. The downside is that it does not detect differences among independent plants. However, both biological and technical variability may be assessed through the more economical method of qRT-PCR.
Our study comprised two conditions (control versus JA), with three pooled biological replicates per condition (i.e., six total samples). Each pooled replicate consisted of equal amounts of RNA from three independent plants; therefore, a total of 18 plants were used. Both small RNA samples and mRNA-enriched samples were prepared, arranged as follows, with replicate number indicated: (C1sRNA: C1mRNA), (C2sRNA: C2mRNA), (C3sRNA: C3mRNA), (J1sRNA: J1mRNA), (J2sRNA: J2mRNA), (J3sRNA: J3mRNA). Here, ‘C’ and ‘J’ indicate control and JA conditions, respectively, and ‘sRNA’ and ‘mRNA’ indicate small RNA and mRNA fractions, respectively. Our sampling design is illustrated in Figure 2.

RNA extraction and enrichment of mRNA or small RNA
The sequencing facility will likely have different options for sample submission that vary in level of downstream processing and associated cost. Commonly accepted submission types include total RNA, enriched RNA (mRNA or small RNA), and ready-to-sequence libraries. For readers who wish to prepare their own small RNA libraries, we recommend the protocol by Mathioni, Kakrana, and Meyers (2017), which covers total RNA extraction from plant tissue, small RNA enrichment, and library construction. Given that very little RNA is required for sequencing (1 to 2 µg of total RNA and 500 ng to 1 µg of small RNA) and that multiple samples need to be processed, the use of RNA isolation kits may be preferred for RNA extraction. We recommend contacting the facility to discuss specific requirements for sample preparation and any recommended kits.
Prior to RNA-seq library preparation, either mRNA enrichment or rRNA depletion of total RNA must be performed. Without this step, most reads would derive from rRNA due to its high relative abundance. The decision to use one strategy over the other depends on the project goal. mRNA enrichment is based on the selection of RNA species containing a poly(A) tail and 5′ cap, whereas rRNA depletion selectively removes rRNA and consequently enriches for all other RNAs, including non-coding RNAs. rRNA depletion involves hybridization of probes containing conserved rRNA sequences, the probes being linked to magnetic beads to facilitate rRNA removal following hybridization. In addition to cytoplasmic rRNA, kits often include probes to remove chloroplast and mitochondrial rRNA. The benefit of rRNA depletion over mRNA selection is that the former results in more information retention. In terms of small RNA characterization, rRNA depletion would enable the identification of both siRNA and miRNA precursors, since the former tend to lack poly(A) tails. We chose mRNA enrichment for our study; not only was it more economical, it was the more conservative approach given the lack of known rRNA sequences for our non-model plant.
Small RNA enrichment involves isolation of the low-weight RNA fraction. As detailed by Mathioni et al. (2017), this can be accomplished by separating total RNA on a urea-polyacrylamide gel and excising the small RNA fraction. This was the chosen strategy for our experiment. As an alternative option, the small RNA fraction can be obtained using dedicated spin kits that employ size-exclusion filters. Such kits are especially convenient in facilitating a paired study design, as one can quickly obtain total RNA, small RNA–enriched, and small RNA–depleted fractions from a single sample.
Library preparation and sequencing
The final step prior to RNA-seq is the preparation of sequence libraries, one per sample. With the mRNA-enriched or rRNA-depleted fraction as input, standard library preparation includes the following steps: RNA fragmentation, cDNA synthesis, adapter ligation, and PCR amplification. For RNA-seq, we recommend the preparation of paired-end, strand-specific libraries. In a paired-end library, the ligated adapters contain an additional priming site to enable sequencing of both ends of the DNA fragment. Size selection is employed to ensure that the majority of DNA fragments are long enough to avoid sequence overlap between the two read-pairs. Although more expensive, paired-end reads provide greater information for downstream transcriptome assembly, since it is known that both pairs derive from a single fragment and are a certain distance apart. This helps resolve regions that are repeat-rich or of low complexity. We also recommend the use of strand-specific library preparation methods to retain information on the orientation of the originally sequenced transcript. Distinguishing between sense and anti-sense transcription of overlapping genes enables more accurate detection of differential expression. For our study, we used strand-specific sequencing for both mRNA and small RNA-seq. Paired-end sequencing was used only for mRNA-seq; it is not necessary for small RNA-seq because miRNAs are short enough to be sequenced completely with one pass.
It is also important to consider the balance between sequencing depth and number of experimental conditions/replicates. Since a single lane provides a fixed output of reads, increasing the number of samples per lane will reduce the amount of reads per sample. The strategy of combining multiple samples in a lane is termed ‘multiplexing’, whereby adapters with sample-specific indexes are used to distinguish independent libraries. Since RNA-seq has fixed costs both per lane and per sample, multiplexing can reduce the overall cost of sequencing at the expense of reduced coverage per sample. If this is the first RNA-seq study for your non-model plant, we suggest that depth take priority; the resulting high-coverage references will serve as a foundation for future work, enabling less deep sequencing in future experiments. In our study, two sequencing lanes were used for both small RNA-seq and mRNA-seq, with respective samples equally distributed between the lanes. The project goals should be discussed in advance with the sequencing facility, as they can provide an expected number of reads given the sequencing platform and number of lanes used.
We note here that two different sequencing platforms were used in our experiment. We chose SOLiD (commercially discontinued since 2016) for small RNA-seq because it had a lower error rate than Illumina at the time of the study, with the latter platform used for mRNA-seq. As miRNAs are ∼20 to 24 nt in length, a low error rate ensures that more reads are retained during alignment steps. Given a fixed proportion of sequence errors, a shorter sequence is more difficult to align to a reference than a longer one. Presently, we suggest the use of Illumina sequencing to perform both mRNA-seq and small RNA-seq. However, we maintain the use of SOLiD data in this procedure because such data are widely available in public sequencing repositories, necessitating an understanding of their special considerations.
Basic Protocol 1: QUALITY CONTROL PROCESSING OF RNA-seq DATA
In this protocol, we cover the format of raw SOLiD and Illumina RNA-seq data, necessary file conversions, and steps to ‘clean’ the reads. Cleaning entails the removal of adapter sequences and low-quality bases. This protocol is designed to be carried out using a public Galaxy account. Galaxy is an open-source, web-based platform that enables researchers without command-line programming experience to perform basic bioinformatic analyses (Giardine et al., 2005). The utility of Galaxy lies with its graphical interface, which allows users to interact with their data, perform large-scale operations, and run standard programs, all without exposure to the underlying computational infrastructure. We introduce Galaxy at this stage to provide the reader with a visual sense of the input data prior to downstream analysis at the terminal. Sample data and final output files are provided in the accompanying Supporting Information.
Materials
- A personal or desktop computer with internet access
- A public Galaxy user account; register at: https://usegalaxy.org/
- Raw SOLiD small RNA-seq dataset: 1.sRNA_reads/1.raw
- Raw Illumina mRNA-seq dataset: 2.mRNA_reads/1.raw
Process the SOLiD small RNA-seq data
1.Upload the small RNA-seq files to Galaxy.
To do this, first navigate to Galaxy at https://usegalaxy.org/ and sign in with your account credentials. From the tool panel on the left, select the category ‘Get Data’. Click on the tool ‘Upload file from computer’. Select the option ‘choose a local file’ to browse your file directory. Upload the 12 small RNA data files. See Figure 3 for an annotated layout of the Galaxy workspace.
>135_886_1067_F3
T32320113231210122000131330201030313
>135_886_1067_F3
31 31 31 31 31 31 31 31 31 31 31 31 31 31 31 31 31 31 31 31 28 31 31 31 31 31 31 31 31 31 31 31 31 31 31

2.For each sample, combine the .qual and .csfasta files into a single fastq file.
Under the tool category ‘NGS: QC and manipulation’, select ‘Convert SOLiD output to fastq’. For the ‘reads’ option, provide the .csfasta file (e.g., sRNA_E1_10k.csfasta), and for the ‘qualities’ option, provide the corresponding .qual file for that sample (e.g., sRNA_E1_10k.qual). Set ‘trim trailing F3 and R3?’ to ‘yes’.
@135_886_1067
T32320113231210122000131330201030313
+
@@@@@@@@@@@@@@@@@@@@=@@@@@@@@@@@@@@
3.Decode the sequence from color space to base space.
Use the tool ‘FASTQ Groomer’ under the category ‘NGS: QC and manipulation’. For ‘file to groom’, select the fastqcssanger file generated from step 2.For ‘input FASTQ quality scores type’, select ‘color space sanger’.
@135_886_1067
AGCTTGTAGCAGTTGAGGGGTACGCCTTGGCCGTA
+
@@@@@@@@@@@@@@@@@@@@=@@@@@@@@@@@@@@
4.Obtain an overview of library quality.
Run the ‘FastQC’ tool under the category ‘NGS: QC and manipulation’. The file to be evaluated is the fastq output from step 3.
5.Remove contaminating adapter sequences.
Select the ‘Clip’ tool from the category ‘NGS: QC and manipulation’. For input, select the fastq file output from step 3.Set ‘minimum sequence length’ to 18, ‘custom clipping sequence’ to CGCCTTGGCCGTACAGCAG, and both ‘discard sequences with unknown bases’ and ‘output only clipped sequences’ to ‘yes’.
6.Remove low-quality bases from reads.
Select ‘Trimmomatic’ from the category ‘NGS: QC and manipulation’. Specify that the data are single-end reads and choose the adapter-trimmed fastq file produced from step 5 as input. Select ‘sliding window trimming’ as the operation to perform, with default parameters.
7.Apply a final length filter.
Use the tool ‘Filter FASTQ’ from the category ‘NGS: QC and manipulation’. Set ‘maximum length’ to 28 nt.
8.Convert the fastq file to fasta format.
Choose the tool ‘FASTQ to FASTA’ from the category ‘Convert Formats’. For input, select the output fastq file from step 7.Set ‘rename sequence names in output file’ to ‘no’.
>135_886_1067
AGCTTGTAGCAGTTGAGGGGTA
Process the Illumina mRNA-seq data
9.Upload the mRNA-seq data to Galaxy.
See the instructions provided in step 1.
@HWI-D00423:99:HB533ADXX:2:1213:12544:78916 1:N:0:CGATGT
TTCCTGTAGGCTGGGAGATTGGAAAGGTACCATGCGATTAATGGGCTGGATGTCACTGGGGTCTCAAGTCCTCCAATGAATGGGTCTCCATTCAAGGGCTGGATTACTTGGTATGTTTGCTTATCAGATTGAATGGCCTTGATGGTAAAGG
+
@CBFFFFFHGHHHJJHIIJIJJIJJJJEFHIIJJJJIJJJIJIJJJJJJJJJJIJIIJJJJHHHHHHHFFFFFFEEEEEEEDDDCDDDDDDFFEEDDDDDDDDDDDDDDDDACDDEEEDDDDDDDEEDCDDDDDDDDDDDDCDDDDEDDDD
@HWI-D00423:99:HB533ADXX:2:1213:12544:78916 2:N:0:CGATGT
CAACAACATCACCAATGGCCAGCCAATTAAAGAGTGGCTTCACCGCAACAATTGTGACTCCCAAGGGCATCTCTGGTCCTGCCTTAAGGCCCTTCCCTTCCATCAGGAGGCACCATTCCTTTACCATCAAGGCCATTCAATCTGATAAGCA
+
CCCFFFFFHHHHHJJJJJJJJJJJJIJJJJIJJJGHJJJJJJJJIJJJJIJJJJIIJIIJJJHHGFFFFEEDEEEECCDDDDDDDDDDDDDDDDDDDDDDDDEDDDDDDDDDDDDDDEEDDDDDDDDDDDDDDDDDDEEEEDDEDEDDDDC
10.Run FastQC on each fastq file to obtain an overview of quality.
The ‘FastQC’ tool can be run as in step 4.Since the raw mRNA-seq reads are already in fastq format, no additional conversions are necessary.
11.Remove adapter sequences and low-quality bases, and apply a minimum length filter.
Use ‘Trimmomatic’ from step 6.Specify that the reads are paired-end and provide the R1 and R2 files. Choose ‘yes’ to ‘perform initial ILLUMINACLIP step’. Choose ‘Truseq3 paired-end adapters’ from the available adapter sequences. Add two additional options: ‘sliding window trimming’ as before and ‘drop reads below a specified read length’ (default is 20 nt).
12.Download and save the clean small RNA-seq and mRNA-seq files.
Basic Protocol 2: TRANSCRIPTOME ASSEMBLY AND ANNOTATION
This protocol covers de novo transcript assembly and annotation using Trinity and Trinotate, respectively. We combine the clean mRNA-seq reads of all samples to produce a single reference transcriptome. From the assembled transcripts, the most likely open reading frames (ORFs) and their translated protein sequences are obtained using TransDecoder. The transcript and protein sequences are used in BLASTX and BLASTP homology searches, respectively, against the curated SwissProt protein database. Additionally, the protein sequences are searched for conserved Pfam protein domains. Using Trinotate, the search results are built into a comprehensive annotation report that provides insight into the function of each transcript. All input and output files available in the Supporting Information are indicated in the code boxes with green and red font, respectively.
Materials
- A computer or high-performance cluster running a UNIX-based operating system
- Clean mRNA-seq dataset from Basic Protocol 1: 2.mRNA_reads/2.clean
- Sample information file: 3.trinity/samples.txt
- The following programs will be run in the terminal through the command line. They must be installed according to the guidelines at the provided URLs, with the base installation directory accessible from the user's $PATH:
- Trinity (v. 2.8.4): https://github.com/trinityrnaseq/trinityrnaseq/wiki
- TransDecoder (v. 5.5.0): https://github.com/TransDecoder/TransDecoder/wiki
- Trinotate (v. 3.1.1): https://github.com/Trinotate/Trinotate.github.io/wiki
- NCBI BLAST+ (v. 2.7.1): http://www.ncbi.nlm.nih.gov/books/NBK52640/
- HMMER (v. 3.2.1): http://hmmer.org/
Assemble the mRNA transcriptome
1.Use Trinity to de novo assemble the clean mRNA-seq reads.
To do this, type the following command into the terminal:
Trinity --seqType fq --max_memory 50G --CPU 16 --samples_filesamples.txt--SS_lib_type RF
E E1 /path/to/mRNA_E1_10k_R1.fastq /path/to/mRNA_E1_10k_R2.fastq
E E2 /path/to/mRNA_E2_10k_R1.fastq /path/to/mRNA_E2_10k_R2.fastq
E E3 /path/to/mRNA_E3_10k_R1.fastq /path/to/mRNA_E3_10k_R2.fastq
J J1 /path/to/mRNA_J1_10k_R1.fastq /path/to/mRNA_J1_10k_R2.fastq
J J2 /path/to/mRNA_J2_10k_R1.fastq /path/to/mRNA_J2_10k_R2.fastq
J J3 /path/to/mRNA_J3_10k_R1.fastq /path/to/mRNA_J3_10k_R2.fastq
>TRINITY_DN66_c0_g1_i1 len=299 path=[0:0-99 2:100-298]
CATTACTCCCAACGTCCATCTCTCTCCGAAGCTCTTTCTCTCTCTTCAACTCCAAGTTCTTTACCTTTTAAGAGAGAGGAAAATGGAGGCCATGAGGATTAGGGTTGCATTCGCCGTCGTGATGATGCTCATCGCCGTCTCCACCGTCCAGAACGCCGCCGCCGCCGATGCTCCGGCGCCGGCACCGACCTCCGACGCCGCCACCTTCCTCCCCGCCGCATTTGTCTCCCTCGTCGCCCTAGCATTCGGCTTTCTCTTCTGAATTTCGAGATTCTCCGTTACCTTTTGTTCAATTTTTG
Identify open reading frames
2.Using TransDecoder, extract all ORFs from the assembled transcripts.
To do this, type the following command into the terminal:
TransDecoder.LongOrfs -tTrinity.fasta-S
3.Use TransDecoder to predict the most likely ORFs.
TransDecoder.Predict -tTrinity.fasta
Create an annotation report
4.Using Trinotate, create and populate a standardized ‘boilerplate’ database and obtain current releases of Swissprot and Pfam.
$TRINOTATE_HOME/admin/Build_Trinotate_Boilerplate_SQLite_db.pl Trinotate
- Trinotate.sqlite (Trinotate boilerplate SQL file)
- uniprot_sprot.pep (Swissprot file to be used for BLAST searches)
- Pfam-A.hmm.gz (Pfam file to be used for protein domain searches)
5.Create a BLAST protein database from the SwissProt file to enable downstream BLAST analyses.
makeblastdb -in uniprot_sprot.pep -dbtype prot
6.Unzip the Pfam file and prepare it for use with Hmmscan.
gunzip Pfam-A.hmm.gz
hmmpress Pfam-A.hmm
7.Conduct a BLASTX search of the Trinity transcripts against the SwissProt database.
blastx -queryTrinity.fasta-db uniprot_sprot.pep -num_threads 16 -max_target_seqs 1 -outfmt 6 -evalue 1e-3 -strand plus >trinity.blastx.outfmt6
8.Similarly, conduct a BLASTP search of the TransDecoder-predicted proteins against the Swissprot database.
blastp -queryTrinity.fasta.transdecoder.pep-db uniprot_sprot.pep -num_threads 16 -max_target_seqs 1 -outfmt 6 -evalue 1e-3 >transdecoder.blastp.outfmt6
9.Use Hmmer to identify Pfam protein domains in the TransDecoder-predicted proteins.
hmmscan --cpu 16 --domtbloutTrinotatePFAM.outPfam-A.hmmTrinity.fasta.transdecoder.pep
10.Now, load all results into the Trinotate database.
- a.First, load the sequences of transcripts and proteins, as well as the gene-transcript relationships specified in the file Trinity.fasta.gene_trans_map. The latter file will be in your Trinity output directory.
Trinotate Trinotate.sqlite init --gene_trans_map
Trinity.fasta.gene_trans_map--transcript_fastaTrinity.fasta--transdecoder_pepTrinity.fasta.transdecoder.pep
- b.Load the BLASTP results.
Trinotate Trinotate.sqlite LOAD_swissprot_blastp
transdecoder.blastp.outfmt6
- c.Load the BLASTX results.
Trinotate Trinotate.sqlite LOAD_swissprot_blastxtrinity.blastx.outfmt6
- d.Load the Pfam results.
Trinotate Trinotate.sqlite LOAD_pfamTrinotatePFAM.out
11.Finally, output the Trinotate annotation report.
Trinotate Trinotate.sqlite report >trinotate_report.xls
C1
Basic Protocol 3: MicroRNA PREDICTION AND ANNOTATION
This protocol covers miRNA prediction and the identification of conserved and novel miRNAs. Plant miRNA precursors tend to be longer and more structurally complex than those of animals; therefore, we use miRDeep-P for miRNA prediction since it adopts plant-specific scoring criteria. First, we process the clean small RNA reads to remove undesired species (rRNA, tRNA, snoRNA, snRNA), preventing their mis-annotation as miRNAs. From the filtered small RNA dataset, we prepare a collapsed small RNA fasta file required by miRDeep-P. miRNAs are then predicted as follows: (1) aligning small RNAs to the reference mRNA transcriptome; (2) extracting and folding the mRNA sequence surrounding aligned regions; (3) evaluating small RNAs within the folded structure, based on experimentally determined features of plant miRNA biogenesis. The ‘evaluation’ step comprises the core algorithm of miRDeep-P. Following prediction, the miRNAs are annotated based on sequence conservation with known plant miRNAs. All input and output files available in the Supporting Information are indicated in code boxes with green and red font, respectively.
Materials
- A computer or high-performance cluster running a UNIX-based operating system
- Clean small RNA-seq dataset from Basic Protocol 1: 1.sRNA_reads/2.clean
- Reference file of plant non-coding RNA contaminants: 6.filter_noncoding_rna/ref_rRNA_tRNA_snoRNA_snRNA.fa
- Perl script used to create a collapsed small RNA fasta file for miRDeep-P: 7.mirdeep.p/collapse.pl
- Assembled mRNA transcriptome from Basic Protocol 2: 3.trinity/Trinity.fasta
- Predicted miRNA sequences supplied from Neller, Klenov, Guzman, and Hudak (2018): 7.mirdeep.p/predicted_miRNAs.fa
- Reference file of known plant miRNAs: 7.mirdeep.p/known_miRNAs.fa
- The following programs will be run in the terminal through the command line. They must be installed according to the guidelines at the provided URLs and accessible from the user's $PATH:
- Bowtie (v. 1.2.2): http://bowtie-bio.sourceforge.net/manual.shtml
- We use Bowtie v.1 rather than v.2 since it is optimized for aligning reads < 50 bp
- miRDeep-P (v. 1.3): https://sourceforge.net/projects/mirdp/
- ViennaRNA (version 2.4.10): https://www.tbi.univie.ac.at/RNA/
- NCBI BLAST+ (v. 2.7.1): http://www.ncbi.nlm.nih.gov/books/NBK52640/
Remove non-coding RNA contaminants
1.Index the contaminant reference file so it can be used with Bowtie.
bowtie-build -f --threads 16ref_rRNA_tRNA_snoRNA_snRNA.fancRNA_junk
2.Using Bowtie, align each clean small RNA file to the indexed reference.
bowtie -f -v 3 --norc --unE1_sRNA_junk_filtered.fa--threads 16 ncRNA_junksRNA_E1_10k_clean.fasta
Prepare the small RNA file for miRDeep-P
3.Combine the six filtered small RNA files (output from step 2) into a single fasta file.
catE1_sRNA_junk_filtered.fa E2_sRNA_junk_filtered.fa
E3_sRNA_junk_filtered.fa J1_sRNA_junk_filtered.fa
J2_sRNA_junk_filtered.fa J3_sRNA_junk_filtered.fa>
combined_E_J_sRNA.fa
4.Create a collapsed fasta file from the newly combined small RNA file.
Use the provided script collapse.pl.
perl collapse.plcombined_E_J_sRNA.faEJ >
combined_E_J_sRNA_collapsed.fa
>EJ_0_x66
TGACAGAAGAGAGTGAGCAC
Predict the miRNAs
5.Perform miRNA prediction using miRDeep-P.
- a.Index the reference transcriptome for use with Bowtie.
bowtie-buildTrinity.fastaTrinity
- b.Align the collapsed small RNA file to the indexed reference transcriptome using Bowtie.
bowtie -f -v 0 -a Trinitycombined_E_J_sRNA_collapsed.fa>
sRNA_aligned.sam
- c.Convert the Bowtie output (.sam) to resemble BLAST output format (.blast).
perl convert_bowtie_to_blast.plsRNA_aligned.sam
combined_E_J_sRNA_collapsed.fa Trinity.fasta>sRNA_aligned.blast
- d.Filter out cases where a small RNA sequence aligned to 15 or more different transcripts.
perl filter_alignments.plsRNA_aligned.blast
-c 15 >sRNA_aligned_15.blast
- e.Obtain the sequences of candidate miRNA precursors.
perl excise_candidate.plTrinity.fasta sRNA_aligned_15.blast250 >precursors.fa
- f.Fold candidate miRNA precursors using the RNAfold program of ViennaRNA.
catprecursors.fa| RNAfold >precursors_struct
- g.
Refine alignments by re-aligning small RNAs to the shorter region excised from potential precursors.
We issue the same commands as above to index the precursor reference file, align small RNAs, and reformat the output:
bowtie-build -fprecursors.faPrecursors
bowtie -f -v 0 -a Precursorscombined_E_J_sRNA_collapsed.fa>
sRNA_precursors_aligned.sam
perl convert_bowtie_to_blast.plsRNA_precursors_aligned.sam
combined_E_J_sRNA_collapsed.fa precursors.fa>
sRNA_precursors_aligned.blast
- h.From the refined alignments produced by the previous step, create a signatures file.
sort +3 -25sRNA_precursors_aligned.blast>signatures
- i.Run miRDeep-P to obtain miRNA predictions.
perl miRDP.plsignatures precursors_struct>miRNA_predictions
Annotate conserved and novel miRNAs
6.Create a BLAST database of known plant miRNAs.
makeblastdb -inknown_miRNAs.fa-dbtype nucl
7.Conduct a BLASTN search of the predicted miRNAs against known plant miRNAs.
blastn -task blastn-short -db known_miRNAs.fa -query
predicted_miRNAs.fa-max_target_seqs 1 -num_threads 16 -outfmt 6 -
strand plus -evalue 1e-3 >blast_annotated_miRNAs.txt
Basic Protocol 4: DIFFERENTIAL EXPRESSION ANALYSIS
This protocol covers mRNA and miRNA differential expression analysis. We begin by quantifying mRNA transcript abundance. The clean RNA-seq reads are aligned back to the reference transcriptome using Bowtie2, then alignments are processed with RSEM to estimate the number of reads derived from each transcript. From these results, we generate a matrix containing the number of reads per transcript for each sample. This count matrix is used as input for differential expression testing with edgeR. miRNA differential expression analysis follows the same procedure, except that quantification with RSEM is not necessary (since one small RNA read = one miRNA). This protocol relies on wrapper scripts packaged with Trinity. Wrapper scripts are helpful for those with limited command-line experience, executing programs ‘behind the scene’ rather than requiring direct interaction from the user. Note that the programs must be installed correctly and be accessible from the user's $PATH in order to work successfully. All input and output files available in Supporting Information are indicated in code boxes with green and red font, respectively.
Materials
- A computer or high-performance cluster running a UNIX-based operating system
- Sample information file from Basic Protocol 2: 3.trinity/samples.txt
- Assembled mRNA transcriptome from Basic Protocol 2: 3.trinity/Trinity.fasta
- Gene-transcript map from Basic Protocol 2: 3.trinity/Trinity.fasta.gene_trans_map
- miRNA count matrix from Neller et al. (2018): 8.edgeR/2.DE_miRNA.counts.matrix
- The following programs will be run in the terminal through the command line. They must be installed according to the guidelines at the provided URLs and be accessible from the user's $PATH:
- Trinity (v. 2.8.4): https://github.com/trinityrnaseq/trinityrnaseq/wiki
- We will use wrapper scripts packaged with Trinity to execute the programs below
- Bowtie2 (v. 2.3.4.3): http://bowtie-bio.sourceforge.net/bowtie2/manual.shtml
- RSEM (v. 1.3.1): http://deweylab.github.io/RSEM/
- R (v. 3.5.2): https://www.r-project.org/
- edgeR package (v. 3.8): https://bioconductor.org/packages/release/bioc/html/edgeR.html
Prepare mRNA expression matrices
1.For each sample, quantify mRNA transcript abundance with RSEM.
Use the Trinity wrapper script align_and_estimate_abundance.pl:
$TRINITY_HOME/util/align_and_estimate_abundance.pl --transcriptsTrinity.fasta--est_method RSEM --aln_method bowtie2 --samples_filesamples.txt--seqType fq --SS_lib_type RF --thread_count 16
2.Create expression matrices from the individual RSEM results of each sample.
Use the Trinity wrapper script abundance_estimates_to_matrix.pl:
$TRINITY_HOME/util/abundance_estimates_to_matrix.pl --est_method RSEM --gene_trans_mapTrinity.fasta.gene_trans_map--name_sample_by_basedir./E1/RSEM.isoforms.results./E2/RSEM.isoforms.results./E3/RSEM.isoforms.results./J1/RSEM.isoforms.results./J2/RSEM.isoforms.results./J3/RSEM.isoforms.results
E1
E1
Identify differentially expressed mRNAs
3.Using the counts matrix from step 2, conduct differential expression testing with edgeR.
Use the Trinity wrapper script run_DE_analysis.pl:
$TRINITY_HOME/Analysis/DifferentialExpression/run_DE_analysis.pl --matrixRSEM.isoform.counts.matrix--method edgeR --samples_filesamples.txt--contrastscontrasts.txt
sampleA
4.Using the normalized expression matrix from step 2, extract and cluster the differentially expressed transcripts.
$TRINITY_HOME/Analysis/DifferentialExpression/analyze_diff_expr.pl --matrixRSEM.isoform.TMM.EXPR.matrix-P 0.05 -C 0 --samplessamples.txt
Identify differentially expressed miRNAs
5.Conduct differential expression testing with edgeR, as shown in step 3.
Use the provided file miRNA.counts.matrix, which contains counts of the 582 predicted miRNAs from Neller et al. (2018).
$TRINITY_HOME/Analysis/DifferentialExpression/run_DE_analysis.pl --matrixmiRNA.counts.matrix--method edgeR --contrastscontrasts.txt
Basic Protocol 5: PREDICTION OF microRNA/TARGET PAIRS
In this protocol, we use the plant small RNA analysis server (psRNAtarget) to predict miRNA/target pairs from the subset of differentially expressed miRNAs and mRNAs identified in Basic Protocol 4. We also create a ‘background’ set for downstream functional enrichment analysis, composed of predicted interactions between all miRNAs and mRNAs. psRNAtarget evaluates sequence complementarity between miRNAs and potential targets using a scoring system benchmarked against experimentally validated plant miRNA/target pairs. All input and output files are available in Supporting Information.
Materials
- A personal computer with internet access
- The Plant Small RNA Target Analysis Server (psRNAtarget): https://plantgrn.noble.org/psRNATarget/home
- Sequences of differentially expressed miRNAs identified in Basic Protocol 4: 9.psRNAtarget/DE_miRNAs.fa
- Sequences of differentially expressed mRNAs identified in Basic Protocol 4: 9.psRNAtarget/DE_mRNAs.fa
- Assembled mRNA transcriptome from Basic Protocol 2: 3.trinity/Trinity.fasta
- Predicted miRNAs from Basic Protocol 3: 7.mirdeep.p/predicted_miRNAs.fa
1.Navigate to psRNAtarget at: https://plantgrn.noble.org/psRNATarget/home.
2.Predict interactions between the differentially expressed miRNAs and mRNAs.
Under the ‘Analysis’ tab, select ‘Submit small RNAs and targets’. Upload the provided files DE_miRNAs.fa and DE_mRNAs.fa for the small RNA file and target file, respectively. Run the job with default parameters. Download the results and open them with Microsoft Excel or another spreadsheet program.
3.Repeat step 2 to predict interactions between all miRNAs and mRNAs.
Upload predicted_miRNAs.fa and Trinity.fa for the small RNA file and target file, respectively.

Basic Protocol 6: FUNCTIONAL ENRICHMENT ANALYSIS OF DIFFERENTIALLY EXPRESSED microRNA/TARGET PAIRS
This protocol covers GO term functional enrichment analysis. GO terms are structured, curated concepts relating to gene functions. A gene product may be annotated in up to three categories: molecular function, cellular component, and biological process. The ontology is structured as a directed graph in which each term has defined relationships to one or more other terms. Lower-level terms have greater specificity than higher-level ones. In this protocol, we use GOseq to identify functional associations that are over-represented among the differentially expressed miRNA/target pairs. GOseq also corrects for transcript length bias: the fact that longer transcripts are more likely to be identified as differentially expressed than shorter transcripts. Note that GO terms are only ascribed to proteins; therefore, this analysis is conducted on targets of miRNAs, not the miRNAs themselves. All input and output files available in Supporting Information are indicated in code boxes with green and red font, respectively.
Materials
- A computer or high-performance cluster running a UNIX-based operating system
- Trinotate report from Basic Protocol 2: 5.trinotate/trinotate_report.xls
- Assembled mRNA transcriptome from Basic Protocol 2: 3.trinity/Trinity.fasta
- Provided target ID file for the test set (differentially expressed miRNA/target pairs) identified in Basic Protocol 5, step 2: 10.goseq/DE_targets_of_DE_miRNAs.txt
- Provided target ID file for the background set (all miRNA/target pairs) identified in Basic Protocol 5, step 3: 10.goseq/targets_of_all_miRNAs.txt
- The following programs will be run in the terminal through the command line. They must be installed according to the guidelines at the provided URLs and accessible from the user's $PATH:
- Trinotate (v. 3.1.1): https://github.com/Trinotate/Trinotate.github.io/wiki
- Trinity (v. 2.8.4): https://github.com/trinityrnaseq/trinityrnaseq/wiki
- We will use a wrapper script packaged with Trinity to execute the programs below:
- R (v. 3.5.2): https://www.r-project.org/
- GOseq package (v. 3.8): https://bioconductor.org/packages/release/bioc/html/goseq.html
1.Extract all GO terms from the Trinotate annotation report.
$TRINOTATE_HOME/util/extract_GO_assignments_from_Trinotate_xls.pl --Trinotate_xlstrinotate_report.xls--trans --include_ancestral_terms >go_annotations.txt
2.Create a transcript lengths file for use with GOseq.
$TRINITY_HOME/util/misc/fasta_seq_length.plTrinity.fasta>
Trinity.fasta.seq_lens
3.Run GOseq to test for enriched GO terms associated with differentially expressed miRNA/target pairs.
Provide the extracted GO terms from step 1, the lengths file from step 2, and the mRNA transcript IDs of targets in the test set (differentially expressed miRNA/target pairs) and reference set (all miRNA/target pairs).
$TRINITY_HOME/Analysis/DifferentialExpression/run_GOseq.pl --genes_single_factorDE_targets_of_DE_miRNAs.txt--GO_assignmentsgo_annotations.txt--lengthsTrinity.fasta.seq_lens--backgroundtargets_of_all_miRNAs.txt
Basic Protocol 7: DETECTION OF microRNA-INDUCED CLEAVAGE USING RNA LIGASE–MEDIATED 5′ RACE
In this protocol, we cover the validation of an miRNA/target pair using RNA ligase–mediated 5′ RACE (RLM-RACE; Fig. 5). Specifically, the 5′ end of the expected cleaved target is cloned and sequenced to assess whether it underwent slicing by a miRNA-guided Argonaute protein. This method takes advantage of the exposed 5′ monophosphate on the target that results from miRNA-induced cleavage. First, the poly(A) mRNA pool (from the Support Protocol) undergoes 5′ ligation with an RNA adapter. Then, the mRNA pool is reverse transcribed into cDNA using a 3′ d(T) adapter. To enrich for low-abundance cleavage products, the cDNA pool is subjected to nonspecific PCR amplification using primers annealing to the RNA adapter and 3′ d(T) adapter. Finally, the specific target is amplified using a 5′ adapter primer and 3′ gene-specific primer. Sequences to facilitate cloning into a vector using Gibson assembly are introduced by a subsequent PCR of the specific target. The cloned target is transformed into E. coli , with successful transformants identified by colony PCR, and plasmid DNA is sequenced. An miRNA/target pair is considered validated if, after alignment, the 5′ end of the cloned sequences lines up with the expected miRNA cut site on the mRNA.

While kits for RLM-RACE are available, they contain reagents not relevant here, which select for full-length, capped mRNAs. Therefore, we did not purchase any kit, though our protocol is based on the GeneRacer RLM-RACE kit (ThermoFisher L1500-01). The RNA adapter and all primers were synthesized separately, and decapping and dephosphorylating steps were omitted.
Materials
-
Poly(A) mRNA pool (Support Protocol)
-
Primers (and RNA adapters) in Table 1
-
10× T4 RNA ligase buffer (NEB B0216L)
-
10 mM ATP (NEB P0756S)
-
40 U/µl murine RNase inhibitor (NEB M0314S)
-
10 U/µl T4 RNA ligase 1 (NEB M0204S)
-
DOW Corning High Vacuum Grease, autoclaved in 5-ml syringe without needle
-
Acid PCI: 25:24:1 (v/v/v) citrate-buffered phenol/chloroform/isoamyl alcohol [25 ml citrate-buffered phenol (Sigma P4682), 24 ml chloroform, 1 ml isoamyl alcohol]
-
Precipitation mix: 50 ml of ethanol, 2 ml of 3 M sodium acetate, pH 5.2
-
5 mg/ml linear acrylamide (ThermoFisher AM9520)
-
10 mM dNTPs (NEB N0446S)
-
5× first-strand buffer (see recipe)
-
0.1 M DTT
-
200 U/µl Superscript III Reverse Transcriptase (ThermoFisher 18080093)
-
RNase H (NEB M0297S)
-
5× Q5 polymerase reaction buffer (NEB B9027S)
-
2 U/µl Q5 DNA polymerase (NEB M0491S)
-
EZ-10 Spin Column Plasmid DNA Miniprep Kit (BioBasic BS413)
-
EZ-10 Spin Column DNA Gel Extraction Kit (Biobasic BS353)
-
Cloning vector (pHSG298; Takara 3298)
-
2× Gibson assembly mastermix (NEB E2611S)
-
5-α competent E. coli (NEB C2987I)
-
LB plates with appropriate antibiotic for cloning vector
-
2 U/µl pfu-sso7d or equivalent polymerase
-
5× pfu-sso7d buffer (see recipe)
-
Thermal cycler with gradient function
-
UV spectrometer
-
0.2-ml PCR tubes
-
Sanger sequencing facility
-
Additional reagents and equipment for agarose gel electrophoresis (Voytas, 2000)
| Primer name | Sequence (5′ to 3′) | Usage |
|---|---|---|
| RNA Adapter | CGACUGGAGCACGAGGACACUGACAUGGACUGAAGGAGUAGAAA | Basic Protocol 7, step 1 |
| 3′ d(T) Adapter | GCTGTCAACGATACGCTACGTAACGGCATGACAGTG-d(T)24 | Basic Protocol 7, step 6 |
| 3′ Adapter Primer (3′AP) | GCTGTCAACGATACGCTACGTAACG | Basic Protocol 7, step 11 |
| 5′ Adapter Primer (5′AP) | CGACTGGAGCACGAGGACACTGA | Basic Protocol 7, step 11, 13 |
| Gene Specific Primer 1 (GSP1) | …XXXXXXXX… | Basic Protocol 7, step 13 |
| 5′ Nested Adapter Primer (5′NAP) | GGACACTGACATGGACTGAAGGAGTA | Basic Protocol 7, step 17 |
| Gene Specific Primer 2 (GSP2) | …XXXXXXXX… | Basic Protocol 7, step 16 |
| 5′ Cloning Nested Adapter Primer | …NNNNNNNN-GGACACTGACATGGACTGAAGGAGTA | Basic Protocol 7, step 20 |
| Cloning Gene Specific Primer 2 | …NNNNNNNN-XXXXXXXX… | Basic Protocol 7, step 20 |
| T7 Promoter Primer | TAATACGACTCACTATAGGG | Basic Protocol 7, step 26 |
| T7 Terminator Primer | GCTAGTTATTGCTCAGCGG | Basic Protocol 7, step 26 |
- a
N's indicate nucleotides added to the primer to facilitate Gibson assembly, and will share ∼15 to 30 bp of sequence complementarity with the cloning vector. X's indicate sequence complementarity with the mRNA under validation.
Ligate the 5′ RNA adapter to the mRNA pool
1.Combine 250 ng of poly(A) mRNA and 250 ng of RNA adapter (sequence provided in Table 1) in a total volume of 7 µl. Incubate at 65°C for 5 min to denature. Chill on ice for 2 min.
2.Add the following reagents to the denatured mRNA:
- 1 µl 10× T4 RNA ligase buffer
- 1 µl 10 mM ATP
- 1 µl 40 U/µl murine RNase inhibitor
- 0.5 µl 10 U/µl T4 RNA ligase 1
- 0.5 µl distilled H2O.
3.Incubate at 37°C for 1 hr to ligate, then place on ice.
4.Into a new 1.5-ml microcentrifuge tube, dispense 50 µl of autoclaved high vacuum grease using a syringe. Microcentrifuge for 30 sec at 16,000 × g , 4°C, to collect grease at the bottom of the tube. Add the ligated mRNA, 90 µl distilled H2O, and 100 µl of acid PCI. Vortex for 30 sec, then microcentrifuge 5 min at 16,000 × g, 4°C.
5.Transfer the supernatant to a new 1.5-ml tube. Add 2.6 volumes of precipitation mix and 4 µl of 5 mg/ml linear acrylamide. Precipitate on dry ice for 10 min or at −20°C overnight.
Centrifuge the ligated mRNA 20 min at 16,000 × g , 4°C. Discard the supernatant and add 1 ml of 75% ethanol. Invert the tube several times, then centrifuge at 16,000 × g , 4°C for an additional minute. Pour off the supernatant, quick-spin once more with the same conditions, and pipette off the remaining supernatant. Resuspend the pellet in 11 µl distilled H2O.
Reverse transcribe the ligated mRNA pool
6.Add the following reagents to the mRNA:
- 1 µl 10 µM 3′ d(T) adapter
- 1 µl 10 mM dNTPs.
7.Denature the mRNA at 65°C for 10 min. Place on ice for at least 2 min. Add the following reagents and mix well:
- 4 µl 5× first-strand buffer
- 1 µl 0.1 M DTT
- 1 µl 40 U/µl murine RNase inhibitor
- 1 µl 200 U/µl Superscript III Reverse Transcriptase.
8.Incubate at 50°C for 1 hr to reverse transcribe the mRNA into cDNA.
9.Incubate at 70°C for 15 min to inactivate the reverse transcriptase.
10.Add 2 U of RNase H to the reverse transcription reaction and incubate at 37°C for 20 min.
Amplify the cDNA pool
11.Using 1 µl of reverse transcription product as template, set up the following reaction to perform nonspecific PCR amplification of the cDNA pool:
- 5 µl 5× Q5 polymerase buffer
- 1.25 µl 10 µM 5′ adapter primer
- 1.25 µl 10 µM 3′ adapter primer
- 0.5 µl 10 mM dNTPs
- 1 µl reverse transcription product as template
- 0.25 µl 2 U/µl Q5 DNA polymerase
- 15.75 µl distilled H2O.
12.Place into a thermal cycler and amplify with the touchdown program shown in Figure 6A.

Amplify the specific cDNA target
13.Using 1 µl of nonspecific PCR product as template, set up the following reaction for the first gene-specific amplification:
- 5 µl 5× Q5 polymerase buffer
- 1.25 µl 10 µM 5′ adapter primer
- 1.25 µl 10 µM gene specific primer 1
- 0.5 µl 10 mM dNTPs
- 1 µl nonspecific PCR product as template
- 0.25 µl 2 U/µl Q5 DNA polymerase
- 15.75 µl distilled H2O.
14.Place into a thermal cycler and amplify as shown in Figure 6B.
15.Separate 8 µl of gene-specific PCR product on a 1.2% agarose gel, then visualize.
16.Dilute the gene-specific PCR product 1:10 with distilled H2O and use 1 µl as template in the following reaction to perform nested PCR. Use the program in Figure 6B.
- 5 µl 5× Q5 polymerase buffer
- 1.25 µl 10 µM 5′ nested adapter primer
- 1.25 µl 10 µM gene specific primer 2
- 0.5 µl 10 mM dNTPs
- 1 µl diluted gene-specific PCR product
- 0.25 µl 2 U/µl Q5 DNA polymerase
- 15.75 µl distilled H2O.
17.Separate the PCR product on a gel as in step 15.
18.If a single band is observed at the expected size, dilute the reaction 1:10 with distilled H2O and proceed to amplification, step 21, for cloning. If multiple bands are observed, separate the remainder of the PCR reaction on a 1% low-melt agarose gel (Voytas, 2000) and excise the band corresponding to the expected product size.
19.Purify the gel fragment with the EZ-10 Spin Column DNA Gel Extraction Kit according to the manufacturer's protocol, then quantify with a UV spectrometer.
Clone the amplified cDNA target using Gibson assembly
20.Use 1 µl of diluted PCR product from step 18 or 1 ng of purified fragment from step 19, and set up the following reaction. Use the PCR program in Figure 6B:
- 5 µl 5× Q5 polymerase buffer
- 1.25 µl 10 µM 5′ cloning nested adapter primer
- 1.25 µl 10 µM cloning gene specific primer 2
- 0.5 µl 10 mM dNTPs
- 1 µl diluted PCR or gel-extracted product (from step 18 or 19)
- 0.25 µl 2 U/µl Q5 DNA polymerase
- 15.75 µl distilled H2O.
21.Gel purify (steps 18 and 19) the resultant PCR product of the expected size and quantify with a UV spectrometer.
22.Combine the cloning vector pHSG298 and insert (i.e., the purified product from step 21) at a 1:2 molar ratio. Also perform a 1:0 reaction as a negative control. Perform Gibson assembly according to manufacturer's protocol.
23.Transform 1 to 5 µl of Gibson assembly product into 50 µl of competent NEB 5-α E. coli , per manufacturer's instructions. Plate onto LB plates with appropriate antibiotics and incubate at 37°C overnight.
Screen transformants using colony PCR
24.Select a 1-mm-diameter colony from the experimental plate and resuspend in 50 µl distilled H2O. Spot 1 µl of the suspension onto an antibiotic patch plate to preserve the clone.
25.Lyse the resuspended E. coli by incubating at 95°C for 5 min. Centrifuge 10 min at 16,000 × g , room temperature, to pellet cell debris. Transfer 10 µl of supernatant to a fresh 0.2-ml PCR tube.
26.Set up the following reaction to perform colony PCR. Use primers that flank the cloning site in your vector, typically the T7 promotor and terminator.
- 5 µl 5× pfu-sso7d buffer
- 1.25 µl 10 mM T7 promotor primer
- 1.25 µl 10 mM T7 terminator primer
- 0.5 µl 10 mM dNTPs
- 10 µl bacterial lysate (from step 26)
- 0.25 µl 2 U/µl pfu-sso7d polymerase
- 15.75 µl distilled H2O.
Amplify with the PCR program illustrated in Figure 6B, using 58°C rather than the gradient annealing step.
27.Separate colony PCR products on a 1% agarose gel, then visualize. For colonies containing the appropriately sized insert, grow liquid cultures using the patch plate from step 24.Extract plasmid DNA from these cultures using a BioBasic EZ-10 Spin Column Plasmid DNA Miniprep Kit and send to a facility for Sanger sequencing with a forward and reverse primer flanking the cloning site.
Analyze the alignments
28.Upon obtaining the sequences, align them to the target mRNA sequence using software. We recommend the online tool Benchling (http://www.benchling.com).
-
Create an account and log on to Benchling. In the tool bar on the left, select the ‘Projects’ tab. Click on the (+) icon to import your mRNA sequence.
-
In the tool bar on the right, click on ‘Alignments’ and select ‘Create New Alignment’. Input the sequences of your individual clones as well as the sequence of the putative miRNA. Choose MAFFT as your alignment algorithm. Click ‘Create Alignment’ and view your results. If your positive control is miR156 with target SPL, the alignment of your colony sequences should resemble Figure7.
miRNAs induce cleavage between the 10thand 11thnucleotides, 5′ to 3′, of the miRNA. 5′ ends of cloned sequences that align to this site indicate successful validation of cleavage for the miRNA/target pair.
Cleavage at unexpected sites suggests that the miRNA is not cleaving the target as anticipated, or that this small RNA is not a functional miRNA (i.e., false positive). In some cases, we observed randomly distributed cleavage surrounding the expected site. This suggested that the limit of detection had been reached: background levels of mRNA degradation were as high as the miRNA cleavage. Thus, it is difficult to obtain trustworthy, reproducible results with RLM-RACE when abundance of the miRNA and/or target is low (e.g., < 10 counts per million and < 100 transcripts per million respectively).

Support Protocol: TOTAL RNA EXTRACTION AND ISOLATION OF SMALL RNA AND mRNA
In this protocol, we cover total RNA extraction from plant tissue and steps to enrich for either small RNA or mRNA. RNAzol reagent is used to enable extraction of both total RNA and small RNA from a single plant sample. Total RNA is obtained first. Plant tissue is ground in liquid nitrogen, followed by addition of RNAzol; DNA and protein contaminants are removed through centrifugation, then total RNA is precipitated with ethanol. Using the supernatant reserved from total RNA extraction, the small RNA fraction is then obtained by isopropanol precipitation. mRNA enrichment is performed using the NEBNext Poly(A) mRNA Magnetic Isolation Module on the previously extracted total RNA. Starting with high-quality RNA is critical for the success of downstream validations. Specifically, total RNA, small RNA, and mRNA will be used for qRT-PCR and stem-loop qRT-PCR (Basic Protocol 8), and RLM-RACE (Basic Protocol 7), respectively. Small-RNA enrichment serves to concentrate low-abundance miRNAs to enable their detection. Similarly, mRNA enrichment concentrates low-abundance targets; it also removes non-polyadenylated transcripts and ribosomal RNA so they will not be cloned inadvertently during RLM-RACE.
Materials
-
Fresh leaf tissue of interest
-
RNAzol reagent (Molecular Research Center RN190)
-
100% and 75% (v/v) ethanol
-
RNA storage buffer [1 mM citrate buffer, pH 6.4 (see recipe) containing 22.5 mM DTT]
-
100% isopropanol
-
NEBNext Poly(A) mRNA magnetic isolation module (NEB E7490S)
Extract total RNA
1.Grind 100 mg of fresh leaf tissue to a fine powder with liquid nitrogen in a 1.5-ml microcentrifuge tube. Add 1 ml RNAzol directly to the tissue and immediately vortex to homogenize.
2.Add 0.4 ml distilled H2O to the RNAzol and vortex briefly. Incubate at room temperature for 10 min.
3.Microcentrifuge the lysate 15 min at 16,000 × g , room temperature, to pellet DNA and protein. Carefully remove 1 ml of the supernatant (75%) and transfer it to a clean 1.5-ml tube.
4.Add 0.4 ml 100% ethanol to the tube and place on ice for 10 min. Microcentrifuge 20 min at 16,000 × g , 4°C, to pellet total RNA. Transfer supernatant to a fresh tube and keep on ice for step 7.
5.Add 1 ml 75% ethanol to the total RNA pellet. Invert the tube several times, then microcentrifuge 1 min at 16,000 × g , 4°C. Pour off the ethanol and microcentrifuge under the same conditions for 30 sec. Pipette off remaining liquid and discard.
6.Resuspend the total RNA pellet in 20 µl of pre-warmed (50°C) RNA storage buffer. Quantify the RNA with UV spectroscopy by measuring the absorbance at 260 nm.
Isolate small RNA
7.To the saved supernatant from step 4, add 0.8 volumes 100% isopropanol. Chill on ice for 30 min. Centrifuge 20 min at 16,000 × g , 4°C, to pellet small RNA. Pour off the supernatant.
8.Repeat steps 5 and 6 to purify and resuspend the small RNA.
Isolate mRNA
9.Using 30 µg of total RNA from step 6, carry out mRNA enrichment according to the product manual of the NEBNext Poly(A) mRNA magnetic isolation module. Upon completion, quantify the RNA with UV spectroscopy by measuring the absorbance at 260 nm.
Basic Protocol 8: QUANTIFICATION OF microRNA AND TARGET EXPRESSION USING qRT-PCR
Here, we cover validation of miRNA and target differential expression results from RNA-seq using stem-loop qRT-PCR and standard qRT-PCR, respectively. Both methods rely on the same underlying principle: a relative fold change is determined by quantifying RNA levels of the species of interest under each condition and normalizing the expression change to an internal control that is stably expressed for the treatment under study. To prepare for qPCR, the RNA of interest and the internal control are both reverse transcribed into cDNA using gene-specific primers. Then, qPCR is conducted to quantify cDNA levels, based on fluorescence occurring from intercalation of dye into the double-stranded PCR product. In terms of primer design for stem-loop and standard qRT-PCR (Fig. 8), the latter is straightforward in that primers must simply produce a product of 150 to 200 bp. In contrast, the short length of miRNAs (∼21 nt) necessitates use of a specialized stem-loop primer during reverse transcription to increase the melting point of the product, followed by PCR using a reverse primer complementary to the stem-loop and a forward primer specific to the miRNA.

Following qPCR, the ΔΔCt formula is used to determine the relative fold change. The Ct value is the PCR cycle at which fluorescence rises above a set threshold during exponential amplification. The lower the Ct value, the higher the abundance of the RNA under study. A statistical test is conducted to determine whether the RNA abundance is significantly different between the experimental and control conditions. Finally, the fold changes obtained by qRT-PCR are correlated with those obtained by RNA-seq to determine whether the two methods of RNA quantification agree. Note that although most miRNA/target pairs under validation should be randomly selected, those of special interest are also tested.
Materials
-
Total RNA or small RNA of interest (Support Protocol)
-
Primers in Table 2
-
10 mM dNTPs (NEB N0446S)
-
5× first-strand buffer (see recipe)
-
0.1 M DTT
-
40 U/µl murine RNase inhibitor (NEB M0314S)
-
200 U/µl Superscript III reverse transcriptase (ThermoFisher 18080093)
-
2× SYBR Green mastermix (Bimake B21202)
-
0.2-ml PCR tubes
-
Rotor-GeneQ qRT-PCR System (Qiagen)
| Primer name | Sequence (5′ to 3′) | Usage |
|---|---|---|
| mRNA/Internal Control Reverse Transcriptase Primer (RTase RP) | …XXXXXXXX… | Basic Protocol 8, step 1 |
| miRNA/Internal Control Stem Loop RTase Primer | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGAC-XXXXXX | Basic Protocol 8, step 1 |
| mRNA/Internal Control Forward Primer (FP) | …XXXXXXXX… | Basic Protocol 8, step 4 |
| mRNA/Internal Control Reverse Primer (RP) | …XXXXXXXX… | Basic Protocol 8, step 4 |
| miRNA/Internal Control Forward Primer | …XXXXXXXX…. | Basic Protocol 8, step 4 |
| Universal Reverse Primer (RP) | CCAGTGCAGGGTCCGAGGTA | Basic Protocol 8, step 4 |
- a
X's indicate sequence complementarity with the miRNA/mRNA under validation.
Reverse transcribe the total RNA and small RNA
1.Dilute 250 ng of total RNA or small RNA (Support Protocol) in 11 µl distilled H2O. Add the following reagents:
- 1 µl 10 µM mRNA reverse transcriptase primer or stem-loop RTase primer
- 1 µl 10 mM dNTPs.
2.Denature the RNA at 65°C for 10 min, then place on ice for at least 2 min. Add the following reagents:
- 4 µl 5× first-strand buffer
- 1 µl 0.1 M DTT
- 1 µl 40 U/µl murine RNase inhibitor
- 1 µl 200 U/µl Superscript III reverse transcriptase.
3.Mix well, then incubate at 50°C for 1 hr to perform reverse transcription. Inactivate the reverse transcriptase at 70°C for 15 min.
Quantify miRNA and target abundance using qPCR
4.For each sample, set up the following reaction in a 1.5-ml microcentrifuge tube to perform qPCR:
- 5 µl reverse transcription product (from step 3)
- 2 µl 10 µM mRNA forward primer or miRNA forward primer
- 2 µl 10 µM mRNA reverse primer or universal reverse primer
- 23 µl distilled H2O
- 33 µl 2× SYBR Green qRT-PCR mastermix.
5.Aliquot the above mixture into three 0.2-ml PCR tubes at 20 µl per tube. Place on ice until you are ready to run the assay in the RotorGene Q system.
6.Insert the tubes into the 36-place rotor and affix the locking ring. Place into the machine and run with the following settings:
- Hold 1: 50°C for 20 sec
- Hold 2: 95°C for 600 sec
- Cycling: 95°C for 15 sec, 60°C for 60 sec
- Melt: Ramp from 58°C to 95°C
- Rising by 1°C each step
- Wait for 90 sec of pre-melt conditioning on first step
- Wait for 5 sec for each step afterwards
- Acquire to Melt A on Green
- Uncheck optimize gain before melt on all tubes.
7.Upon completion of program, remove PCR tubes from machine. Download the run file (.rex) and the Rotor-Gene Q Series software; run in virtual mode with serial number 123456 to analyze data on different computers.
Analyze the qPCR results
8.Use the ΔΔCt equation below to determine relative fold change:
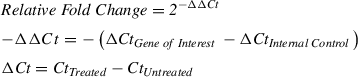
9.After calculation of fold change, test for differential expression using the Student's t -test on Ct values.
10.Create a scatterplot that compares the fold change by RNA-seq against the fold change by qRT-PCR. Make two separate plots, one for miRNAs and one for targets, with each point on the graph representing a single miRNA or target. Calculate the R 2 correlation value to quantify the strength of the linear relationship between the two methods. An example of such a plot comparing the fold change of eight JA-responsive miRNAs measured by RNA-seq and qRT-PCR is illustrated in Neller et al. (2018).
REAGENTS AND SOLUTIONS
First-strand buffer, 5×
- 375 mM KCl
- 15 mM MgCl2
- 250 mM Tris·Cl, pH 8.3
- Store up to 1 year at −20°C
pfu-sso7d buffer, 5×
- 0.5 mg/ml bovine serum albumin (BSA; Sigma B6917)
- 50 mM KCl
- 10 mM MgSO4
- 50 mM ammonium acetate
- 150 mM Tris·Cl, pH 10
- 0.5% (v/v) Triton X-100
- Store up to 1 year at −20°C
Sodium citrate buffer, pH 6.4, 0.1 M
Purchase citric acid monohydrate (210.14 g/mol) and sodium citrate dihydrate (294.12 g/mol). Prepare 80 ml of distilled water in a suitable container. Add 0.21 g of citric acid monohydrate to the solution. Add 2.65 g of sodium citrate dihydrate to the solution. Adjust solution to final desired pH using HCl or NaOH. Add distilled water until volume is 0.1 liter.
COMMENTARY
Background Information
Background on miRNA biogenesis and function
miRNAs are non-coding RNAs of ∼20 to 24 nt that regulate post-transcriptional gene expression of targets (reviewed in Yu, Jia, & Chen, 2017). As shown in Figure 9, miRNA biogenesis begins with transcription of a MIR gene by RNA polymerase II to produce a primary miRNA (pri-miRNA), consisting of a stem-loop region flanked by unstructured arms. In sequential steps, dicer-like 1 RNase (DCL1) excises the stem-loop to form the miRNA precursor (pre-miRNA), then generates a duplex comprising the miRNA and its opposing strand, classically termed microRNA (miRNA). Once in the cytoplasm, the RNA-induced silencing complex (RISC) is established upon association of the miRNA with Argonaute 1 (AGO1). Plant miRNAs tend to have high sequence complementarity with targets and act primarily through target mRNA cleavage rather than translational inhibition, the latter being more common in animals. Although the high sequence complementarity of miRNA/target pairs observed in plants is conducive to RNA cleavage, this is not always the case, as miRNA-mediated translational inhibition has been shown in plants for highly complementary miRNA/target pairs.

Intent of the article
The outcome of this article is a candidate set of biologically relevant miRNA/target pairs, along with validations of differential expression and target cleavage. This information may be sought for the purpose of basic research, i.e., to uncover a novel layer of gene expression regulation for the pathway under study. Indeed, plant miRNAs are implicated in numerous abiotic and biotic stress responses, as well as maintenance of normal growth and development (Noman et al., 2017). Alternatively, one may wish to leverage these results in a more applied manner. For example, the use of miRNAs to manipulate transcript abundance is a popular strategy for crop genetic engineering, with several reports demonstrating its successful application in agronomic trait improvement (Djami-Tchatchou, Sanan-Mishra, Ntushelo, & Dubery, 2017).
This article is intended for researchers studying non-model plants. These species tend to lack public genomic resources, most notably a reference genome or transcriptome. Ideally, miRNA prediction is performed at the genomic level, with several prediction algorithms operating solely on the genome sequence (Rajendiran, Chatterjee, & Pan, 2018). We provide a strategy that considers the unique challenges associated with large-scale bioinformatic analysis of a non-model plant. Specifically, assembly and annotation of a reference transcriptome is easier, faster, and less expensive than that of a genome. The former also requires less specialized knowledge, due in large part to the availability of integrated, user-friendly tools aimed at biologists with limited bioinformatic experience. It is important to note that the use of genome or transcriptome references from related plant species is not recommended. This is because plant miRNAs tend to be highly species-specific, and unlike animals, miRNA precursors are generally not conserved across plants (Bartel, 2004). For plant miRNAs that are conserved, this usually occurs at the level of the mature miRNA (Bartel, 2004). Even so, we advise against miRNA prediction based solely on sequence comparison against mature miRNAs of related species, as the identification of a suitable miRNA precursor is an important feature used to reduce false positive predictions. Therefore, the approach of this article is to create and use resources that are specific to the plant under study.
Strengths and limitations of the procedure
Due to the tendency of plant miRNAs to be species-specific, an advantage of our procedure is that miRNAs are predicted using a transcriptomic reference generated for the species under study. Additionally, the integration of both miRNA and target differential expression in the procedure described in this article provides a filter to identify the most biologically relevant interactions for downstream analysis. This approach assumes that miRNAs and targets whose expression levels change in response to the treatment variable are more likely to be important mediators of the response than those pairs whose expression levels do not change significantly. This is a reasonable assumption given that the expression of an miRNA and its mRNA target tend to be correlated, an attribute that has been exploited successfully by us and others to discover relevant miRNA/target pairs in plants (Ji et al., 2018; Ma et al., 2018; Neller et al., 2018; Ye, Wang, & Wang, 2016). Therefore, an investigation of differentially expressed miRNAs without considering target expression only reveals half of the story. Our use of paired small RNA and mRNA samples in this procedure enables downstream application of correlation methods to further refine miRNA/target relationships.
Restriction of the analysis to targets that are differentially expressed has a limitation in that it may filter out miRNAs acting through translational inhibition rather than transcript cleavage. However, since this mechanism of action is not observed frequently in plants, the limitation is not critical unless the reader is specifically interested in miRNAs that operate in this manner. Furthermore, the two modes of miRNA action may be difficult to distinguish in plants, as miRNA-induced cleavage occurs on targets undergoing active translation (reviewed in Yu et al., 2017). Another limitation of our procedure is that validation by the modified RLM-RACE used in this article only provides information on the presence of cleaved targets, not their relative abundance, and it may be unsuccessful in the case of low-abundance miRNAs or targets, where less cleavage product is available for detection. A high-throughput equivalent to this procedure is degradome sequencing (Lin, Chen, & Lu, 2019). By incorporating RNA-seq, this method enables detection of all cleaved targets in a sample and their relative abundance. It is less economical and requires extensive data analysis, but the combination of small RNA-seq and degradome-seq is highly informative; see Ji et al. (2018) for a recent implementation of this strategy.
Comparison of the bioinformatic workflow with current methods
The workflow presented in this article is based on our experience with available software options. We have prioritized qualities of open access, user friendliness, in-depth documentation, and smooth integration. Some components of our workflow are more advanced than others. For the tasks of de novo transcriptome assembly and annotation, we highly recommend Trinity and its companion software Trinotate and TransDecoder. Additionally, we suggest the use of scripts packaged with Trinity to facilitate differential expression and GO enrichment analyses. Each of these tasks requires an advanced level of expertise that can overwhelm a novice user, resulting in incorrect application of methods. For this reason, we view the integration and guidance provided by Trinity developers as highly advantageous. However, other options do exist for de novo transcriptome assembly. For a plant-focused summary of these tools and other resources, see Geniza and Jaiswal (2017). Additionally, refer to Honaas et al. (2016) for a comparative analysis of transcriptomes generated from different assemblers for the model plants rice and Arabidopsis. It is also possible to bypass de novo transcriptome assembly completely by performing Iso-seq, a relatively new implementation of long-read technology that has been used in plants (An, Cao, Li, Humbeck, & Wang, 2018). Regardless of the chosen strategy, note that Trinotate and TransDecoder can be used for annotating any transcriptome as long as the required inputs are provided.
Other aspects of the workflow are more amenable to user customization. For example, there are numerous options available for miRNA prediction and target identification (reviewed in Rajendiran et al., 2018). Recently, supervised machine learning was used to predict miRNAs in a reference-free manner (Vitsios et al., 2017). Although promising, this approach was more successful in predicting miRNAs for animals than plants. Furthermore, it is unlikely to outperform reference-based prediction for a non-model plant, as the user must provide training data consisting of known miRNAs or instead use the ‘universal plant’ model. Other options for user customization of our workflow are at the level of transcript quantification and differential expression analysis. We used the alignment-based method RSEM for transcript quantification, but alignment-free methods such as Kallisto and Salmon are also popular due to their improved speed. Although alignment-free and alignment-based methods are comparable in accuracy for standard investigations like protein-coding mRNA quantification, note that alignment-based methods perform better when quantifying small or low-abundance RNAs (Wu, Yao, Ho, Lambowitz, & Wilke, 2018). The reader may also wish to investigate alternatives for differential expression analysis, with popular options including DESeq2 and Voom/Limma. The Trinity accessory scripts used in our workflow support these various programs/packages for transcript quantification and differential expression analysis, thereby accommodating differences in experimental design and user preference. For a comparison of RNA-seq mapping methods (both alignment-free and alignment-based) and differential expression tools, see Costa-Silva, Domingues, and Lopes (2017).
Extended bioinformatic analysis
With the paired small RNA and mRNA samples as used in our workflow, the investigator is equipped to perform advanced correlation analysis for obtaining greater insight into miRNA/target interactions. In our study, we computed the Pearson correlation coefficient (PCC) for the expression of each miRNA and its target(s) and imposed a PCC cut-off to filter the set of candidate pairs. The PCC ranges from −1 to +1, indicating perfect negative and positive linear association, respectively. There is a tendency in literature to retain only negatively correlated miRNA/target pairs. This derives from the rationale that a cleavage-inducing miRNA acting on its target in absence of other influences leads to reduced target expression due to RNA degradation. However, we and others have observed and validated positively correlated miRNA/target pairs. This dynamic can arise from miRNA-mediated spatial restriction of the target (Kawashima et al., 2009; Kidner & Martienssen, 2004; Levine, McHale, & Levine, 2007; Nikovics et al., 2006). It may also indicate that the miRNA functions in a ‘buffering’ capacity, minimizing changes in target expression caused by other interacting factors (Wu, Shen, & Tang, 2009). For these reasons, we do not recommend restricting analysis to negatively correlated miRNA/target pairs. If readers are interested in generating an advanced miRNA/target interaction network, we direct them to reviews on the various mathematical models and integrated approaches used (Carroll, Goodall, & Liu, 2014; Muniategui, Pey, Planes, & Rubio, 2013). For simple visualization of predicted miRNA/target interactions, we recommend import of results from this workflow into Cytoscape (Shannon et al., 2003).
Wet-lab procedures
RNA extraction
High-quality RNA is essential for both RNA-seq and the validations used in this article. It consists of RNA that is primarily free of degradation by cellular nucleases and lacks contamination by genomic DNA. Extraction of RNA begins by grinding the tissue sample and solubilizing its contents. Solubilization buffers containing guanidinium compounds protect against nucleases and aid in breakdown of the cell membrane (Chomczynski, 1993). Following solubilization, the user can continue with a reagent-based extraction, such as with RNAzol, or switch to purification with silica columns. RNAzol is advantageous as it enables convenient isolation of the small RNA fraction and contains additives to reduce genomic DNA contamination. Alternatively, silica-based purification allows much faster RNA isolation and on-column DNase treatment to remove genomic DNA; however, a major drawback of this technology is its often-poor yield.
RLM-RACE
5′ RACE was originally developed to map the +1 transcription start site of mRNAs (Sambrook & Russell, 2006). In this application, the mRNA was reverse transcribed with an oligo d(T) primer, then terminal deoxytransferase (TdT) was used to add multiple nucleotides to the 3′ end of the cDNA (known as ‘tailing’) to produce an adapter sequence. RLM-RACE forgoes tailing and instead ligates a 5′ RNA adapter directly to an mRNA pool that has been phosphatase-treated and decapped to select for full-length mRNAs. This is ideal for the original application of the method but not for validating miRNA-induced cleavage events, since the targeted mRNA is sliced. To modify this procedure for detecting cleaved products, we omitted the selection of full-length mRNAs, resulting in any exposed 5′ phosphate in the mRNA pool becoming ligated to the RNA adapter. Subsequent amplification with PCR and cloning with Gibson assembly allows the identification of 5′ cut sites of specific mRNA targets.
RLM-RACE is superior to 5′ RACE for detecting miRNA-induced cleavage. T4 RNA ligase is efficient at adding an RNA adapter to the 5′ end of mRNA, while TdT used to tail the cDNA in 5′ RACE can add nucleotides to ssDNA, dsDNA, and, at a lower efficiency, RNA, meaning there is less specificity for the cDNA of cleaved mRNA. Additionally, RLM-RACE avoids potential artifacts caused by the reverse transcriptase stalling during cDNA synthesis. As described above, degradome-seq is a high-throughput equivalent to the modified RLM-RACE used in this article, enabling relative quantification of all cleaved targets. To quantify miRNA-directed repression regardless of whether it derives from transcript cleavage or translational inhibition, a transient dual luciferase assay has been optimized for use in Nicotiana benthamiana (Moyle et al., 2017).
qRT-PCR and stem-loop qRT-PCR
qRT-PCR is a common and rapid method for relative RNA quantification. The transcript of interest is quantified in both the treated and untreated sample, with the change in expression normalized to that of an internal control transcript in each sample to account for differences in amount of starting RNA. Internal controls are often referred to as ‘housekeeping’ genes for their stable expression across various treatments. In this article, we use qRT-PCR and stem-loop qRT-PCR to validate expression of target mRNAs and miRNAs, respectively. Design of primers for standard qRT-PCR is relatively straightforward: the two primers should produce a product of 150 to 200 bp. To identify and reduce the impact of genomic DNA contamination in the RNA sample, primers should span an intronic region if one is known to exist. Stem-loop qRT-PCR, developed by Chen et al. (2005), uses a stem-loop primer complementary to the last six bases on the 3′ end of the miRNA. This increases the length and melting point of the PCR product to make it compatible with standard cycling. Therefore, specificity of the PCR reaction is conferred mainly by the forward primer, which spans most of the miRNA sequence.
We use SYBR Green to measure fluorescence during qPCR, but specificity can be increased by substituting sequence-specific hydrolysis probes such as TaqMan (Applied Biosystems) or Universal ProbeLibrary (UPL, Roche Diagnostics). The probes anneal to single-stranded DNA and emit fluorescence only upon DNA polymerase-induced cleavage, which results from 5′ to 3′ exonuclease activity of the polymerase as it extends the primer. Use of these probes reduces background fluorescence due to primer dimers and increases specificity, as the probe binds between primer annealing sites. UPL probes provide increased specificity by incorporating locked nucleic acids, which are modified nucleotides with ribose rings stabilized in an ideal conformation for Watson-Crick base pairing. For a protocol utilizing UPL stem-loop qRT-PCR to quantify low-abundance plant miRNAs, see Varkonyi-Gasic, Wu, Wood, Walton, and Hellens (2007). Although beneficial, the high cost of hydrolysis probes likely excludes them from use in initial screening validations, but their incorporation may be worthwhile for characterization of a few key miRNAs.
Critical Parameters
High-quality RNA is an essential input for both RNA-seq and the validations performed in this article. General plant health is an important contributor to RNA quality, with lack of light or nutrients resulting in lesser-quality RNA. If the treatment under study is intended to elicit a stress response, the strength and duration of treatment must be optimized to avoid impacting overall RNA quality. When harvesting plants, tissue should be flash-frozen in liquid nitrogen and processed immediately. Use of pre-chilled tools and tubes prevents frozen tissue from melting, thereby limiting nuclease activity. If liquid nitrogen is unavailable, tissue can be preserved in saturated ammonium sulfate solution, such as RNAlater (Ambion). Note that the lysate is stable upon solubilization in RNAzol and can be stored long-term at −20°C. Once extracted, RNA is susceptible to degradation, by nucleases and resulting from hydrolysis under basic conditions. Both factors can be controlled by resuspending RNA pellets in RNA storage buffer, which contains DTT to inactivate ribonucleases and citrate buffer to reduce pH and chelate metal ions.
Sufficient computational power and memory are required to perform RNA-seq analysis. Raw data and output files must be stored, and the workflow must be able to complete in a reasonable timeframe. We performed all analysis for the full-scale job using a personal server with 64 Gb RAM and a 4 Tb hard drive. If suitable infrastructure is not available, public options for researchers include Galaxy and CyVerse (originally iPlant Collaborative; Merchant et al., 2016) for both high-performance computing and data storage. We introduce the user to Galaxy in this article.
Certain aspects of the RNA-seq experimental design and bioinformatic workflow are essential to include. If differential expression analysis is performed, both biological replicates and strand-specific reads must be incorporated. The former allow for an assessment of sample variability, while the latter ensures accurate transcript quantification. Note that many peer-reviewed journals now require a minimum of three biological replicates for inclusion of RNA-seq differential expression analysis. It is also essential to quality control the raw reads prior to analysis. Contaminating adapter sequences create artifacts in both the assembled transcriptome and small RNA sequences, and low-quality bases introduce sequence errors. Both issues interfere with transcriptome assembly, differential expression analysis, and miRNA prediction.
Factors affecting success of wet-lab validations are also important to consider. Low-abundance miRNAs (<10 CPM) and targets (<100 TPM) are difficult to detect. Cleaved mRNA is quickly degraded in the cell, so a low initial abundance further inhibits detection by RLM-RACE. Similarly, low abundance results in undetectable or variable Ct values during qRT-PCR. Another critical factor for a successful qRT-PCR experiment is the selection of appropriate internal controls. A preliminary test should be performed to ensure that expression is stable for the treatment under study. Two popular programs that aid in the selection of internal controls are NormFinder (Aanes et al., 2014) and geNorm (Vandesompele et al., 2002). These programs use pairwise comparisons of expression data to rank the stability of reference genes. Both are available as Microsoft Excel plugins, which allows analysis without extensive bioinformatics knowledge. However, these programs are unable to process large transcriptome datasets with ease. We reduced the list of input reference genes by calculating the mean, standard deviation, and relative standard deviation of abundance for each transcript across all samples using Excel. Relative standard deviation is calculated by multiplying standard deviation by 100 and dividing by the mean. Transcripts were sorted first by lowest relative standard deviation and then by highest mean expression level. Such transcripts would vary little in level across different treatments and would be of sufficient abundance to detect reliably. The reduced gene list was used as input for NormFinder. The final list of candidate reference genes was verified with qRT-PCR across the different treatments tested.
Troubleshooting
For problems concerning any portion of the bioinformatic workflow, we refer the reader to the appropriate program guides at the links provided. Ensure that each program is installed correctly, including any required dependencies, and that it is accessible from the user's $PATH. Additionally, ensure that the full file path (/path/to/file.ext) is provided for input on the command line; otherwise, the file may not be located by the program. For troubleshooting common issues that may be experienced during wet-lab procedures, refer to Tables 3 to 5.
| Problem | Possible cause | Solution | Notes |
|---|---|---|---|
|
|
|
RNA extraction requires a level of manual dexterity and practice to achieve optimal results. Degradation is evident if the 28S rRNA band appears fainter than the 18S on an agarose gel or if the RIN is <8. At the initial stage, degradation can occur if the ground leaf powder is not kept at liquid nitrogen temperatures and if the powder is not solubilized in the RNAzol fast enough. |
| Genomic DNA contamination |
|
|
Genomic DNA contamination can be detected as high-molecular-weight bands or smears when total RNA is separated on an agarose gel. As well, qRT-PCR reactions can be separated on an agarose gel after the reaction has finished; products of unexpected size indicate genomic contamination if qRT-PCR primers were designed to span intron/exon junctions. |
| Problem | Possible cause | Solution | Notes |
|---|---|---|---|
| No bands on agarose gel after nested PCR |
|
|
It is imperative to have a working positive control for RLM-RACE. For plants, miR156 and its target, SQUAMOSA-PROMOTER BINDING PROTEIN-LIKE (SPL) are both abundant and easy to detect. If the positive control gives the product of expected size on a gel, then errors in adapter ligation and losses in mRNA purification can be ruled out. |
| Unexpected and random cleavage sites in target mRNA |
|
|
Cleavage at unexpected sites suggests that the miRNA is not cleaving the expected target, or that this small RNA is not a functional miRNA (i.e., false positive). In some cases, we observed randomly distributed cleavage surrounding the expected site. This suggested that the limit of detection had been reached; background levels of mRNA degradation were as likely as miRNA cleavage. |
| Problem | Possible cause | Solution | Notes |
|---|---|---|---|
| No amplification |
|
|
No amplification is often a case of technical error, either from missing reagents or incorrectly used equipment |
| Variable/unpredictable amplification |
|
Increase amount of starting RNA | Increasing input of RNA by 50%-100% can help detect low-abundance miRNA/mRNA pairs; however, this may cause the internal control to amplify earlier as well |
| Very early amplification |
|
|
|
Anticipated Results
Bioinformatic analysis
For an overview of expected results, we refer the reader to the accompanying Supporting Information, which provides all output files referenced in this article. Beginning with raw RNA-seq data, the user will perform read cleaning, de novo transcriptome assembly and annotation, miRNA prediction and annotation, differential expression analysis, miRNA/target prediction, and functional enrichment analysis.
Wet-lab procedures
When visualizing extracted RNA on a gel, the following should be observed. For total RNA, a distinctive pattern forms due to the high abundance of rRNA: 28S produces the brightest band, followed by 18S and 5S. Equal intensity of 28S and 18S is acceptable, while a brighter 18S indicates degradation. Enriched mRNA should appear as banding or smearing above the 1 Kb marker of an RNA-specific ladder, while the small RNA-enriched fraction produces a smear below the 200 bp marker. In RLM-RACE, nonspecific amplification of the cDNA pool should produce an indistinct smear on an agarose gel. The first round of gene-specific amplification may produce multiple bands, a smear, or no visible product. Following nested PCR, the expected band should be observed, either alone or easily distinguished from a small number of nonspecific bands. For qRT-PCR, relative fold changes between treated and untreated samples may not coincide 1:1 with the RNA-seq results. However, there should be a strong linear correlation between the two methods (R 2 > 0.8).
Time Considerations
Bioinformatic analysis
Using the sample data and reference files provided in this article, each protocol can be completed comfortably within an hour. Tasks requiring extensive computational resources and run time are transcriptome de novo assembly, BLAST homology searches, and any read-mapping procedures. Completion times will vary depending on the user's resources. Completion times for the full-scale job on our relatively modest server (64 Gb RAM, 16 CPUs) ranged from several hours (read-mapping) to several days (de novo assembly and BLAST).
Wet-lab procedures
We recommend implementing a consistent planting schedule so that minimal time is spent waiting for plants to grow. Once plants are available and the experimental treatment applied, samples should be flash-frozen in bulk and processed in smaller batches. The RNA extraction protocol requires 1 to 2 days for a single batch of 8 to 16 samples. Validation of 5 to 10 targets with RLM-RACE will require at least 4 weeks. The limiting step is PCR optimization, which may require testing of multiple primer pairs. Colony screening should take 1 to 2 days, with Sanger sequencing results likely available in 2 to 3 days. For qRT-PCR, reverse transcription can be performed comfortably on 12 to 24 samples within 1 day. Assuming use of a 36-well rotor, 36 samples can be assessed by qPCR in one batch. As an example, this could be split between six ‘gene targets’ (e.g., 5 miRNAs + 1 internal control miRNA) × 3 technical replicates × 2 conditions. Sample setup and qPCR can be performed within 1 day for a single batch. Note that optimization of starting RNA amount and primer design will likely be required for low-abundance miRNA/target pairs.
Acknowledgements
The authors gratefully acknowledge Ms. Camille Diaz, York University, for Figure 2. The authors thank Dr. Yutaka Amemiya, Manager, Genomics Core Facility, Next Generation Sequencing Lab, Sunnybrook Research Institute, for small RNA sequencing and for his advice with sequence analysis. This work was funded by a Discovery Grant to KH from the Natural Sciences and Engineering Research Council of Canada and PGS-D Scholarships to KN and AK.
Supporting Information
| Filename | Description |
|---|---|
| cppb20090-sup-0001-SuppInfofiles.zip26 MB | Supporting Information |
Please note: The publisher is not responsible for the content or functionality of any supporting information supplied by the authors. Any queries (other than missing content) should be directed to the corresponding author for the article.
Literature Cited
- Aanes, H., Winata, C., Moen, L. F., Østrup, O., Mathavan, S., Collas, P., … Aleström, P. (2014). Normalization of RNA-sequencing data from samples with varying mRNA levels. PloS One , 9, 1–7. doi: 10.1371/journal.pone.0089158.
- An, D., Cao, H., Li, C., Humbeck, K., & Wang, W. (2018). Isoform sequencing and state-of-art applications for unravelling complexity of plant transcriptomes. Genes (Basel) , 9, 43. doi: 10.3390/genes9010043.
- Bartel, D. P. (2004). MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell , 116, 281–297. doi: 10.1016/S0092-8674(04)00045-5.
- Carroll, A. P., Goodall, G. J., & Liu, B. (2014). Understanding principles of miRNA target recognition and function through integrated biological and bioinformatics approaches. Wiley Interdisciplinary Reviews, RNA , 5, 361–379. doi: 10.1002/wrna.1217.
- Chen, C., Ridzon, D. A., Broomer, A. J., Zhou, Z., Lee, D. H., Nguyen, J. T., … Guegler, K. J. (2005). Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Research , 33, 1–9. doi: 10.1093/nar/gni178.
- Chomczynski, P. (1993). A reagent for the single-step simultaneous isolation of RNA, DNA and proteins from cell and tissue samples. Biotechniques , 15, 532–4, 536–537.
- Costa-Silva, J., Domingues, D., & Lopes, F. M. (2017). RNA-Seq differential expression analysis: An extended review and a software tool. PloS One , 12, e0190152. doi: 10.1371/journal.pone.0190152.
- Djami-Tchatchou, A. T., Sanan-Mishra, N., Ntushelo, K., & Dubery, I. A. (2017). Functional roles of microRNAs in agronomically important plants—Potential as targets for crop improvement and protection. Frontiers in Plant Science , 8, 378. doi: 10.3389/fpls.2017.00378.
- Geniza, M., & Jaiswal, P. (2017). Tools for building de novo transcriptome assembly. Current Plant Biology , 11–12, 41–45. doi: 10.1016/j.cpb.2017.12.004.
- Giardine, B., Riemer, C., Hardison, R. C., Burhans, R., Elnitski, L., Shah, P., … Nekrutenko, A. (2005). Galaxy: A platform for interactive large-scale genome analysis. Genome Research , 15, 1451–1455. doi: 10.1101/gr.4086505.
- Honaas, L. A., Wafula, E. K., Wickett, N. J., Der, J. P., Zhang, Y., Edger, P. P., … dePamphilis, C. W. (2016). Selecting superior de novo transcriptome assemblies: Lessons learned by leveraging the best plant genome. PloS One , 11, e0146062. doi: 10.1371/journal.pone.0146062.
- Ji, Y., Chen, P., Chen, J., Pennerman, K., Liang, X., Yan, H., … Huang, L. (2018). Combinations of small RNA, RNA, and degradome sequencing uncovers the expression pattern of microRNA–mRNA pairs adapting to drought stress in leaf and root of Dactylis glomerata L. International Journal of Molecular Sciences , 19, 3114. doi: 10.3390/ijms19103114.
- Kawashima, C. G., Yoshimoto, N., Maruyama-Nakashita, A., Tsuchiya, Y. N., Saito, K., Takahashi, H., & Dalmay, T. (2009). Sulphur starvation induces the expression of microRNA-395 and one of its target genes but in different cell types. Plant Journal , 57, 313–321. doi: 10.1111/j.1365-313X.2008.03690.x.
- Kidner, C. A., & Martienssen, R. A. (2004). Spatially restricted microRNA directs leaf polarity through ARGONAUTE1. Nature , 428, 81–84. doi: 10.1038/nature02366.
- Levine, E., McHale, P., & Levine, H. (2007). Small regulatory RNAs may sharpen spatial expression patterns. PloS Computational Biology , 3, 2356–2365. doi: 10.1371/journal.pcbi.0030233.
- Lin, S. S., Chen, Y., & Lu, M. J. (2019). Degradome sequencing in plants. Methods in Molecular Biology , 1932, 197–213. doi: 10.1007/978-1-4939-9042-9_15.
- Ma, C., Yang, J., Cheng, Q., Mao, A., Zhang, J., Wang, S., … Wen, C. (2018). Comparative analysis of miRNA and mRNA abundance in determinate cucumber by high-throughput sequencing. PloS One , 13, e0190691. doi: 10.1371/journal.pone.0190691.
- Mathioni, S. M., Kakrana, A., & Meyers, B. C. (2017). Characterization of plant small RNAs by next generation sequencing. Current Protocols in Plant Biology , 2, 39–63. doi: 10.1002/cppb.20043.
- Merchant, N., Lyons, E., Goff, S., Vaughn, M., Ware, D., Micklos, D., & Antin, P. (2016). The iPlant collaborative: Cyberinfrastructure for enabling data to discovery for the life sciences. PloS Biology , 14, e1002342. doi: 10.1371/journal.pbio.1002342.
- Moyle, R. L., Carvalhais, L. C., Pretorius, L.-S., Nowak, E., Subramaniam, G., Dalton-Morgan, J., … Schenk, P. M. (2017). An optimized transient dual luciferase assay for quantifying MicroRNA directed repression of targeted sequences. Frontiers in Plant Science , 8, 1631. doi: 10.3389/fpls.2017.01631.
- Muniategui, A., Pey, J., Planes, F. J., & Rubio, A. (2013). Joint analysis of miRNA and mRNA expression data. Briefings in Bioinformatics , 14, 263–278. doi: 10.1093/bib/bbs028.
- Neller, K. C. M., Klenov, A., Guzman, J. C., & Hudak, K. A. (2018). Integration of the pokeweed miRNA and mRNA transcriptomes reveals targeting of jasmonic acid-responsive genes. Frontiers in Plant Science , 9, 589. doi: 10.3389/fpls.2018.00589.
- Neller, K. C. M., Klenov, A., & Hudak, K. A. (2016). The pokeweed leaf mRNA transcriptome and its regulation by jasmonic acid. Frontiers in Plant Science , 7, 1–13. doi: 10.3389/fpls.2016.00283.
- Nikovics, K., Blein, T., Peaucelle, A., Ishida, T., Morin, H., Aida, M., & Laufs, P. (2006). The balance between the MIR164A and CUC2 genes controls leaf margin serration in Arabidopsis. Plant Cell , 18, 2929–2945. doi: 10.1105/tpc.106.045617.
- Noman, A., Fahad, S., Aqeel, M., Ali, U., Amanullah Anwar, S., … Zainab, M. (2017). miRNAs: Major modulators for crop growth and development under abiotic stresses. Biotechnology Letters , 39, 685–700. doi: 10.1007/s10529-017-2302-9.
- Rajendiran, A., Chatterjee, A., & Pan, A. (2018). Computational approaches and related tools to identify microRNAs in a species: A bird's eye view. Interdisciplinary Sciences: Computational Life Sciences , 10, 616–635. doi: 10.1007/s12539-017-0223-x.
- Sambrook, J., & Russell, D. W. (2006). Rapid amplification of 5′ cDNA Ends (5′-RACE). CSH Protocols , 1, pii: Pdb.prot3989. doi: 10.1101/pdb.prot3989.
- Shannon, P., Markiel, A., Ozier, O., Baliga, N. S., Wang, J. T., Ramage, D., … Ideker, T. (2003). Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Research , 13, 2498–2504. doi: 10.1101/gr.1239303.
- Vandesompele, J., De Preter, K., Pattyn, F., Poppe, B., Van Roy, N., De Paepe, A., & Speleman, F. (2002). Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biology , 3, RESEARCH0034. doi: 10.1186/gb-2002-3-7-research0034.
- Varkonyi-Gasic, E., Wu, R., Wood, M., Walton, E. F., & Hellens, R. P. (2007). Protocol: A highly sensitive RT-PCR method for detection and quantification of microRNAs. Plant Methods , 3, 12. doi: 10.1186/1746-4811-3-12.
- Vitsios, D. M., Kentepozidou, E., Quintais, L., Benito-Gutiérrez, E., Van Dongen, S., Davis, M. P., … Enright, A. J. (2017). Mirnovo: Genome-free prediction of microRNAs from small RNA sequencing data and single-cells using decision forests. Nucleic Acids Research , 45, e177. doi: 10.1093/nar/gkx836.
- Voytas, D. (2000). Agarose gel electrophoresis. Current Protocols in Molecular Biology , 51, 2.5A.1–2.5A.9.
- Wu, C. I., Shen, Y., & Tang, T. (2009). Evolution under canalization and the dual roles of microRNAs—A hypothesis. Genome Research , 19, 734–743. doi: 10.1101/gr.084640.108.
- Wu, D. C., Yao, J., Ho, K. S., Lambowitz, A. M., & Wilke, C. O. (2018). Limitations of alignment-free tools in total RNA-seq quantification. BMC Genomics , 19, 510. doi: 10.1186/s12864-018-4869-5.
- Ye, B., Wang, R., & Wang, J. (2016). Correlation analysis of the mRNA and miRNA expression profiles in the nascent synthetic allotetraploid Raphanobrassica. Scientific Reports , 6, 37416. doi: 10.1038/srep37416.
- Yu, Y., Jia, T., & Chen, X. (2017). The “how” and “where” of plant microRNAs. The New Phytologist , 216, 1002–1017. doi: 10.1111/nph.14834.

