Preclinical Models of Neuroendocrine Prostate Cancer
Alessia Cacciatore, Alessia Cacciatore, Domenico Albino, Domenico Albino, Carlo V. Catapano, Carlo V. Catapano, Giuseppina M. Carbone, Giuseppina M. Carbone
genetically engineered mouse model (GEMM)
neuroendocrine prostate cancer (NEPC)
patient-derived xenografts (PDX)
preclinical models
Abstract
Prostate cancer (PCa) is the most common malignancy and the second leading cause of cancer-related death amongst men in the United States. Neuroendocrine prostate cancer (NEPC) can either arise de novo or emerge as a consequence of therapy. De novo NEPC is rare, with an incidence of <2% of all PCa cases. In contrast, treatment-induced NEPC is frequent with >20% of patients with metastatic castration-resistant prostate cancer (CRPC) reported to progress to neuroendocrine (NE) differentiation. The emergence of treatment-induced NEPC is linked to the increased therapeutic pressure, due to the broad application of androgen deprivation therapy (ADT) for PCa management and the development of novel more potent androgen receptor (AR) pathway inhibitors. NEPC is a high-grade tumor type characterized by aggressive phenotype and clinical behavior. Patients affected by NEPC frequently develop visceral metastases and have a poor prognosis. The molecular mechanisms underlying the development and progression of NEPC are still poorly understood. Transcriptional and epigenetic reprogramming appears to be involved in NE progression. In this review, we aim to provide a comprehensive view of the available models for NEPC detailing their strengths and limitations. Moreover, we describe novel approaches to expand the repertoire of preclinical models to better study, prevent, or reverse NEPC. The integration of multiple preclinical models along with molecular and omics approaches will provide important insights to understand disease progression and to devise novel therapeutic strategies for the management of NEPC in the near future. © 2023 The Authors. Current Protocols published by Wiley Periodicals LLC.
Basic Protocol 1 : Generation of organoids starting from the prostate gland of a GEMM or a human PDX
Basic Protocol 2 : Ex vivo tumor sphere formation
INTRODUCTION
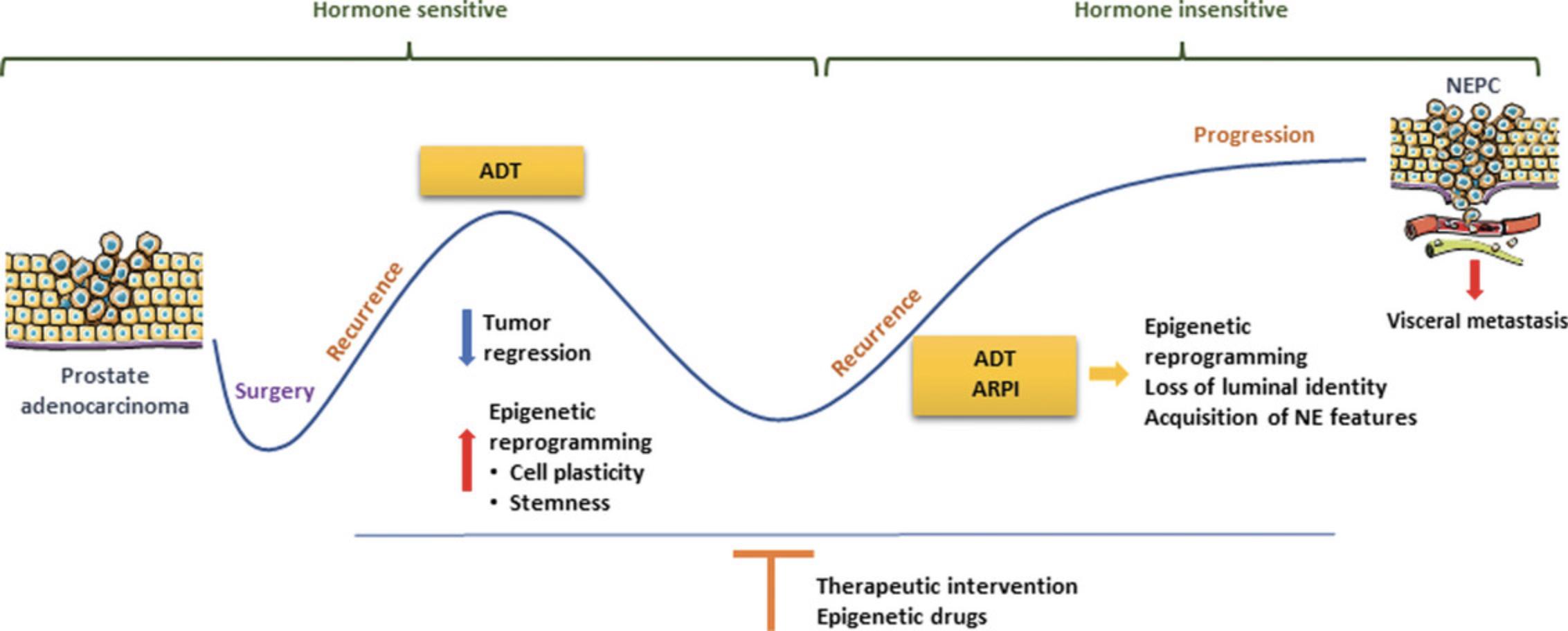
Prostate cancer (PCa) is the most common malignancy and the second leading cause of cancer-related death among men in the United States (Siegel et al., 2022). Recent advances in PCa treatment have significantly improved disease management (Litwin & Tan, 2017). Nevertheless, disease progression occurs in a significant proportion of PCa patients, with poor prognosis. Neuroendocrine prostate cancer (NEPC) can either arise de novo or emerge as a consequence of therapy and particularly androgen deprivation therapy (ADT; Merkens et al., 2022). De novo NEPC is rarely diagnosed and presents an incidence of <2% of all PCa cases (Nadal et al., 2014). By contrast, diagnosis of treatment-induced NEPC has increased over the years, as >20% of patients with metastatic castration-resistant prostate cancer (CRPC) have been reported to progress to neuroendocrine (NE) differentiation (Aggarwal et al., 2018; Desai et al., 2021; Zhang et al., 2020). The emergence of treatment-induced NEPC has been linked to the increased therapeutic pressure, due to the broad application of ADT for PCa management. Development of novel and more potent AR pathway inhibitors, such as abiraterone acetate and enzalutamide, have improved the management of disease progression but enhanced the frequency of acquisition of neuroendocrine features (Chen et al., 2022; Davis et al., 2019; Merkens et al., 2022). NEPC is a high-grade tumor type characterized by aggressive phenotype and clinical behavior (Cejas et al., 2021). Treatment induced NEPC arises from a complex process of neuroendocrine transdifferentiation of prostate adenocarcinoma (Parimi et al., 2014). ADT results in epigenetic reprogramming with induced cell plasticity. Multiple waves of ADT [extensive ADT and androgen receptor pathway inhibitor (ARPI)] culminate in a loss of luminal epithelial cell identity with the acquisition of stem-like and neuroendocrine features (Fig. 1). The degree of epithelial dedifferentiation is associated with increased aggressiveness. The acquisition of aggressive features results in a high rate of visceral metastases and poor prognosis (Conteduca et al., 2019). Neuroendocrine tumors are generally defined by the expression of chromogranin A (CHGA), synaptophysin (SYP), neural cell adhesion molecule 1 (NCAM1), and enolase 2 (ENO2; Epstein et al., 2014; Su et al., 2022). In recent years, an increasing effort is being made to understand the specific markers that characterize neuroendocrine prostate tumors. To this end, the work by Beltran et al. (2016) identified a signature of 164 genes that differentiate NEPC from adenocarcinoma. The molecular mechanisms underlying the development and progression of NEPC are still poorly understood, highlighting the need for a deeper understanding of the genomic aberrations and the epigenetic changes occurring during disease progression. The use of preclinical models can therefore be of great value in order to expand the knowledge on the drivers of NEPC differentiation. In this hybrid review and protocols-based article, we aim to provide a comprehensive view of the available models for NEPC, detailing their strengths and limitations. Additionally, we detail two protocols for the generation and culture of NEPC organoids and NEPC tumor spheres ex vivo. We also describe novel approaches to expand the repertoire of NEPC models to better study, prevent, or reverse NEPC.
In Vitro and In Vivo Models for the Evaluation of Experimental Agents Against NEPC
In this section, we describe several models available to date and suitable for the study of NEPC (Table 1). Strengths and limitations of the models described are also highlighted (Table 2). These models can be implemented for understanding the underlying biology and for testing therapeutic strategies to reverse this phenotype. Each model displays unique features and, for a comprehensive understanding of the disease, we encourage the implementation and integration of multiple models.
| Model | Origin | Features |
|---|---|---|
| NCI-H660 (Beltran et al., 2011) | Cell line |
|
| LASCPC-01 (Lee et al., 2016) | Cell line |
|
| 22Rv1 (Sramkoski et al., 1999) | Cell line |
|
| p53PE−/−; RbPE−/− (Miao et al., 2022) | GEMM |
|
| PBCre4; Ptenf/f; Rb1f/f; Trp53f/f (Ku et al., 2017; Mu et al., 2017) | GEMM |
|
| TRAMP (C57BL/6) (Gingrich & Greenberg, 1996; Greenberg et al., 1995) | GEMM |
|
| PBCre4; Ptenf/f; Rb1f/f (Ku et al., 2017; Mu et al., 2017) | GEMM |
|
| Nkx3.1CreERT2/+; Ptenf/f; Trp53f/f (Zou et al., 2017) | GEMM |
|
| MYCN-myrAKT1 (Lee et al., 2016) | GEMM |
|
| T2-Cre+/+; Ptenf/f; LSL-; MYCN+/+ (Dardenne et al., 2016) | GEMM |
|
| Ptenf/f; R26ERG (Chen et al., 2013) | GEMM |
|
LuCaP series (Nguyen et al., 2017; Shi et al., 2022; True et al., 2002)
|
PDX |
|
LTL series (Lin et al., 2014)
|
PDX |
|
| WISH-PC2 (Pinthus et al., 2000) | PDX |
|
MDA PCa series (Palanisamy et al., 2020)
|
PDX |
|
- a Abbreviations: AR, androgen receptor; NE, neuroendocrine; PSA, prostate-specific antigen; PCa, prostate cancer; GEMM, genetically engineered mouse model; NEPC, neuroendocrine prostate cancer; EMT, epithelial-to-mesenchymal transition; PSCA, prostate stem cell antigen; PSMA, prostate-specific membrane antigen; PDX, patient-derived xenograft.
| Model | Strengths | Limitations |
|---|---|---|
| Cell lines |
|
|
| GEMM |
|
|
| PDX |
|
|
| Organoids |
|
|
| Spheroids |
|
|
- a Abbreviations: TME, tumor microenvironment; NEPC, neuroendocrine prostate cancer; GEMM, genetically engineered mouse model; PDX, patient-derived xenograft.
NEPC Cell Models
In vitro NEPC cell models including NCI-H660 and LASCPC-01 are relatively well-characterized (Lee et al., 2016; Singh et al., 2021; VanDeusen et al., 2020). NCI-H660 cells are AR negative, express high levels of neuroendocrine markers, and grow as suspending cell clusters, a morphology distinct from adherent luminal epithelial cells. When grafted in immune comprised mice, the NCI-H660 cell line form NEPC xenografts that progress rapidly (Beltran et al., 2011). For this reason, NCI-H660 cells can be considered a good in vitro model that can be used for mechanistic studies to understand NEPC alterations. The LASCPC-01 cells grow rapidly in suspension as floating and attached clusters, resembling small-cell lung cancer cells. These cells are characterized by the expression of N-MYC, ASCL1, and neuron specific enolase (NSE), and show activated AKT1 and AURKA. Moreover, engraftment of LASCPC-01 cells in immune compromised mice leads to development of xenograft tumors that exhibit both neuroendocrine and adenocarcinoma phenotypes (Lee et al., 2016). 22Rv1 is a cell line derived by serial passaging of the xenograft CWR22R (Sramkoski et al., 1999). These cells were isolated from a patient with prostate adenocarcinoma that developed bone metastases after repeated tumor regressions and relapsed under castrated condition (Sramkoski et al., 1999). The 22Rv1 cell line displays some overlapping features of NEPC. Even though 22Rv1 cells are androgen sensitive, when maintained under hypoxia conditions for few days up-regulate the NE marker NSE and develop neurite-like structures (Chlenski et al., 2001). Additionally, 22Rv1 cells are resistant to enzalutamide treatment in vitro and, when implanted in mice, express NE markers within highly hypoxic tumorigenic regions (Shiota et al., 2018).
To expand the repertoire of cell lines with NE phenotype, an alternative approach is to generate transgenic cell lines with loss (by knockout/knockdown) or gain (by overexpression) of genes putatively responsible for NE deviation. Transgenic cells with overexpression of multiple oncogenic proteins can be a valid model to understand progression and test therapeutic strategies against NEPC.
Genetically Engineered Mouse Models of NEPC
Over the years, the use of genetically engineered mouse models (GEMMs) significantly contributed to understanding the complex mechanisms that underlie cancer biology. Moreover, these GEMMs provide models to understand the interactions between cancer cells and tumor microenvironment, drug responses, and emergence of therapy resistance (Kersten et al., 2017). GEMMs suitable for the study of NE dedifferentiation are listed in Table 1. Due to the specific genetic alterations implemented, each model has its own strengths and weaknesses in recapitulating the disease progression. Human NEPC is often characterized by the combinatorial loss of p53 and Rb1 , two key tumor suppressors (Merkens et al., 2022; The Cancer Genome Atlas Research Network, 2015). Therefore, the p53PE−/−; RbPE−/− GEMM appears to be a valid model to gain insights into the management of human NE tumors (Cejas et al., 2021; Yamada & Beltran, 2021). Indeed, double knockout (KO) of TP53 and RB1 (p53PE−/−; RbPE−/−) develops NE features by facilitating lineage plasticity (Miao et al., 2022; Zhou et al., 2006). However, while loss of these tumor suppressors is crucial in driving lineage plasticity, they may not be sufficient to induce a neuroendocrine transformation (Beltran et al., 2020). To overcome this issue, the triple KO model (PBCre4; Ptenf/f; Rb1f/f; Trp53f/f), which includes also PTEN loss, displayed a more aggressive disease, characterized by the development of numerous metastases (Ku et al., 2017; Mu et al., 2017). Interestingly, while PTEN alone is not sufficient to lead to NE alterations, it has been shown to synergistically cooperate with additional alterations in critical oncogenes or tumor suppressor genes. For this reason, several models combine PTEN loss with other tumor suppressor or oncogenes in order to establish NE models, consistent with the relevant tumor suppressor role of PTEN. GEMMs that combine PTEN loss with other oncogenic hits include: Combination with Rb1 loss PBCre4; Ptenf/f; Rb1f/f (Ku et al., 2017); Nkx3.1 and Trp53f/f in the GEMM Nkx3.1CreERT2/+; Ptenf/f; Trp53f/f (Zou et al., 2017) and combination with LSL-MYCN in mice T2-Cre+/+; Ptenf/f; LSL-; MYCN+/+(Dardenne et al., 2016). Specifically, the GEMM PBCre4; Ptenf/f; Rb1f/f (Ku et al., 2017) double KO is initially characterized by the development of aggressive PCa that later progresses in NE features, such as low expression of AR, castration-resistance and high expression of SYP. The Nkx3.1CreERT2/+; Ptenf/f; Trp53f/f mice develop prostatic intraepithelial neoplasia (PIN) at early stages and are initially sensitive to castration, but usually relapse and progress to more severe disease (Zou et al., 2017). The T2-Cre+/+; Ptenf/f; LSL-; MYCN+/+ (Dardenne et al., 2016) mouse model is characterized by an overexpression of mesenchymal markers, an absence of pan-cytokeratin expression, and variable levels of AR expression. These features of epithelial-to-mesenchymal transition (EMT) are also accompanied by the presence of NEPC foci.
A GEMM of NEPC differentiation that does not comprise loss of PTEN combines MYCN with AKT (MYCN-myrAKT1; Lee et al., 2016). Concomitant overexpression of MYCN and myrAKT1 results in the development of invasive and metastatic castration-resistant tumors. Eventually these tumors become positive for NE markers, including low expression of AR-regulated genes and epithelial markers. Finally, a relevant GEMM that recapitulates the features of NEPC is represented by the TRAMP (C57BL/6) model (Gingrich & Greenberg, 1996; Gingrich et al., 1996; Greenberg et al., 1995). The transgenic adenocarcinoma mouse prostate (TRAMP) mouse model mimics the histopathology of human PCa development, from prostate hyperplasia to CRPC and NEPC disease. It occasionally develops metastases to the lymph nodes, lung, and bone. Intriguingly, in a previous study, by applying the luminal metagene, we found that the transcriptome of TRAMP mice was significantly low in luminal markers, consistent with an aggressive NE androgen resistant tumor sub-type (Mapelli et al., 2020).
Overall, the implementation of GEMMs has the advantage of reproducing human disease more faithfully than other models. Accordingly, the interaction with the tumor microenvironment (TME) along with hormonal manipulations—castration, stimulation with dihydrotestosterone (DHT)—and the possibility to study metastasis formation enlarge the opportunities for discovery and therapy implementation. Therefore, GEMMs represent to date valuable models to study different aspects of the evolution of NEPC ranging from lineage plasticity to frank metastatic disease. Moreover, GEMMs offer advantages for understanding the impact of the TME and a robust platform to test new therapies in vivo in specifically defined genetic backgrounds.
Patient-Derived Xenograft Models of NEPC
Patient-derived xenografts (PDXs) are established by transplantation of tumor fragments of patients into immunodeficient mice (Shi et al., 2022). PDXs provide a significant advantage to study PCa development and progression as they accurately outline the tumor of origin. PDXs, because they have the ability to partly recreate the complexity of the tumor microenvironment and the systemic interactions occurring during disease progression, can provide a more reliable and clinically predictive model for drug response testing (Risbridger et al., 2018). The availability of patient-derived preclinical models that recapitulate the entire transdifferentiation process of NEPC development is limited. By contrast, several established NEPC PDX models and patient-derived organoids are available for NEPC research (Puca et al., 2018). For instance, the LuCaP PDX series was derived from both primary and metastatic tumors, and consisted of five NEPC models, named LuCaP 49, LuCaP 93, LuCaP 145.1, LuCaP 145.2, and LuCaP 173.1 (Nguyen et al., 2017; Shi et al., 2022). In this regard, LuCaP 49 is an androgen insensitive model that was derived from an omental metastasis of a patient with poorly differentiated neuroendocrine cancer. It is characterized by the absence of expression of prostate-specific antigen (PSA) and AR, and overexpression of neuroendocrine markers (Nguyen et al., 2017; True et al., 2002). Additionally, two extensively used models of this series are LuCaP 145.1 and 145.2, which were derived from liver and node metastases of the same patient, respectively. Interestingly, while these two models present the same histopathological NE features, they are characterized by different genomic status, in particular in TP53, PTEN, and BRCA1. Therefore, these two LuCaP models reflect the high degree of heterogeneity of PCa (Nguyen et al., 2017; Shi et al., 2022). Another example of NEPC PDX is the LTL series, established in the Living Tumor Laboratory. The growth of these LTL PDXs in vivo is androgen independent, and they retain NEPC histopathological features (Lin et al., 2014). Of note, LTL352 and LTL370 xenografts originated by engrafting prostate tumors from a urethral (LTL352) and penile metastasis (LTL370) under the kidney capsule of non-obese diabetic (NOD)/severe combined immunodeficiency (SCID) mice. Tumors engrafted in mice retained the expression of neuroendocrine markers, were negative for AR and PSA expression, and the TMPRSS2:ERG gene fusion was detectable (Lin et al., 2014).
Notably, a similar adenocarcinoma PDX model, LTL331, after host castration, initially regresses but relapses at a later stage and transdifferentiates to LTL331R, mimicking the development and progression of clinical NEPC (Lin et al., 2014; Shi et al., 2022). Interestingly, LTL331R is characterized by histologic changes of NEPC, expression of NE markers, and loss of AR and AR signaling, including PSA expression. Similar to patients with NEPC, LTL331R showed upregulation of neuronal transcription factors (TF), membrane ion receptors, secreted peptides, and upregulation of epigenetic regulators (e.g., EZH2, CBX2) compared with the pre-castration adenocarcinoma LT331 model (Lin et al., 2014). In a recent study to identify mechanisms of resistance in a cohort of 21 patients with CRPC, biopsies were collected prior to enzalutamide treatment and at the time of tumor progression, and transcriptional studies were carried out by RNA sequencing (Westbrook et al., 2022). Interestingly, from these data, a lineage plasticity risk signature was defined. This signature included a total of fourteen genes related to the WNT pathway and the spliceosome, as well as MYC and E2F targets. Interestingly, the PDX LTL331 was the only PDX with activation of this specific signature. These data confirmed the aggressive phenotype of the PDX LTL331 and support the notion that PDXs retain with high fidelity the transcriptional state of the patient of origin (Westbrook et al., 2022).
Another interesting PDX model was described by Pinthus et al. (2000) named WISH-PC2, derived from a transurethral resection of the prostate from a patient with prostatic adenocarcinoma. This tumor was serially passaged in vivo and was able to grow after castration. Interestingly, WISH-PC2 tumors occasionally metastasize to different sites, including lymph nodes, lungs, and liver. In particular, they manifest NE differentiation features and expression of NE tumor markers, including CHGA, NSE, and SYP. Moreover, the WISH-PC2 xenograft is characterized by the absence of AR, PSA, prostate stem cell antigen (PSCA), and prostate-specific membrane antigen (PSMA; Pinthus et al., 2000). Of note, Palanisamy and colleagues (2020) reported the generation of a large number of patient-derived xenografts (MDA PCa PDX series) representative of the clinical spectrum of PCa. The MD Anderson (MDA) PCa PDX series is a resource tool that captures the molecular landscape of PCa, providing insight into the biological basis for heterogeneity. It serves as an invaluable resource for discovery, therapy development, and optimization of personalized therapy targeting prostate cancer-specific molecular markers. As an example, the MDA PCA 144 PDX was derived from the radical resection of the rectum, bladder, and reproductive organs of a patient with castrate-resistant prostate carcinoma with small cell carcinoma (Palanisamy et al., 2020). Particularly, multiple PDXs were generated from different areas of the same MDA PCa 144 PDX tumor and five cell lines with NE features were obtained from the same PDX. More recently, a new cohort of PDX models of PCa is available through the Melbourne Urological Research Alliance (MURAL; Risbridger et al., 2021). This large collection accounts for 59 serially transplantable PDXs that, based on histopathology characteristics, can be distinguished into three groups: adenocarcinomas, neuroendocrine tumors, and mixed phenotypes, reflecting the clinical heterogeneity of prostate tumors (Risbridger et al., 2021). This collection represents a valuable portfolio of models to address the multifaceted aspect of prostate cancer progression with neuroendocrine features.
Additional Models to Study NEPC
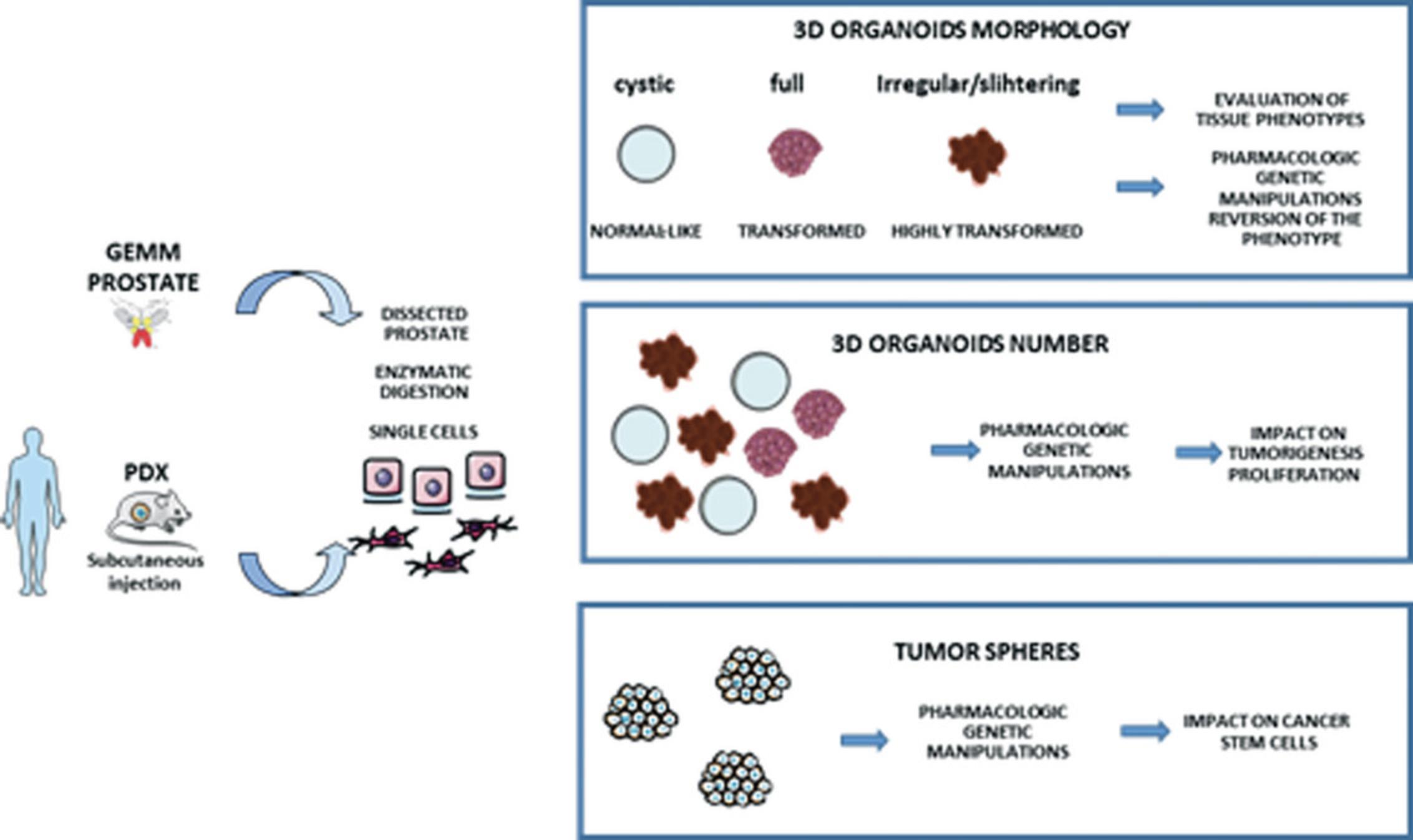
Additional strategies can be implemented to enlarge the repertoire of available NE models. Figure 2 shows a schematic of these alternative models. Ex vivo strategies present multiple advantages, including flexibility for the testing of multiple oncogenic hits and the evaluation of experimental agents (Karthaus et al., 2014; Lawson et al., 2007). These strategies include the isolation of single cells from PDX models or from mouse prostate glands and establishment of organoids and tumor spheres. These models are suitable for further manipulations by applying both loss and gain of function strategies (including ablation of selected tumor suppressor and/or overexpression of oncogenic drivers).
3D Organoids
In recent years, the transition from monolayer to 3D culture methods represents an important tool to advance both basic and translational research (Hofer & Lutolf, 2021; Kim et al., 2020). Organoids are in vitro self-organizing 3D culture systems. They represent miniaturized and simplified model systems of both human and murine organs. Most 3D culture methods for prostate tissue commonly use Matrigel for the extracellular matrix (ECM) component in which a liquid medium overlay covers prostate epithelial cells (Kleinman & Martin, 2005). Some limitations in the use of 3D organoids exist, as the TME is usually lacking, thereby preventing the system to recapitulate entirely the complex tumor-microenvironment interactions. However, 3D structures are composed of fully differentiated basal and luminal cells, and cells with stem cells features (Drost et al., 2016; Gao et al., 2014; Puca et al., 2018). Additionally, 3D organoids are genetically stable, and can reconstitute the prostate gland when dissociated and implanted into recombination assays. Importantly, organoids can be genetically manipulated for gain and loss of function studies that can be supportive to understand the biology of NE tumors and to test therapeutic approaches. Organoids provide a promising tool to advance the personalized medicine field and next-generation drug screening (Drost & Clevers, 2018; Elbadawy et al., 2020; Goldstein et al., 2008; Kim et al., 2020). Moreover, an increasing number of scientific works have employed organoid models for high-throughput drug screening. A paper from Puca et al. (2018) generated a cohort of patient-derived organoids (PDO) that shared the expression of NEPC signature genes. Intriguingly, they evaluated the activity of a drug library of 129 chemotherapeutics, and targeted agents in the four NEPC organoids generated in the study and compared them with adenocarcinoma CRPC organoids. Through this study, they were able to define drugs that, either alone or in combination, were more efficacious in NEPC organoids, compared to the CRPC organoids. Overall, these analyses indicated that NEPC organoids are a clinically relevant model for the discovery and evaluation of novel treatment options.
Ex Vivo Tumor Sphere Formation
Spheroids are spherical cellular units that are generally cultured as floating 3D multicellular structures. Tumor derived spheroids can be prepared from mechanical or enzymatic dissociation of bulk tissue into a single cell suspension, followed by culture in serum or serum-free medium. Formation of spheroids can be monitored in real time. Tumor sphere number and size allow for the evaluation of the cancer stem cells in a given tissue. Moreover, spheroids represent useful models for drug screening with specific impact on the cancer stem cell compartment (Albino et al., 2021; Zoma et al., 2021).
NOTE : Federal regulations require that research projects involving human subjects be reviewed by an Institutional Review Board (IRB). The IRB must approve or determine the project to be exempt prior to the start of any research activities.
Basic Protocol 1: GENERATION OF ORGANOIDS STARTING FROM THE PROSTATE GLAND OF A GEMM OR A HUMAN PDX
This protocol describes and provides a detailed procedure for the generation of organoids (Fig. 2). Tissue from a human PDX or prostate tissue from a GEMM serve as source material to obtain individual cells. Next, single cells are mixed with Matrigel matrix and seeded as a drop in desired plates to generate 3D organoids.
About 7 to 10 days post seeding, 3D organoids are well formed and reach a dimension of 150 to 300 µm in diameter.
End points to be evaluated:
1.Number of organoids 2.Phenotype of organoids
The number of organoids formed reflects the proliferative index of the tissue of origin and is an important parameter to be evaluated. Depending on the tissue of origin, 3D organoids can show different morphologies and sizes. Recent advances in the assessment of tumor-derived organoids indicate that prostate epithelial normal cells give rise to 3D cystic-like organoids with a regular shape and lumen (Fig. 1). By contrast, transformed cells form full hyperplastic or irregular structures (Karthaus et al., 2020). 3D organoid cultures are a suitable model to test drugs that can affect their number, morphology, and shape.
NOTE : All protocols using live animals must first be reviewed and approved by an Institutional Animal Care and Use Committee (IACUC) and must follow officially approved procedures for the care and use of laboratory animals.
Materials
-
Gibco Dulbecco's modified Eagle medium (DMEM; Thermo Fisher Scientific, cat. no. 12100-061)
-
Gibco RPMI-1640 medium (Thermo Fisher Scientific, cat. no. 31800-089)
-
FBS (Capricorn Scientific, cat. no. FBS-11A)
-
Mouse organoid medium (Drost et al., 2016)
-
PBS, 1× (Chemie Brunschwig, cat. no. CS1PBS01-01)
-
Matrigel Growth Factor Reduced (GFR), Phenol red-free (BD Biosciences, cat. no. 356231)
-
L-Glutamine (Thermo Fisher Scientific, cat. no. 25030032)
-
Collagenase type II (Thermo Fisher Scientific, cat. no. 17101-015)
-
Y-27632 dihydrochloride (MilliporeSigma, cat. no. SCM075)
-
TrypLE™ Express Enzyme (1×), phenol red, 100 ml (Thermo Fisher Scientific, cat. no. 12605-010)
-
Trypan blue, 0.4% (MilliporeSigma, cat. no. T8154)
-
Penicillin-Streptomycin (10,000 U/ml; Thermo Fisher Scientific, cat. no. 15-140-122)
-
Dimethyl sulfoxide (DMSO; MilliporeSigma, cat. no. D2650)
-
Dissecting medium: DMEM supplemented with 10% (v/v) FBS, 1% (v/v) 100× glutamine, and 1% (v/v) 100× penicillin/streptomycin solution
-
12- to 36-week-old male Ptenflox/flox; R26ERG mice, housed in individually ventilated microisolator cages
-
Dissecting tools (sterilized):
- Dissecting scissors
- Micro scissors
- Dissecting forceps
- Scalpel
- Dissecting microscope (Olympus/Leica)
-
Cell strainer 40-μm pore size (Falcon; Corning, cat. no. 352340)
-
Cell culture centrifuge (Beckman Coulter)
-
Shaking platform
-
Tissue culture hood
-
Cell incubator set to 5% CO2 and 37°C
-
Cell culture disposables:
- Petri dishes (BD Flacon)
- Centrifuge tubes (Eppendorf)
- Pipets
- Pipet tips
- Filter units (Millipore Sigma)
-
96-well non-tissue culture plate (BD Falcon; Corning, cat. no. 351172)
-
Inverted microscope
1.Sacrifice male mice according to specific animal licenses.
2.Place mice with the abdomen in upside position. Spray abdomen with 70% ethanol.
3.Cut the skin with the dissecting scissors vertically along the abdomen from proximal to distal to open the peritoneum.
4.Using forceps, carefully secure the bladder and pull it gently in order to remove the entire urogenital apparatus consisting of clearly visible seminal vesicles and prostate.
5.Place seminal vesicles, prostate, and bladder in a 10-cm Petri dish containing 3 ml of ice-cold dissecting medium (Lukacs et al., 2010).
6.Remove blood vessels and connective tissue, and make an incision (0.5 cm) at the base of the urethra.
7.Remove bladder, seminal vesicles, vas deferens, and fat tissue covering the prostate lobes by gentle cuttings.
8.Remove urethra and ampullary gland, and carefully move the whole prostate into a new clean 10-cm Petri dish.
9.Mince the prostate lobes into small pieces (∼1 mm3) in the 10-cm Petri dish by using a clean scalpel, keeping tissue in 3 ml of ice-cold dissecting medium.
10.Centrifuge tissue chunks at 300 × g for 5 min at 25°C.
11.Transfer minced prostate into a 15-ml tube containing 4-5 ml of 5 mg/ml collagenase type II with Y-27632 (10 µM), mix well with a P1000 pipet, and incubate at 37°C for 1.5-2 hr on a rotator.
12.Centrifuge at 300 × g for 5 min at 4°C.
13.Decant supernatant and resuspend pellet in 500 µl TrypLE with Y-27632 (10 µM) and incubate 15 min at 37°C on a shaking platform. Pipet up and down with a P1000 pipet to ensure efficient digestion.
14.Put a 40-µm cell strainer on the top of a 15-ml Falcon tube and load digestion through the strainer.
15.Add 2 ml ice-cold sterile 1× PBS and centrifuge at 300 × g for 5 min at 4°C.
16.Decant supernatant and resuspend pellet in 2 ml ice-cold sterile 1× PBS and centrifuge at 300 × g for 5 min at 4°C.
17.Decant supernatant and resuspend pellet in 1 ml ice-cold sterile 1× PBS.
18.Take an aliquot of 10 or 20 µl of cell suspension and add it to an equal volume of 0.4% trypan blue for counting of viable cells by using an automatic counter.
19.Combine 5,000 single cells with ice-cold sterile Matrigel (previously thawed at 4°C overnight) in a 1.5-ml Eppendorf tube in ice bucket.
20.Mix well combined cells and Matrigel with a 20-µl pipet and pipet 10 µl into the middle of one well of a pre-warmed (37°C) non-tissue treated 96-well plate to form a dome (Fig. 3).
21.Invert the 96-well plate to prevent adherence of cells to the plate bottom and carefully place it upside down into the 37°C incubator for at least 30-45 min to allow the Matrigel to solidify.
22.Transfer the 96-well plate carefully to the biological hood and gently add to each dome 100 µl of pre-warmed (37°C) Mouse organoid medium (Drost et al., 2016).
23.Optional: Drug as a single agent or combination can be diluted directly into mouse organoid medium for treatment during 3D organoid formation assay.
24.Evaluate organoid formation with an inverted microscope after 7-12 days post seeding (see Fig. 2). Culture organoids in tissue culture incubator at 37°C and 5% CO2.
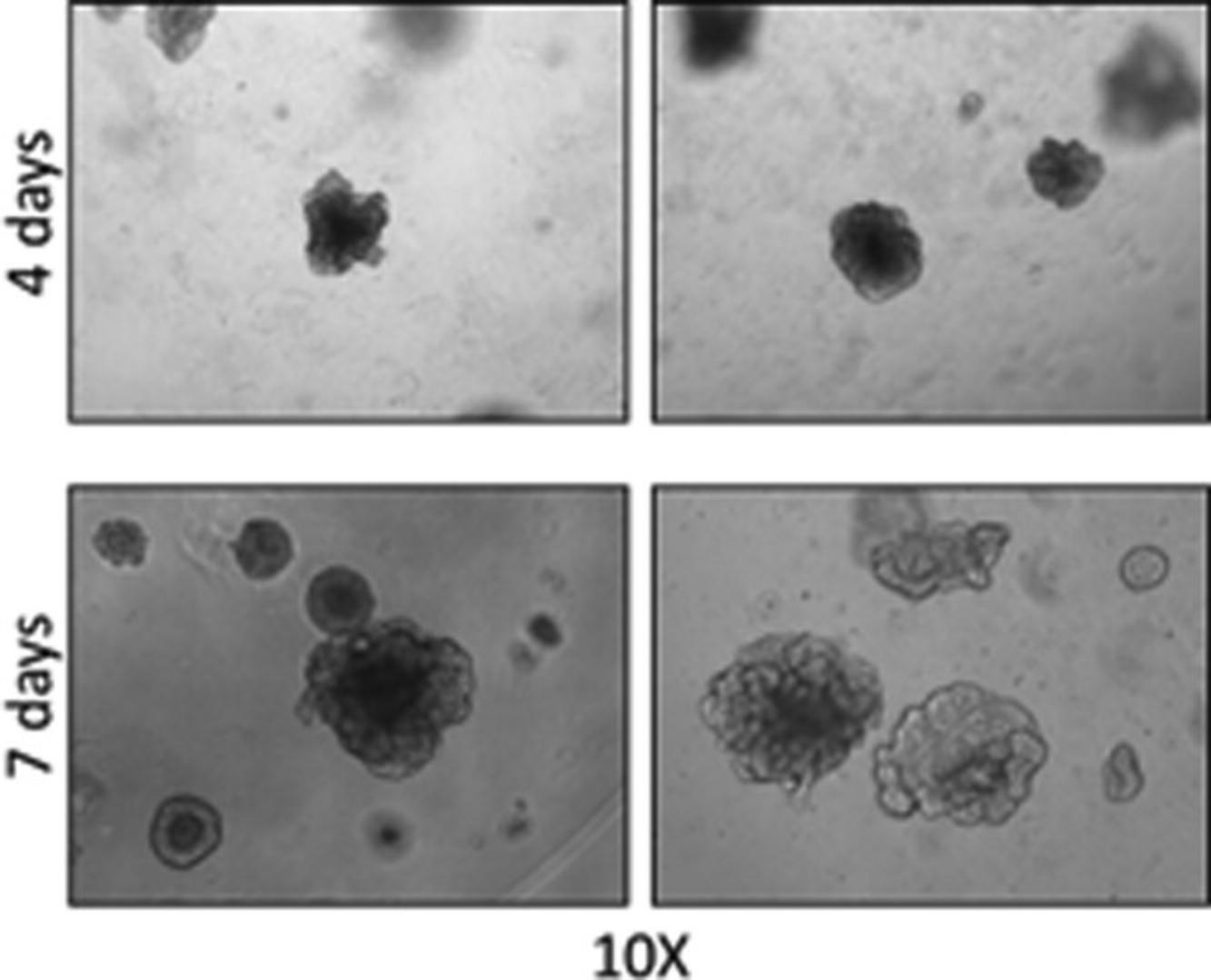
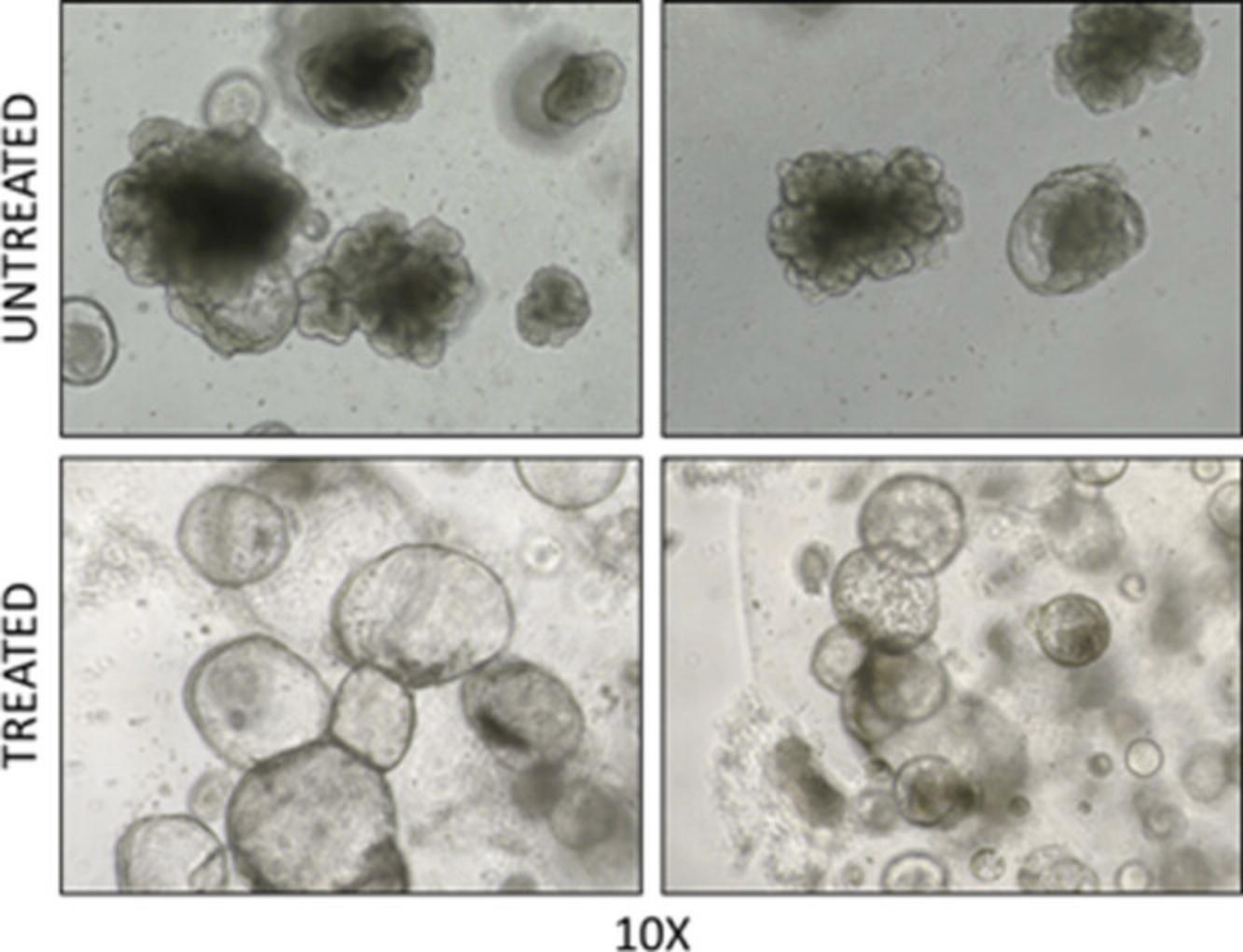
25.Take images of 3D organoids with inverted microscope and count organoids, evaluating only the organoids with a diameter >50 or >100 µm (Fig. 4).
Optional: Reversion of aggressive phenotype
Follow steps 1-19 for the first part of 3D organoid establishment.
26.Evaluate organoid formation and morphology after 7 days in culture.
27.Count and take images of organoid morphology.
28.Remove mouse organoid medium (Drost et al., 2016) and gently add to each well 100 µl of pre-warmed (37°C) mouse drug treatment medium (Chan et al., 2022) containing the drug or vehicle to be tested at the desired range of concentrations.
29.Evaluate reversion of aggressive organoid morphology after 4 days of treatment. Count and take images of organoids with reverted morphology (Fig. 5).
Basic Protocol 2: EX VIVO TUMOR SPHERE FORMATION
This protocol provides a detailed procedure for the generation and culture of tumor spheres derived from human (e.g., PDX or cancer cell xenograft) or from GEMM prostates. First, single cells are obtained after enzymatic digestion of minced xenografts; tumor spheres are formed by plating single cells in low attachment, low density, and serum-free conditions. Tumor spheres can be incubated in vitro with specific drugs for the evaluation of the impact of treatment on tumor spheres morphology, size, and number.
Materials
-
LuCaP 145.2 PDX tumors (Nguyen et al., 2017)
-
Collagenase D (MilliporeSigma, cat. no. 11088858001)
-
Gibco RPMI-1640 medium (Thermo Fisher Scientific, cat. no. 31800-089)
-
PBS, 1× (Chemie Brunschwig, cat. no. CS1PBS01-01)
-
Trypan blue, 0.4% (MilliporeSigma, cat. no. T8154)
-
B27 supplement, 50× (Life Technologies, cat. no. 17504-044)
-
hBFGF (MilliporeSigma, cat. no. F0291-25UG)
-
EGF (LubioScience, cat. no. AF-100-15-100UG)
-
Insulin (MilliporeSigma, cat. no. 12585-014)
-
DNase I Solution (1 unit/µl), RNase-free (Thermo Fisher Scientific, cat. no. 89836)
-
Red blood lysis buffer (Roche, cat. no. 11814389001)
-
Mammary Epithelial Basal Medium (MEBM; Lonza, cat. no. CC-3151)
-
Poly-2-hydroxyethyl methacrylate, 1× (MilliporeSigma, cat. no. P3932), dissolved in 95% ethanol (12 mg/ml) for coating 12-well tissue culture plates
-
Dissecting tools (sterilized):
- Dissecting scissors
- Dissecting forceps
- Dissecting microscope (Olympus/Leica)
-
Cell strainer 40-μm pore size (Falcon; Corning, cat. no. 352340)
-
Cell culture centrifuge (Beckman Coulter)
-
CO2 incubator set to 5% CO2 and 37°C
-
Cell culture disposables:
- Petri dishes (BD Flacon)
- Centrifuge tubes (Eppendorf)
- Pipets
- Pipet tips
- Filter units (MilliporeSigma)
-
12-well plates (VWR, cat. no. 10062-894)
1.Remove subcutaneous LuCaP 145.2 tumor xenografts, separating them from mouse skin by using scissors and forceps.
2.Put xenograft in a 10-cm Petri dish containing 3 ml ice-cold 1× PBS.
3.Mince xenograft into small pieces (∼1 mm3) in a 10-cm Petri dish by using a scalpel.
4.Centrifuge tissue chunks at 300 × g for 5 min at 25°C.
5.Transfer minced xenograft into a 15-ml tube containing 5 ml RPMI-1640 with collagenase D (5 mg/ml) and DNase I (100 U/ml).
6.Mix well with a P1000 pipet and incubate at 37°C for 45 min on a rotator.
7.Centrifuge at 300 × g for 5 min at 4°C.
8.Decant supernatant and resuspend cell pellet in 5 ml of 1× PBS. Pipet up and down with a P1000 pipet to ensure efficient resuspension.
9.Centrifuge at 300 × g for 5 min at 4°C.
10.Decant supernatant and resuspend cell pellet in 5 ml of 1× PBS.
11.Put a 40-µm cell strainer on the top of a 15-ml Falcon tube and load resuspended cells trough the strainer.
12.Centrifuge at 300 × g for 5 min at 4°C.
13.Decant supernatant and resuspend pellet in 2-5 ml of red blood cell lysis buffer for 5 min at room temperature.
14.Add 5 ml of 1× PBS and centrifuge at 300 × g for 5 min at 4°C.
15.Decant supernatant and resuspend pellet in 1× PBS.
16.Take an aliquot of 10 or 20 µl of cell suspension and add to an equal volume of 0.4% trypan blue for counting of viable cells by using an automatic hemacytometer.
17.Prepare a mix of 5,000 cells in 2 ml of serum-free MEBM with specific supplements: 1× B27; 20 ng/ml hBFGF; 20 ng/ml EGF; 0.4 µg/ml insulin; 1% pen/strep.
18.Seed cells in pre-coated poly-2-hydroxyethyl methacrylate 12-well plates.
19.Add therapeutic agent of interest at desired concentration(s) as a single agent or in combinations for treatment during tumor sphere formation assay. Culture tumor spheres in tissue culture incubator at 37°C and 5% CO2.
20.Replenish 2 ml of fresh serum-free MEBM with specific supplements after 3 days.
21.Evaluate tumor sphere formation after 14 days in culture (see Fig. 2).
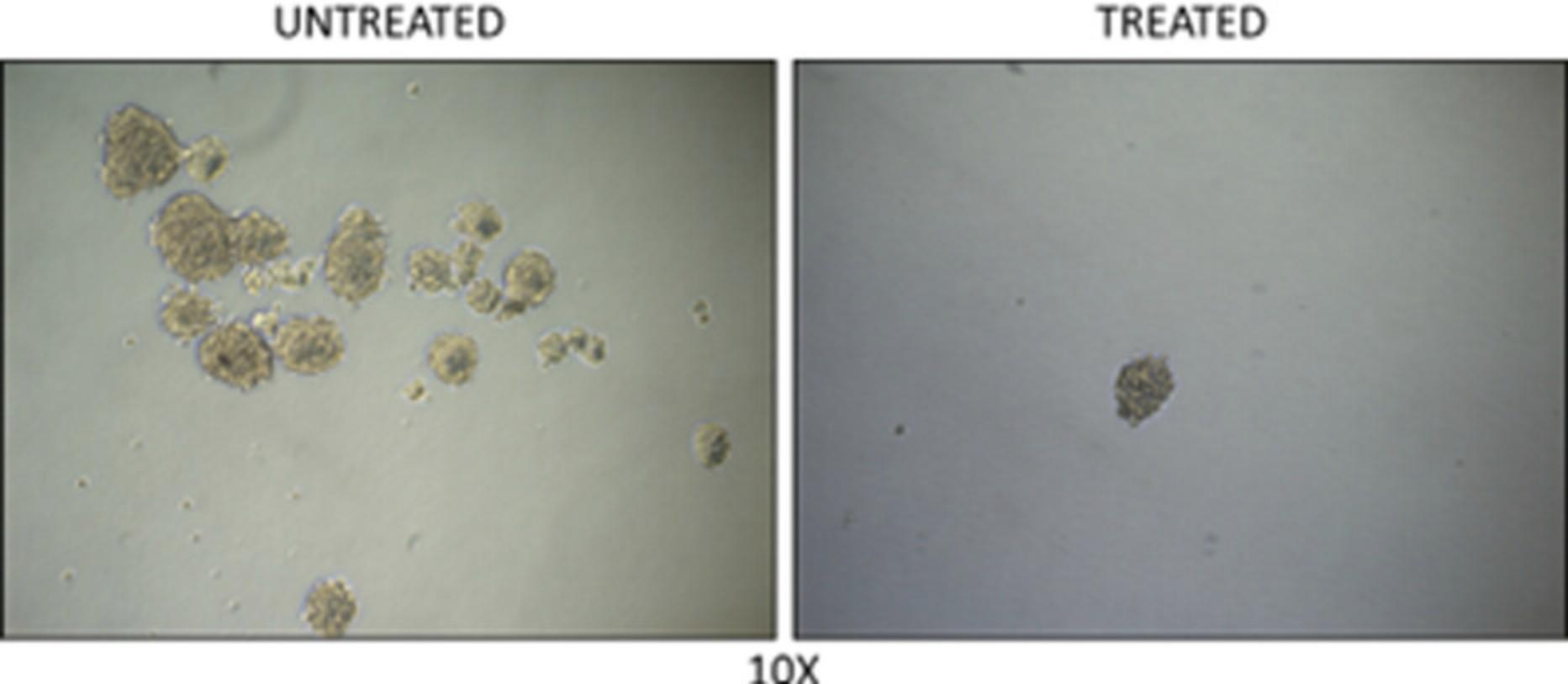
22.Count and picture tumor spheres with a diameter >50 µm (Fig. 6).
COMMENTARY
Background Information
The basic protocols described in this article are a useful tool to assess the therapeutic potential of new drugs in the treatment of NEPC. As described in Basic Protocol 1, 3D organoids can be used to evaluate the effect of a drug treatment on growth (Fig. 4) and morphology of organoids (Fig. 5). Specifically, while the evaluation of organoid growth assesses the anti-proliferative effects of a given therapeutic, the evaluation of organoids morphology can provide additional information of the impact on phenotypic characteristics more strictly related to differentiation/dedifferentiation state of the tumor cells. This strategy is particularly useful to test drugs that might impact directly on the transcriptome and epigenome, thus preventing or reversing the reprogramming and progression towards a more aggressive phenotype. To this end, this approach could be refined with assessment of specific markers by immunostaining and fluorescence microscopy. Similarly, the method described in Basic Protocol 2, based on the in vitro /ex vivo tumor sphere formation assay, may address specifically the effect of a treatment on the survival, self-renewal capability, and tumorigenic potential of the stem-like tumor-initiating cancer cells (Fig. 6).
The protocols described in this review were optimized based on previous works for 3D organoid formation (Drost & Clevers, 2018; Drost et al., 2016) and for sphere formation (Albino et al., 2021; Zoma et al., 2021). These protocols can provide preclinical models for the development of new therapeutic approaches for the treatment of NEPC.
Critical Parameters
In the two protocols described above, critical factors that can influence the results are the following: (1) during dissection of prostate glands, PDX, and tumor xenografts it is critical to avoid contamination from other tissues; (2) keeping Matrigel matrix on ice is extremely critical to avoid solidification.
Troubleshooting
The troubleshooting guide is highlighted in Table 3.
| Problem | Possible cause | Solution |
|---|---|---|
| Matrigel matrix solidification | Not appropriate storage conditions | Always keep on ice and avoid repeated thaw and freezing cycles |
| Low yield cell viability | Increased time of tissue digestion | Reduce the time for enzymatic digestion step |
| Low yield in 3D structure numbers |
Not appropriate calculation of cell viability Number of cells seeded is too low |
Confirm cell viability with automated and manual counting Increase number of cells |
Understanding Results
These protocols allow the evaluation of NEPC features by employing preclinical models, such as 3D organoids and tumor spheres. As a general consideration, these preclinical models represent a valuable source for downstream applications. These include, but are not limited to, multi-omics studies. Genetic manipulation of these models can provide a greater insight into the mechanisms that lead to NEPC emergence and progression.
Time Considerations
Establishment of 3D organoids, as described in Basic Protocol 1, requires 7 to 12 days. For the assessment of reversed phenotype in organoids, an additional 10 days are required. Ex vivo tumor spheres formation, as described in Basic Protocol 2, requires up to 14 days.
Acknowledgments
We acknowledge support from the Krebsliga Schweiz (grant KLS-4569-08-2018 and KLS-4899-08-2019), Swiss National Science Foundation (grant SNSF-310030_189081, SNSF-IZLSZ3_170898, SNSF-IZCOZO_189862, and SNSF-310030L_170182), Foundation Nelia et Amadeo Barletta (FNAB), Fondazione Ticinese Ricerca sul Cancro, and Fondazione San Salvatore.
Author Contribution
Alessia Cacciatore : Conceptualization, methodology, writing original draft, writing review and editing. Domenico Albino : Conceptualization, methodology, writing original draft, writing review and editing. Carlo Catapano : Conceptualization, investigation, methodology, resources, supervision, validation, writing original draft, writing review and editing. Giuseppina Carbone : Conceptualization, data curation, funding acquisition, methodology, resources, supervision, writing original draft, writing review and editing.
Conflict of Interest
Authors declare no conflict of interest.
Open Research
Data Availability Statement
Data sharing not applicable to this article as no datasets were generated or analyzed during the current study.
Literature Cited
- Aggarwal, R., Huang, J., Alumkal, J. J., Zhang, L., Feng, F. Y., Thomas, G. V., Weinstein, A. S., Friedl, V., Zhang, C., Witte, O. N., Lloyd, P., Gleave, M., Evans, C. P., Youngren, J., Beer, T. M., Rettig, M., Wong, C. K., True, L., Foye, A., … Small, E. J. (2018). Clinical and genomic characterization of treatment-emergent small-cell neuroendocrine prostate cancer: A multi-institutional prospective study. Journal of Clinical Oncology , 36(24), 2492–2503. https://doi.org/10.1200/JCO.2017.77.6880
- Albino, D., Falcione, M., Uboldi, V., Temilola, D. O., Sandrini, G., Merulla, J., Civenni, G., Kokanovic, A., Stürchler, A., Shinde, D., Garofalo, M., Mestre, R. P., Constâncio, V., Wium, M., Burrello, J., Baranzini, N., Grimaldi, A., Theurillat, J.- P., Bossi, D., … Carbone, G. M. (2021). Circulating extracellular vesicles release oncogenic miR-424 in experimental models and patients with aggressive prostate cancer. Communications Biology , 4(1), 119. https://doi.org/10.1038/s42003-020-01642-5
- Beltran, H., Prandi, D., Mosquera, J. M., Benelli, M., Puca, L., Cyrta, J., Marotz, C., Giannopoulou, E., Chakravarthi, B. V. S. K., Varambally, S., Tomlins, S. A., Nanus, D. M., Tagawa, S. T., Van Allen, E. M., Elemento, O., Sboner, A., Garraway, L. A., Rubin, M. A., & Demichelis, F. (2016). Divergent clonal evolution of castration-resistant neuroendocrine prostate cancer. Nature Medicine , 22(3), 298–305. https://doi.org/10.1038/nm.4045
- Beltran, H., Rickman, D. S., Park, K., Chae, S. S., Sboner, A., Macdonald, T. Y., Wang, Y., Sheikh, K. L., Terry, S., Tagawa, S. T., Dhir, R., Nelson, J. B., De La Taille, A., Allory, Y., Gerstein, M. B., Perner, S., Pienta, K. J., Chinnaiyan, A. M., Wang, Y., … Rubin, M. A. (2011). Molecular characterization of neuroendocrine prostate cancer and identification of new drug targets. Cancer Discovery , 1(6), 487–495. https://doi.org/10.1158/2159-8290.CD-11-0130
- Beltran, H., Romanel, A., Conteduca, V., Casiraghi, N., Sigouros, M., Franceschini, G. M., Orlando, F., Fedrizzi, T., Ku, S.-Y., Dann, E., Alonso, A., Mosquera, J. M., Sboner, A., Xiang, J., Elemento, O., Nanus, D. M., Tagawa, S. T., Benelli, M., & Demichelis, F. (2020). Circulating tumor DNA profile recognizes transformation to castration-resistant neuroendocrine prostate cancer. Journal of Clinical Investigation , 130(4), 1653–1668. https://doi.org/10.1172/JCI131041
- The Cancer Genome Atlas Research Network. (2015). The molecular taxonomy of primary prostate cancer. Cell , 163(4), 1011–1025. https://doi.org/10.1016/j.cell.2015.10.025
- Cejas, P., Xie, Y., Font-Tello, A., Lim, K., Syamala, S., Qiu, X., Tewari, A. K., Shah, N., Nguyen, H. M., Patel, R. A., Brown, L., Coleman, I., Hackeng, W. M., Brosens, L., Dreijerink, K. M. A., Ellis, L., Alaiwi, S. A., Seo, J.-H., Baca, S., & … Long, H. W. (2021). Subtype heterogeneity and epigenetic convergence in neuroendocrine prostate cancer. Nature Communications , 12(1), 5775. https://doi.org/10.1038/s41467-021-26042-z
- Chan, J. M., Zaidi, S., Love, J. R., Zhao, J. L., Setty, M., Wadosky, K. M., Gopalan, A., Choo, Z. - N., Persad, S., Choi, J., Laclair, J., Lawrence, K. E., Chaudhary, O., Xu, T., Masilionis, I., Linkov, I., Wang, S., Lee, C., Barlas, A., … Sawyers, C. L. (2022). Lineage plasticity in prostate cancer depends on JAK/STAT inflammatory signaling. Science , 377(6611), 1180–1191. https://doi.org/10.1126/science.abn0478
- Chen, Y., Chi, P., Rockowitz, S., Iaquinta, P. J., Shamu, T., Shukla, S., Gao, D., Sirota, I., Carver, B. S., Wongvipat, J., Scher, H. I., Zheng, D., & Sawyers, C. L. (2013). ETS factors reprogram the androgen receptor cistrome and prime prostate tumorigenesis in response to PTEN loss. Nature Medicine , 19(8), 1023–1029. https://doi.org/10.1038/nm.3216
- Chen, Y., Zhou, Q., Hankey, W., Fang, X., & Yuan, F. (2022). Second generation androgen receptor antagonists and challenges in prostate cancer treatment. Cell Death & Disease, 13(7), 632. https://doi.org/10.1038/s41419-022-05084-1
- Chlenski, A., Nakashiro, K. - I., Ketels, K. V., Korovaitseva, G. I., & Oyasu, R. (2001). Androgen receptor expression in androgen-independent prostate cancer cell lines. Prostate , 47(1), 66–75. https://doi.org/10.1002/pros.1048
- Conteduca, V., Oromendia, C., Eng, K. W., Bareja, R., Sigouros, M., Molina, A., Faltas, B. M., Sboner, A., Mosquera, J. M., Elemento, O., Nanus, D. M., Tagawa, S. T., Ballman, K. V., & Beltran, H. (2019). Clinical features of neuroendocrine prostate cancer. European Journal of Cancer , 121, 7–18. https://doi.org/10.1016/j.ejca.2019.08.011
- Dardenne, E., Beltran, H., Benelli, M., Gayvert, K., Berger, A., Puca, L., Cyrta, J., Sboner, A., Noorzad, Z., Macdonald, T., Cheung, C., Yuen, K. S., Gao, D., Chen, Y., Eilers, M., Mosquera, J. - M., Robinson, B. D., Elemento, O., Rubin, M. A., … Rickman, D. S. (2016). N-Myc induces an EZH2-mediated transcriptional program driving neuroendocrine prostate cancer. Cancer Cell , 30(4), 563–577. https://doi.org/10.1016/j.ccell.2016.09.005
- Davis, I. D., Martin, A. J., Stockler, M. R., Begbie, S., Chi, K. N., Chowdhury, S., Coskinas, X., Frydenberg, M., Hague, W. E., Horvath, L. G., Joshua, A. M., Lawrence, N. J., Marx, G., McCaffrey, J., McDermott, R., McJannett, M., North, S. A., Parnis, F., Parulekar, W., … for the ENZAMET Trial Investigators and the Australian and New Zealand Urogenital and Prostate Cancer Trials Group. (2019). Enzalutamide with standard first-line therapy in metastatic prostate cancer. New England Journal of Medicine , 381(2), 121–131. https://doi.org/10.1056/NEJMoa1903835
- Desai, K., Mcmanus, J. M., & Sharifi, N. (2021). Hormonal therapy for prostate cancer. Endocrine Reviews , 42(3), 354–373. https://doi.org/10.1210/endrev/bnab002
- Drost, J., & Clevers, H. (2018). Organoids in cancer research. Nature Reviews Cancer , 18(7), 407–418. https://doi.org/10.1038/s41568-018-0007-6
- Drost, J., Karthaus, W. R., Gao, D., Driehuis, E., Sawyers, C. L., Chen, Y., & Clevers, H. (2016). Organoid culture systems for prostate epithelial and cancer tissue. Nature Protocols , 11(2), 347–358. https://doi.org/10.1038/nprot.2016.006
- Elbadawy, M., Abugomaa, A., Yamawaki, H., Usui, T., & Sasaki, K. (2020). Development of prostate cancer organoid culture models in basic medicine and translational research. Cancers , 12(4), 777. https://doi.org/10.3390/cancers12040777
- Epstein, J. I., Amin, M. B., Beltran, H., Lotan, T. L., Mosquera, J.-M., Reuter, V. E., Robinson, B. D., Troncoso, P., & Rubin, M. A. (2014). Proposed morphologic classification of prostate cancer with neuroendocrine differentiation. American Journal of Surgical Pathology , 38(6), 756–767. https://doi.org/10.1097/PAS.0000000000000208
- Gao, D., Vela, I., Sboner, A., Iaquinta, P. J., Karthaus, W. R., Gopalan, A., Dowling, C., Wanjala, J. N., Undvall, E. A., Arora, V. K., Wongvipat, J., Kossai, M., Ramazanoglu, S., Barboza, L. P., Di, W., Cao, Z., Zhang, Q. F., Sirota, I., Ran, L., … Chen, Y. (2014). Organoid cultures derived from patients with advanced prostate cancer. Cell , 159(1), 176–187. https://doi.org/10.1016/j.cell.2014.08.016
- Gingrich, J. R., Barrios, R. J., Morton, R. A., Boyce, B. F., DeMayo, F. J., Finegold, M. J., Angelopoulou, R., Rosen, J. M., & Greenberg, N. M. (1996). Metastatic prostate cancer in a transgenic mouse. Cancer Research , 56(18), 4096–4102. https://www.ncbi.nlm.nih.gov/pubmed/8797572
- Gingrich, J. R., & Greenberg, N. M. (1996). A transgenic mouse prostate cancer model. Toxicologic Pathology , 24(4), 502–504. https://doi.org/10.1177/019262339602400414
- Goldstein, A. S., Lawson, D. A., Cheng, D., Sun, W., Garraway, I. P., & Witte, O. N. (2008). Trop2 identifies a subpopulation of murine and human prostate basal cells with stem cell characteristics. Proceedings of the National Academy of Sciences of the United States of America , 105(52), 20882–20887. https://doi.org/10.1073/pnas.0811411106
- Greenberg, N. M., Demayo, F., Finegold, M. J., Medina, D., Tilley, W. D., Aspinall, J. O., Cunha, G. R., Donjacour, A. A., Matusik, R. J., & Rosen, J. M. (1995). Prostate cancer in a transgenic mouse. Proceedings of the National Academy of Sciences of the United States of America , 92(8), 3439–3443. https://doi.org/10.1073/pnas.92.8.3439
- Hofer, M., & Lutolf, M. P. (2021). Engineering organoids. Nature Reviews Materials , 6(5), 402–420. https://doi.org/10.1038/s41578-021-00279-y
- Karthaus, W. R., Hofree, M., Choi, D., Linton, E. L., Turkekul, M., Bejnood, A., Carver, B., Gopalan, A., Abida, W., Laudone, V., Biton, M., Chaudhary, O., Xu, T., Masilionis, I., Manova, K., Mazutis, L., Pe'er, D., Regev, A., & Sawyers, C. L. (2020). Regenerative potential of prostate luminal cells revealed by single-cell analysis. Science , 368(6490), 497–505. https://doi.org/10.1126/science.aay0267
- Karthaus, W. R., Iaquinta, P. J., Drost, J., Gracanin, A., Van Boxtel, R., Wongvipat, J., Dowling, C. M., Gao, D., Begthel, H., Sachs, N., Vries, R. G. J., Cuppen, E., Chen, Y., Sawyers, C. L., & Clevers, H. C. (2014). Identification of multipotent luminal progenitor cells in human prostate organoid cultures. Cell , 159(1), 163–175. https://doi.org/10.1016/j.cell.2014.08.017
- Kersten, K., Visser, K. E., Miltenburg, M. H., & Jonkers, J. (2017). Genetically engineered mouse models in oncology research and cancer medicine. EMBO Molecular Medicine , 9(2), 137–153. https://doi.org/10.15252/emmm.201606857
- Kim, J., Koo, B.-K., & Knoblich, J. A. (2020). Human organoids: Model systems for human biology and medicine. Nature Reviews Molecular Cell Biology , 21(10), 571–584. https://doi.org/10.1038/s41580-020-0259-3
- Kleinman, H. K., & Martin, G. R. (2005). Matrigel: Basement membrane matrix with biological activity. Seminars in Cancer Biology , 15(5), 378–386. https://doi.org/10.1016/j.semcancer.2005.05.004
- Ku, S. Y. U., Rosario, S., Wang, Y., Mu, P., Seshadri, M., Goodrich, Z. W., Goodrich, M. M., Labbé, D. P., Gomez, E. C., Wang, J., Long, H. W., Xu, B., Brown, M., Loda, M., Sawyers, C. L., Ellis, L., & Goodrich, D. W. (2017). Rb1 and Trp53 cooperate to suppress prostate cancer lineage plasticity, metastasis, and antiandrogen resistance. Science , 355(6320), 78–83. https://doi.org/10.1126/science.aah4199
- Lawson, D. A., Xin, L., Lukacs, R. U., Cheng, D., & Witte, O. N. (2007). Isolation and functional characterization of murine prostate stem cells. Proceedings of the National Academy of Sciences of the United States of America , 104(1), 181–186. https://doi.org/10.1073/pnas.0609684104
- Lee, J. K., Phillips, J. W., Smith, B. A., Park, J. W., Stoyanova, T., Mccaffrey, E. F., Baertsch, R., Sokolov, A., Meyerowitz, J. G., Mathis, C., Cheng, D., Stuart, J. M., Shokat, K. M., Gustafson, W. C., Huang, J., & Witte, O. N. (2016). N-Myc drives neuroendocrine prostate cancer initiated from human prostate epithelial cells. Cancer Cell , 29(4), 536–547. https://doi.org/10.1016/j.ccell.2016.03.001
- Lin, D., Wyatt, A. W., Xue, H., Wang, Y., Dong, X., Haegert, A., Wu, R., Brahmbhatt, S., Mo, F., Jong, L., Bell, R. H., Anderson, S., Hurtado-Coll, A., Fazli, L., Sharma, M., Beltran, H., Rubin, M., Cox, M., Gout, P. W., … Wang, Y. (2014). High fidelity patient-derived xenografts for accelerating prostate cancer discovery and drug development. Cancer Research , 74(4), 1272–1283. https://doi.org/10.1158/0008-5472.CAN-13-2921-T
- Litwin, M. S., & Tan, H.-J. (2017). The diagnosis and treatment of prostate cancer: A Review. JAMA , 317(24), 2532–2542. https://doi.org/10.1001/jama.2017.7248
- Lukacs, R. U., Goldstein, A. S., Lawson, D. A., Cheng, D., & Witte, O. N. (2010). Isolation, cultivation and characterization of adult murine prostate stem cells. Nature Protocols , 5(4), 702–713. https://doi.org/10.1038/nprot.2010.11
- Mapelli, S., Albino, D., Mello-Grand, M., Shinde, D., Scimeca, M., Bonfiglio, R., Bonanno, E., Chiorino, G., Garcia-Escudero, R., Catapano, C., & Carbone, G. (2020). A novel prostate cell type-specific gene signature to interrogate prostate tumor differentiation status and monitor therapeutic response. Cancers , 12(1), 176. https://doi.org/10.3390/cancers12010176
- Merkens, L., Sailer, V., Lessel, D., Janzen, E., Greimeier, S., Kirfel, J., Perner, S., Pantel, K., Werner, S., & von Amsberg, G. (2022). Aggressive variants of prostate cancer: Underlying mechanisms of neuroendocrine transdifferentiation. Journal of Experimental & Clinical Cancer Research, 41(1), 46. https://doi.org/10.1186/s13046-022-02255-y
- Miao, C., Tsujino, T., Takai, T., Gui, F., Tsutsumi, T., Sztupinszki, Z., Wang, Z., Azuma, H., Szallasi, Z., Mouw, K. W., Zou, L., Kibel, A. S., & Jia, L. (2022). RB1 loss overrides PARP inhibitor sensitivity driven by RNASEH2B loss in prostate cancer. Science Advances , 8(7), eabl9794. https://doi.org/10.1126/sciadv.abl9794
- Mu, P., Zhang, Z., Benelli, M., Karthaus, W. R., Hoover, E., Chen, C.-C., Wongvipat, J., Ku, S-Y., Gao, D., Cao, Z., Shah, N., Adams, E. J., Abida, W., Watson, P. A., Prandi, D., Huang, C-H., De Stanchina, E., Lowe, S. W., Ellis, L., … Sawyers, C. L. (2017). SOX2 promotes lineage plasticity and antiandrogen resistance in TP53 - and RB1 -deficient prostate cancer. Science , 355(6320), 84–88. https://doi.org/10.1126/science.aah4307
- Nadal, R., Schweizer, M., Kryvenko, O. N., Epstein, J. I., & Eisenberger, M. A. (2014). Small cell carcinoma of the prostate. Nature Reviews Urology , 11(4), 213–219. https://doi.org/10.1038/nrurol.2014.21
- Nguyen, H. M., Vessella, R. L., Morrissey, C., Brown, L. G., Coleman, I. M., Higano, C. S., Mostaghel, E. A., Zhang, X., True, L. D., Lam, H-M., Roudier, M., Lange, P. H., Nelson, P. S., & Corey, E. (2017). LuCaP prostate cancer patient-derived xenografts reflect the molecular heterogeneity of advanced disease an–d serve as models for evaluating cancer therapeutics. Prostate , 77(6), 654–671. https://doi.org/10.1002/pros.23313
- Palanisamy, N., Yang, J., Shepherd, P. D. A., Li-Ning-Tapia, E. M., Labanca, E., Manyam, G. C., Ravoori, M. K., Kundra, V., Araujo, J. C., Efstathiou, E., Pisters, L., Wan, X., Wang, X., Vazquez, E. S., Aparicio, A. M., Carskadon, S. L., Tomlins, S. A., Kunju, L. P., Chinnaiyan, A. M., … Navone, N. M. (2020). The MD Anderson prostate cancer patient-derived xenograft series (MDA PCa PDX) captures the molecular landscape of prostate cancer and facilitates marker-driven therapy development. Clinical Cancer Research , 26(18), 4933–4946. https://doi.org/10.1158/1078-0432.CCR-20-0479
- Parimi, V., Goyal, R., Poropatich, K., & Yang, X. J. (2014). Neuroendocrine differentiation of prostate cancer: A review. American Journal of Clinical and Experimental Urology , 2(4), 273–285. https://www.ncbi.nlm.nih.gov/pubmed/25606573
- Pinthus, J. H., Waks, T., Schindler, D. G., Harmelin, A., Said, J. W., Belldegrun, A., Ramon, J., & Eshhar, Z. (2000). WISH-PC2: A unique xenograft model of human prostatic small cell carcinoma. Cancer Research , 60(23), 6563–6567. https://www.ncbi.nlm.nih.gov/pubmed/11118033
- Puca, L., Bareja, R., Prandi, D., Shaw, R., Benelli, M., Karthaus, W. R., Hess, J., Sigouros, M., Donoghue, A., Kossai, M., Gao, D., Cyrta, J., Sailer, V., Vosoughi, A., Pauli, C., Churakova, Y., Cheung, C., Deonarine, L. D., McNary, T. J., … Beltran, H. (2018). Patient derived organoids to model rare prostate cancer phenotypes. Nature Communications , 9(1), 2404. https://doi.org/10.1038/s41467-018-04495-z
- Risbridger, G. P., Clark, A. K., Porter, L. H., Toivanen, R., Bakshi, A., Lister, N. L., Pook, D., Pezaro, C. J., Sandhu, S., Keerthikumar, S., Quezada Urban, R., Papargiris, M., Kraska, J., Madsen, H. B., Wang, H., Richards, M. G., Niranjan, B., O'Dea, S., Teng, L., … Taylor, R. A. (2021). The MURAL collection of prostate cancer patient-derived xenografts enables discovery through preclinical models of uro-oncology. Nature Communications , 12(1), 5049. https://doi.org/10.1038/s41467-021-25175-5
- Risbridger, G. P., Toivanen, R., & Taylor, R. A. (2018). Preclinical models of prostate cancer: Patient-derived xenografts, organoids, and other explant models. Cold Spring Harbor perspectives in medicine , 8(8), a030536. https://doi.org/10.1101/cshperspect.a030536
- Shi, M., Wang, Y., Lin, D., & Wang, Y. (2022). Patient-derived xenograft models of neuroendocrine prostate cancer. Cancer Letters , 525, 160–169. https://doi.org/10.1016/j.canlet.2021.11.004
- Shiota, M., Dejima, T., Yamamoto, Y., Takeuchi, A., Imada, K., Kashiwagi, E., Inokuchi, J., Tatsugami, K., Kajioka, S., Uchiumi, T., & Eto, M. (2018). Collateral resistance to taxanes in enzalutamide-resistant prostate cancer through aberrant androgen receptor and its variants. Cancer Science , 109(10), 3224–3234. https://doi.org/10.1111/cas.13751
- Siegel, R. L., Miller, K. D., Fuchs, H. E., & Jemal, A. (2022). Cancer statistics, 2022. CA: A Cancer Journal for Clinicians , 72(1), 7–33. https://doi.org/10.3322/caac.21708
- Singh, N., Ramnarine, V. R., Song, J. H., Pandey, R., Padi, S. K. R., Nouri, M., Olive, V., Kobelev, M., Okumura, K., Mccarthy, D., Hanna, M. M., Mukherjee, P., Sun, B., Lee, B. R., Parker, J. B., Chakravarti, D., Warfel, N. A., Zhou, M., Bearss, J. J., … Kraft, A. S. (2021). The long noncoding RNA H19 regulates tumor plasticity in neuroendocrine prostate cancer. Nature Communications , 12(1), 7349. https://doi.org/10.1038/s41467-021-26901-9
- Sramkoski, R. M., Pretlow, T. G. 2nd., Giaconia, J. M., Pretlow, T. P., Schwartz, S., Sy, M. S., Marengo, S. R., Rhim, J. S., Zhang, D., & Jacobberger, J. W. (1999). A new human prostate carcinoma cell line, 22R v1. In Vitro Cellular & Developmental Biology Animal, 35(7), 403–409. https://doi.org/10.1007/s11626-999-0115-4
- Su, R., Chen, L., Jiang, Z., Yu, M., Zhang, W., Ma, Z., Ji, Y., Shen, K., Xin, Z., Qi, J., Xue, W., & Wang, Q. (2022). Comprehensive analysis of androgen receptor status in prostate cancer with neuroendocrine differentiation. Frontiers in Oncology , 12, 955166. https://doi.org/10.3389/fonc.2022.955166
- True, L. D., Buhler, K., Quinn, J., Williams, E., Nelson, P. S., Clegg, N., Macoska, J. A., Norwood, T., Liu, A., Ellis, W., Lange, P., & Vessella, R. (2002). A neuroendocrine/small cell prostate carcinoma xenograft-LuCaP 49. American Journal of Pathology , 161(2), 705–715. https://doi.org/10.1016/S0002-9440(10)64226-5
- VanDeusen, H. R., Ramroop, J. R., Morel, K. L., Bae, S. Y. I., Sheahan, A. V., Sychev, Z., Lau, N. A., Cheng, L. C., Tan, V. M., Li, Z., Petersen, A., Lee, J. K., Park, J. W., Yang, R., Hwang, J. H., Coleman, I., Witte, O. N., Morrissey, C., Corey, E., … Drake, J. M. (2020). Targeting RET kinase in neuroendocrine prostate cancer. Molecular Cancer Research , 18(8), 1176–1188. https://doi.org/10.1158/1541-7786.MCR-19-1245
- Westbrook, T. C., Guan, X., Rodansky, E., Flores, D., Liu, C. J., Udager, A. M., Patel, R. A., Haffner, M. C., Hu, Y. -M., Sun, D., Beer, T. M., Foye, A., Aggarwal, R., Quigley, D. A., Youngren, J. F., Ryan, C. J., Gleave, M., Wang, Y., Huang, J., … Alumkal, J. (2022). Transcriptional profiling of matched patient biopsies clarifies molecular determinants of enzalutamide-induced lineage plasticity. Nature Communications , 13(1), 5345. https://doi.org/10.1038/s41467-022-32701-6
- Yamada, Y., & Beltran, H. (2021). Clinical and Biological features of neuroendocrine prostate cancer. Current Oncology Reports , 23(2), 15. https://doi.org/10.1007/s11912-020-01003-9
- Zhang, Q., Han, Y., Zhang, Y., Liu, D., Ming, J., Huang, B., & Qiu, X. (2020). Treatment-emergent neuroendocrine prostate cancer: A clinicopathological and immunohistochemical analysis of 94 cases. Frontiers in Oncology , 10, 571308. https://doi.org/10.3389/fonc.2020.571308
- Zhou, Z., Flesken-Nikitin, A., Corney, D. C., Wang, W., Goodrich, D. W., Roy-Burman, P., & Nikitin, A. Y. U. (2006). Synergy of p53 and Rb deficiency in a conditional mouse model for metastatic prostate cancer. Cancer Research , 66(16), 7889–7898. https://doi.org/10.1158/0008-5472.CAN-06-0486
- Zoma, M., Curti, L., Shinde, D., Albino, D., Mitra, A., Sgrignani, J., Mapelli, S. N., Sandrini, G., Civenni, G., Merulla, J., Chiorino, G., Kunderfranco, P., Cacciatore, A., Kokanovic, A., Rinaldi, A., Cavalli, A., Catapano, C. V., & Carbone, G. M. (2021). EZH2-induced lysine K362 methylation enhances TMPRSS2-ERG oncogenic activity in prostate cancer. Nature Communications , 12(1), 4147. https://doi.org/10.1038/s41467-021-24380-6
- Zou, M., Toivanen, R., Mitrofanova, A., Floch, N., Hayati, S., Sun, Y., Le Magnen, C., Chester, D., Mostaghel, E. A., Califano, A., Rubin, M. A., Shen, M. M., & Abate-Shen, C. (2017). Transdifferentiation as a mechanism of treatment resistance in a mouse model of castration-resistant prostate cancer. Cancer Discovery , 7(7), 736–749. https://doi.org/10.1158/2159-8290.CD-16-1174
Citing Literature
Number of times cited according to CrossRef: 4
- Emran Askari, Kamran Aryana, Amir Hosein Jafarian, Alireza Bari, Somaye Barashki, Quadruplet Therapy in De Novo High-Volume Mixed Neuroendocrine Prostate Cancer Using 177Lu-PSMA, Clinical Nuclear Medicine, 10.1097/RLU.0000000000005046, 49 , 3, (e120-e122), (2024).
- Naz Chaudary, E. Wiljer, Warren Foltz, Pratibha Thapa, Richard P. Hill, Michael Milosevic, An orthotopic prostate cancer model for new treatment development using syngeneic or patient‐derived tumors, The Prostate, 10.1002/pros.24701, 84 , 9, (823-831), (2024).
- Denisse Mena-Silva, Aline Alfaro, Andrea León, Emanuel Guajardo-Correa, Estefania Elgueta, Patricia Diaz, Cristian Vilos, Hugo Cardenas, Juliano C. Denardin, Pedro A. Orihuela, Zeolite Nanoparticles Loaded with 2-Methoxystradiol as a Novel Drug Delivery System for the Prostate Cancer Therapy, International Journal of Molecular Sciences, 10.3390/ijms241310967, 24 , 13, (10967), (2023).
- Vittoria Rago, Anna Perri, Silvia Di Agostino, New Therapeutic Perspectives in Prostate Cancer: Patient-Derived Organoids and Patient-Derived Xenograft Models in Precision Medicine, Biomedicines, 10.3390/biomedicines11102743, 11 , 10, (2743), (2023).

