Photorepair Fluence Response Protocol
Daniel Ma, NATALIE HULL, David McDonald
Disclaimer
DISCLAIMER – FOR INFORMATIONAL PURPOSES ONLY; USE AT YOUR OWN RISK
The protocol content here is for informational purposes only and does not constitute legal, medical, clinical, or safety advice, or otherwise; content added to protocols.io is not peer reviewed and may not have undergone a formal approval of any kind. Information presented in this protocol should not substitute for independent professional judgment, advice, diagnosis, or treatment. Any action you take or refrain from taking using or relying upon the information presented here is strictly at your own risk. You agree that neither the Company nor any of the authors, contributors, administrators, or anyone else associated with protocols.io, can be held responsible for your use of the information contained in or linked to this protocol or any of our Sites/Apps and Services.
Abstract
Photorepair light exposures are performed on microorganism samples following UV light exposure (i.e. UV disinfection) to determine microorganism regrowth and repair kinetics. Photorepair is a light-dependent, enzyme mediated DNA repair process in bacteria that may reverse UV-induced DNA damage and decrease UV disinfection efficacy. Previously, photorepair kinetics were determined on a time-basis (i.e. exposure times). However, time-based photorepair determinations are not standardized. A photorepair fluence response protocol was developed here according to a standardized protocol proposed by Bohrerova and Linden (2007). If you cite your use of this protocol, you should also cite the relevant original literature.
Before start
For microbial samples, sanitize bench top surfaces with 70% ethanol.
Steps
Introduction and Pre-Experiment
The photorepair fluence response protocol is based on Bohrerova and Linden (2007).
Polychromatic UV Fluence (Dose) Response Determination
Before the Photorepair Experiment
UV Irradiation : Perform the UV fluence response experiment to prepare the UV irradiated samples for the photorepair fluence response experiment. Experiments must be performed in a dark environment with minimal red light for visibility.
After UV irradiation : Immediately remove sample from UV exposure. Keep samples in the dark and covered. Perform sample handling and plating in the dark with only red light for visibility. Take sample for microorganism enumeration and molecular analyses. Dilute samples and plate. Set aside samples for molecular analysis and store at -80°C.
Transfer the desired of the irradiated sample for photorepair and the remaining sample for dark repair into two identical, sterile Petri dishes (60 mm dishes).
Preparing the Photorepair Apparatus
Before performing the photorepair fluence response experiment, set up the photorepair fluence response experiment apparatus. A light source emitting the desired photorepair light (minimum 300-500 nm emission spectrum) should be suspended above a magnetic stirrer and dish containing the sample.
- Warm up the photorepair light source (e.g. 30 minutes before performing exposures).
- Start up spectroradiometer and acquisition software.
- If your spectroradiometer has not been set up to obtain spectral irradiance measurements, set it up, load calibration data (if necessary), and obtain background reading as part of the set up.
- Obtain spectra irradiance measurements at the surface of the water sample (Figure 1).
Use the Photorepair Fluence spreadsheet to determine the photorepair fluence: PhotorepairFluence_Calculation.xlsx. The contents of each sheet are summarized in Table 1.
| A | B |
|---|---|
| Sheet Name | Description |
| Fluence Calculations | Primary sheet where fluence calculations are performed. Values must be entered by user for volume, diameter, water path length, distance from light source to sample, weighting factor, and radiometer reading. |
| Absorbance | UV-VIS spectrophotometer measurements for the microorganism working stock solution is imported into this sheet. |
| Reflection Factor | Pre-calculated reflection factor based on personal communications with Professor Jim Bolton (2016). |
| Weighting | Sheet that accounts for the relative lamp emission of the photorepair lamp, radiometer factor, sample absorbance, Beer Lambert law, photolyase absorbance, and weighting factors to calculate the average intensity. |
| Lamp Spectra | Sheet that depicts the lamp spectra, average wavelength, and peak wavelength. |
| Irradiance Interpolation | Sheet that performs interpolation function in Excel to output spectral irradiance values for integer wavelengths so that values can be used in the Weighting tab. |
| Photolyase Spect Interpolation | Sheet that performs interpolation function in Excel to output photolyase action spectra values for integer wavelengths so that values can be used in the Weighting tab. |
Table 1. Description of contents for the sheets included in PhotorepairFluence_Calculations.xlsx
Data for the photolyase action spectra was digitized from Payne and Sancar (1990):
Optional : To compensate for the change in sample distance from the light source with each subsequent volume withdrawal for sample analysis by calculating the height differential between sampling events, and measuring the light intensity at the center of the dish at the expected level at the start of the experiment to account for the changes in fluence calculation. The change in distance can be calculated using the volume withdrawn and the geometry of the dish (e.g. inner diameter and surface area). The number of measurements will be determined by the number of sampling events that should be pre-determined at the start of the experiment.
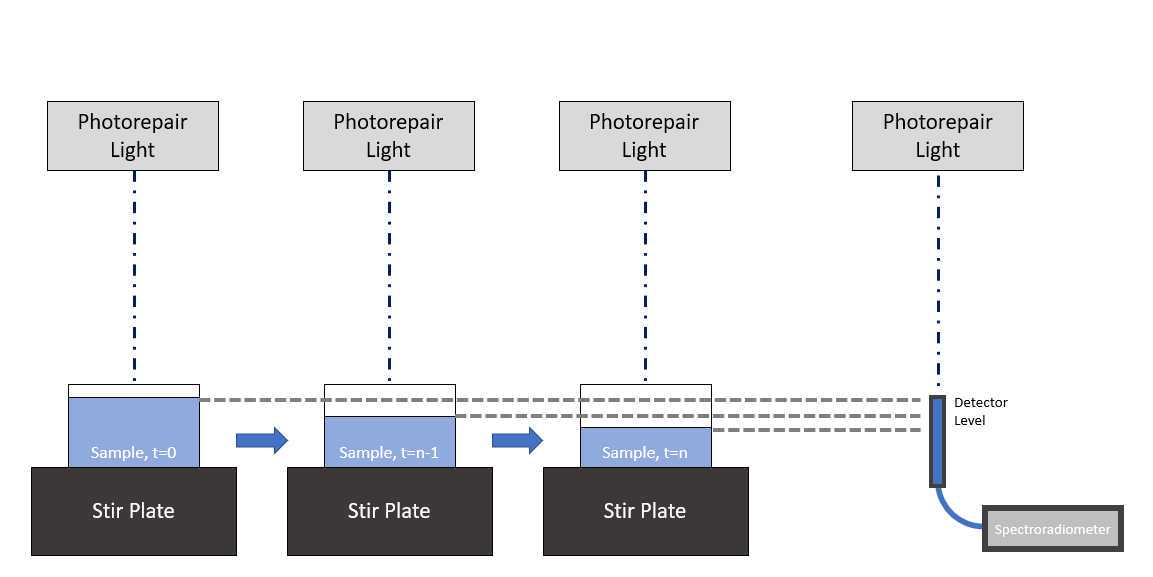
Photorepair Irradiance Example Data : Measurements obtained using the method described in Figure 1 are shown in this example: PhotorepairIrradiance.xlsx . A summary of the contents of this spreadsheet is provided in Table 2 below.
| A | B |
|---|---|
| Sheet Name | Description |
| Plot | Shows a plot of all spectral irradiance measurements obtained at different distances from the light source |
| RawData | Contains all the raw measurements, including background measurements (i.e., dark controls with the cap on the spectroradiometer), and measurements taken at different distances |
| SpecIrr_BkgdCorrected | Spectral irradiance data with background corrections (i.e., background spectra subtracted from light spectra measurements) |
| Tabs named 12 to 12.35 | Each tab has the data for measurements obtained at the various heights (units in cm). The sheets interpolate the measurements to integer wavelength values which can then be used in the Photorepair Fluence Calculation spreadsheet. The total irradiance from 320-475 nm is calculated in cell M2 as the sum of the spectral irradiance values between 320-475 nm. |
Table 2. Description of contents for the sheets included in PhotorepairIrradiance.xlsx
Performing the Photorepair Fluence Response Experiment
After obtaining measurements ( optional : for each height required), export spectral irradiance data.
Photorepair : Set the distance from the sample surface to the light source. Place the sample for photorepair exposure under the photorepair light at the pre-determined distance. Stir the sample continuously with the magnetic stirrer.
Dark Repair : Place the dark repair sample on magnetic stirrer in a dark environment.
Sampling : After each time interval, remove the pre-determined sampling volume from the photorepair and dark repair samples.
Handling : Transfer samples to empty, sterile microcentrifuge tubes.
Dilute and plate organisms immediately : Perform serial dilutions with 1X PBS of samples and plate. Incubate plates under the desired growing conditions for the microorganism. Incubation times will vary per microorganism.
Post-Experiment
Optional if users account for the height adjustment and spectra measurements at different heights : After the experiment, calculate the photorepair fluence for each time interval. Use the irradiance measurements taken at each height for the corresponding time interval to account for the differences in light intensity at different distances from the lamp: PhotorepairFluence_Post-Calculation.xlsx
Perform enumeration of microorganism and calculate concentrations. The reference list below provides some sample equations used in the literature for determining repair kinetics.
Percent (Photo)reactivation
Perform molecular analyses or molecular processing such as DNA extraction or protein extraction and perform further desired downstream molecular quantification methods, such as examples referenced below.
ATP Quantification

