Optimizing Genetic Modification in Agrobacterium rhizogenes K599 through Electroporation Method
Manoj-Kumar Arthikala, Kalpana Nanjareddy, Mariana López-Sámano
Agrobacterium rhizogenes K599
Electrocompetent cells
Transformation Protocol
Transformation efficiency
Disclaimer
DISCLAIMER – FOR INFORMATIONAL PURPOSES ONLY; USE AT YOUR OWN RISK
The protocol content here is for informational purposes only and does not constitute legal, medical, clinical, or safety advice, or otherwise; content added to protocols.io is not peer reviewed and may not have undergone a formal approval of any kind. Information presented in this protocol should not substitute for independent professional judgment, advice, diagnosis, or treatment. Any action you take or refrain from taking using or relying upon the information presented here is strictly at your own risk. You agree that neither the Company nor any of the authors, contributors, administrators, or anyone else associated with protocols.io, can be held responsible for your use of the information contained in or linked to this protocol or any of our Sites/Apps and Services.
Abstract
Electroporation has emerged as a highly effective method for swiftly and proficiently introducing exogenous plasmid DNA into various cell types, particularly those lacking natural competence. This protocol article delineates a streamlined approach for the transformation of Agrobacterium rhizogenes K599 using electroporation. This method, while necessitating purified plasmid DNA, competent bacteria, and standard electroporation equipment such as gene pulse controllers and cuvettes, offers notable advantages in terms of transformation efficiency and speed. The protocol detailed herein not only outlines the procedural steps but also underscores the significance of efficient transformation in the context of A. rhizogenes K599 research. Moreover, it provides insights into the achieved transformation rates, thereby offering researchers a benchmark for assessing the efficacy of this method. By elucidating the equipment requirements and procedural nuances, this protocol aims to empower researchers in adopting electroporation as a reliable tool for genetic manipulation in A. rhizogenes K599, facilitating advancements in various biotechnological applications.
Before start
Prepare all necessary reagents, including LB medium, sterile 10% glycerol, sterile water, and LB Petri dishes with or without antibiotics.
-
Ensure access to a pre-cooled centrifuge (4°C) capable of accommodating 50 ml falcon tubes.
-
Prepare sterile centrifuge tubes (0.2 ml and 1.5 ml Eppendorf tubes) in advance and keep them chilled (4°C).
-
Create a fresh petri dish plate containing the strain.
Steps
Preinoculum preparation
Begin by inoculating 10 ml of LB medium with a single colony of Agrobacterium rhizogenes K599 from a freshly streaked plate. Incubate the culture overnight at 28°C with agitation at 180 rpm, which typically takes approximately 12 hours.
Preparation of Electrocompetent cells
Inoculate 100 ml of LB medium with 1 ml of the preceding pre-culture, maintaining a 1:100 ratio for inoculation. Incubate the culture at 28°C with agitation at 180 rpm until reaching an optical density (OD600) of ~ 0.5-0.6, typically requiring 4-5 hours.
Determine the optical density at the 600nm wavelength.
Chill the cell culture on ice for 15 minutes.On ice
Decant the supernatant, then add 40 ml of sterile cold water to the cell pellet and gently resuspend. Centrifuge the mixture at 3,000 g for 10 minutes at 4°C.
Decant the supernatant, then add 20 ml of sterile cold water to the cell pellet and thoroughly resuspend. Subsequently, centrifuge the suspension at 3,000 g for 10 minutes at 4°C.
Decant the supernatant, then resuspend the cell pellet in 30 ml of sterile 10% glycerol. Centrifuge the suspension at 3,000 g for 10 minutes at 4°C.
Decant the supernatant by decantation, then resuspend the cell pellet in 20 ml of sterile 10% glycerol. Centrifuge the suspension at 3,000 g for 10 minutes at 4°C.
Repeat the wash 3 times
Dispense 1 mL of sterile 10% glycerol into each falcon tube, then gently resuspend the cell pellet in the solution.
Store electrocompetent cells at -80°C for up to one year.
Transformation protocol
Place an eppendorf tube of electrocompetent cells on ice and allow them to chill for 5 minutes.
Add 1 pg to 100 ng of plasmid DNA (1-5 µl) to the cells, gently mix, and incubate on ice for 30 minutes.
Caution: Avoid vortexing the cells during this step.
For a control experiment, prepare another tube of competent cells devoid of plasmid DNA and allow it to rest on ice for 30 minutes.
Immediately transfer the cuvette onto ice and allow it to incubate for 2 minutes.
Apply the same procedure to the control tube of electrocompetent cells.
Incubate the Eppendorf tubes at 28 °C, 180 rpm for 2 hours.
Spread 30 μl of either the 'control' or 'plasmid DNA' transformed cells onto separate LB agar plates, with LB agar supplemented with kanamycin (50 mg/ml) for the latter.
Incubate the plates at 28 °C for two days.

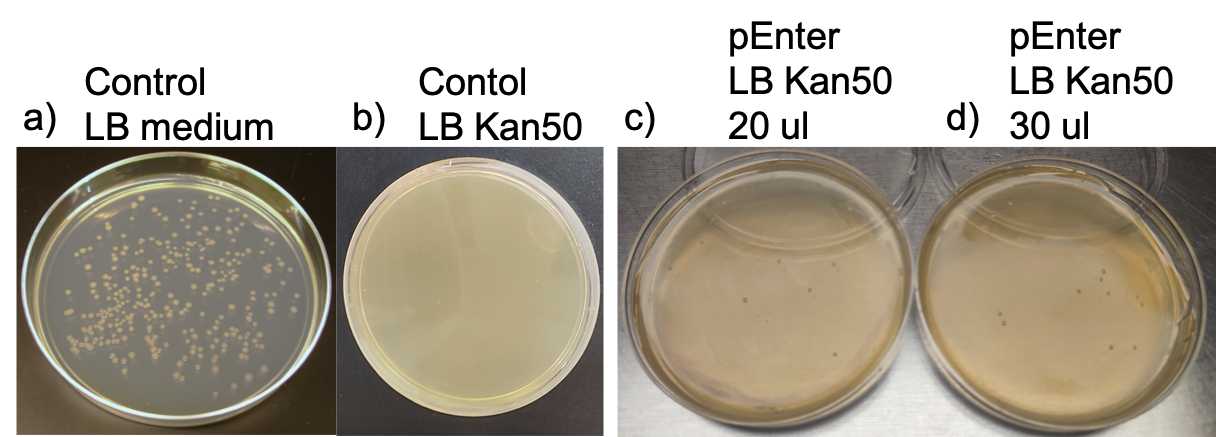
The equation for calculating Transformation Efficiency (TE) is: TE = Colonies/µg/Dilution.
Applications:
The efficient transformation of Agrobacterium rhizogenes K599 using electroporation can open up various applications in biotechnology, agriculture, and research. Here are some potential applications:
Plant Genetic Engineering : A. rhizogenes K599 is widely used in plant genetic engineering due to its ability to induce hairy root formation. Efficient transformation of this bacterium can facilitate the introduction of desired genes into plant genomes, leading to the development of transgenic plants with improved agronomic traits such as disease resistance, herbicide tolerance, and enhanced nutritional content.
Production of Bioactive Compounds : Hairy roots induced by A. rhizogenes K599 have been utilized as efficient platforms for the production of various bioactive compounds such as pharmaceuticals, nutraceuticals, and natural products.
Functional Genomics Studies : The ability to efficiently introduce foreign DNA into A. rhizogenes K599 enables functional genomics studies aimed at understanding gene function and regulatory networks in plants.
Rhizosphere Engineering : A. rhizogenes K599-mediated hairy root formation can influence the rhizosphere environment and plant-microbe interactions.
Bioremediation : Hairy roots produced by A. rhizogenes K599 have been explored for their potential in bioremediation applications.
Functional Expression Studies : A. rhizogenes K599 can serve as a host for the functional expression of heterologous proteins.






