Optimized Workflow for Whole Genome and Transcriptome Next-Generation Sequencing of Single Cells or Limited Nucleic Acid Samples
Ioanna Andreou, Ioanna Andreou, Markus Storbeck, Markus Storbeck, Peter Hahn, Peter Hahn, Samuel Rulli, Samuel Rulli, Eric Lader, Eric Lader
Abstract
Whole genome and whole transcriptome sequencing require orders of magnitude more of starting nucleic acid than what is found in single cells or other extremely limited samples. High fidelity amplification of this minute amount of nucleic acids is essential to overcome the limitations caused by the low input, degradation and contamination, and to ensure a sufficient amount of DNA for preparation of high complex and high quality next-generation sequencing (NGS) libraries. Recent technical advances in multiple displacement amplification (MDA) enable studies of rare cell types, heterogeneity of body fluids, tissues, environmental samples, and organisms that cannot be cultured. Several strategies for amplification of limiting amounts of nucleic acid have been described, with PCR being popular. However, PCR-based methods result in high error rates, lower library complexity, and lower coverage uniformity. In this article, a HiFi MDA is used to accurately amplify the limited material and to allow library preparation starting from high input, while reducing PCR cycling to achieve sufficient library yields. This article describes a complete workflow from cells and small quantities of DNA or RNA to NGS libraries for Illumina sequencing instruments. © 2023 QIAGEN GmbH. Current Protocols published by Wiley Periodicals LLC.
Basic Protocol 1 : Whole genome amplification from single cells
Support Protocol 1 : PicoGreen™ quantification of MDA amplified DNA
Support Protocol 2 : Purification of amplified DNA after MDA
Basic Protocol 2 : Whole transcriptome amplification from single cells
Alternate Protocol : Whole transcriptome amplification from purified RNA
Basic Protocol 3 : Enrichment of complete small genomes using target-specific primers in MDA
Basic Protocol 4 : Complete viral RNA amplification using target-specific primers in MDA
Basic Protocol 5 : Enzymatic fragmentation and adapter ligation of MDA amplified material
Basic Protocol 6 : Normalization of library concentration using magnetic beads
INTRODUCTION
Next-generation sequencing (NGS) capacities have been enormously increased, while sequencing costs have dropped. This allows studies of a high number of samples and even studies of more difficult samples such as single cells, rare cells, and other limited material samples. All of these samples have different characteristics and require optimized conditions to process them and generate high quality libraries. The most important requirement when working with such limited materials is the amplification of all nucleic acids, without the introduction of amplification bias. In this way library generation starts with sufficient input, so that no excessive PCR cycling is necessary to achieve the required library yields for next-generation sequencing. The resulting libraries have a higher complexity due to lower duplication rate, which is relative to the PCR cycles used for library amplification, and result in higher coverage.
This protocol describes, in detail, the methods to generate and normalize libraries suitable for next-generation sequencing from pre-amplified single cells, or other limited samples using whole genome amplification (WGA) and whole transcriptome amplification (WTA).
Basic Protocol 1 starts with efficient cell lysis and denaturation of duplex DNA. Subsequently, whole genomes are amplified in an isothermal reaction using the REPLI-g® multiple displacement amplification (MDA) technology (Dean et al., 2002). During this method repeated annealing of random oligonucleotide primers and HiFi amplification ensure high yields with low amplification bias.
Basic Protocol 2 can start with total RNA, poly A+ mRNA, or single cells. In the first step, cells are lysed and RNA is denatured under conditions that do not impact RNA integrity. Following cell lysis, genomic DNA (gDNA) is removed, to eliminate any sequence contribution caused by gDNA contamination, and allows accurate measurement of transcript levels. In the subsequent Quantiscript reverse transcription (RT), random oligonucleotides or oligo-dT can be used to either process total RNA or mRNA, respectively, and generate long cDNAs. For NGS applications, we recommend using oligo-dT primers to generate cDNA and avoid obtaining reads of ribosomal RNA (rRNA) to efficiently utilize sequencing capacity. Following RT, the synthesized cDNA is ligated using a high-efficiency ligation mix. During this ligation step, cDNA fragments are assembled randomly with each other. These will be then amplified in an isothermal reaction similar to that previously described for WGA, using the HiFi REPLI-g® SensiPhi DNA polymerase to generate enough material for library construction. During this amplification, the ligated cDNAs will be concatemerized, resulting in very long cDNA fragments. The chemistry is designed to process long mRNAs and efficiently ligate long double stranded cDNA fragments, so that the formed cDNA concatamers will be dissolved during library generation and the number of chimeric reads after sequencing will be low. Caution is required when working with fragmented RNA. This will be processed equally well in the WTA protocol but will generate a higher number of chimeric sequencing reads, which will reduce the output of paired end sequencing runs. Statistically the number of undesirable chimeric cDNAs is kept low when starting with high integrity RNA, so that analysis of nucleic acid sequences is not complicated in downstream NGS applications.
For accurate single-cell analysis, complete and uniform coverage is dependent on efficient cell lysis and release of high integrity DNA and RNA. This is one of the most critical steps in these workflows. The lysis conditions for the WTA procedure need to minimize RNase activity to prevent RNA degradation. The described protocols use nuclease free reagents to allow efficient cell lysis and nucleic acid denaturation, while maintaining nucleic acid integrity. When high molecular weight nucleic acid is used as starting material for the REPLI-g based MDA, it typically will yield micrograms of amplified DNA or cDNA in only 2 to 3 hr. In addition, the reagents used for both WGA and WTA are decontaminated from any nucleic acid that will compete with the low input template during REPLI-g amplification.
Basic Protocols 3 and 4 describe the amplification of small genomes such as mitochondrial DNA and viral DNA or RNA in the presence of high gDNA or RNA background. Mitochondrial DNA or viral DNA and RNA are only an exceedingly small portion of the usually purified total nucleic acid sample. Sequencing of the complete sample leads to a high waste of sequencing reads, which belong to host nucleic acids or other contaminating nucleic acids and increase the cost of sequencing. For these samples, enrichment of the corresponding sequences is required. Conventional enrichment strategies using PCR amplification require a high number of PCR primers, which will generate PCR errors and increase the cost of library generation. Alternatively, hybrid capture strategies require a high amount of starting material so that samples of 1 ng and lower will not be sufficient for enrichment using this method. We have modified the MDA chemistry to use a significant small number of target-specific primers to facilitate enrichment of small genomes from limited samples.
Basic Protocol 5 describes the generation of an NGS library using enzymatic fragmentation of amplified DNA or cDNA, and subsequent ligation of sequencing adapters. This method uses reagents provided by the QIAseq single cell DNA or QIAseq single cell RNA library unique dual-index (UDI) kits. During this streamlined protocol, all steps take place in one single tube, by sequential addition of components at each step after completion of each reaction. Finally, libraries can be individually qualified and quantified for NGS on the Illumina platforms. Alternatively, the libraries can be normalized using QIAseq normalizer kits, a workflow described in Basic Protocol 6.
Basic Protocol 6 describes the normalization of the generated NGS libraries using magnetic beads, provided by the QIAseq library normalizer kits, which use modified magnetic beads to bind and release a specific amount of library in a strictly controlled mode. This procedure eliminates the need of time-consuming and material-consuming library quantification. Finally, all normalized libraries can be pooled and qualified on electrophoretic devices as a pool.
Summarizing the above in this article we provide protocols for whole genome amplification (Basic Protocol 1) and whole transcriptome amplification from single cells or purified nucleic acid (Basic Protocol 2 and Alternate Protocol) using MDA, altogether with support protocols (Support Protocols 1 and 2) for quantification and purification of the amplified DNA. Furthermore, protocols for enrichment of small DNA or RNA genomes using target-specific primers are described (Basic Protocols 3 and 4). The workflows are completed by protocols for library preparation (Basic Protocol 5) and normalization (Basic Protocol 6).
CAUTION : When working with chemicals, always wear a suitable lab coat, disposable gloves, and protective goggles. For more information, consult the appropriate safety data sheets (SDSs) of the material used.
CAUTION : When working with single cell and other low concentrated samples, always use low binding tubes and avoid prolonged storage and vigorous vortexing of the sample because this will lead to material loss. Maintain a clean, nucleic acid-free and nuclease-free environment and use freshly opened pipet tips and tubes.
Basic Protocol 1: WHOLE GENOME AMPLIFICATION FROM SINGLE CELLS
The protocol describes the QIAseq single cell procedure for WGA from single cells, or very small quantities of purified genomic DNA using MDA. The reaction is facilitated by an optimized phi29 polymerase together with an optimized buffer composition. All reagents used in the MDA reaction are provided with the QIAseq single cell DNA library kit and are specially treated to eliminate contaminating DNA, which is critical to achieve reliable results in single cell or ultra-low input applications.
This protocol is optimized for single cells without a cell wall from eukaryotic species, for example, mammalian sorted cells or plant protoplasts, micro dissected cells from frozen sections, and tissue cultured cells, as well as for nuclei and gram-positive bacteria. Cells that are treated with formalin or other cross-linking agents cannot be amplified by this method.
An input of 1 to 1,000 intact cells (e.g., human or bacterial cells) or purified DNA >6 pg is optimal for WGA reactions. Caution is required when working with purified eukaryotic DNA; in order to achieve complete genome coverage input, >1 ng is required to compensate DNA losses during the purification. Before starting with the WGA, carefully decontaminate the workbench with nucleic acid and nuclease eliminating agents. To avoid any DNA contamination of reagents, use separate laboratory equipment (e.g., pipets, filter pipet tips, reaction vials). It is recommended that the WGA reaction be set up in a location that minimizes the possibility of exogenous DNA contamination. Figure 1 illustrates the workflow of the whole genome amplification. This starts with the cell lysis and DNA denaturation under mild alkaline conditions, followed by neutralization and by isothermal multiple displacement amplification using Phi29 polymerase. All these steps take place in one single tube by sequential addition of the corresponding reagents.
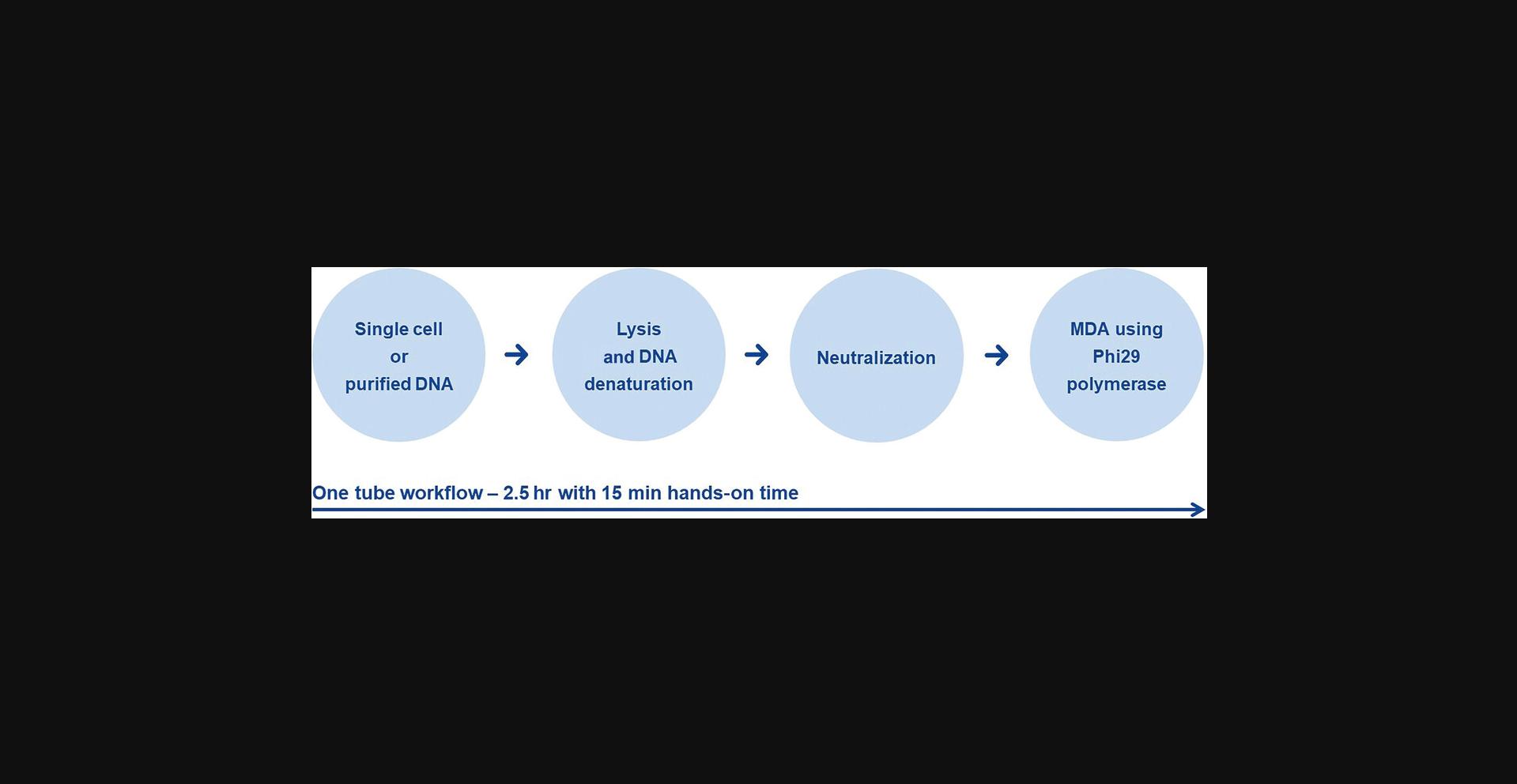
NOTE : This protocol may also be used with purified DNA from enriched circulating tumor cells (CTCs) using AdnaTest (QIAGEN, cat. no. 395092), followed by nucleic acid purification using AllPrep DNA/mRNA Nano kit (QIAGEN, cat. no. 80272) and the protocol Simultaneous Purification of Genomic DNA and mRNA from Low-Biomass Samples in the AllPrep DNA/mRNA Nano kit Handbook.
Materials
-
QIAseq Single Cell DNA Library Kit Unique Dual-Index (UDI; QIAGEN, cat. no. 181703, 181705, or 181707):
- PBS single cell (sc), 1× (PBS buffer specially treated to eliminate any DNA contamination)
- Buffer DLB
- DTT, 1 M
- Stop Solution
- H2O sc
- REPLI-g sc DNA Polymerase
- REPLI-g sc Dilution Buffer
- REPLI-g sc Advanced Oligo for human cells or DNA or REPLI-g sc Universal Oligo for all species
- QIAseq Human Control DNA
-
Isolated cells or purified DNA in <4 µl PBS sc
-
Vortexer
-
Microcentrifuge tubes, PCR strips, or plates
-
Thermal cycler
-
Microcentrifuge
-
Pipets: 1-10 µl, 20-200 µl, and 100-1,000 µl and pipet tips
1.Vortex all buffers and reagents except the enzyme REPLI-g sc polymerase before use to ensure thorough mixing.
2.Thaw H2O sc, DTT, and REPLI-g sc dilution buffer at room temperature, vortex, and then centrifuge briefly. The REPLI-g sc Reaction Buffer may form a precipitate after thawing; dissolve precipitate by vortexing 10 s.
3.Set thermal cycler to 30°C and enter the program listed in Table 1.If a thermal cycler with a heated lid is used, the temperature of the lid should be set to 70°C.
| Time | Temperature | Comment |
|---|---|---|
| 2 hr | 30°C | Multiple displacement amplification |
| 3 min | 65°C | Enzyme inactivation |
| Hold | 4°C | Cooling of amplified material |
4.Prepare Buffer DLB by adding 500 μl H2O sc to the DLB tube provided. Mix thoroughly and centrifuge briefly to dissolve.
5.Prepare sufficient Buffer D2 (denaturation buffer; Table 2) and DTT dilution (Table 3) for the total number of whole genome amplification reactions.
| Component | Volume for 12 reactions (plus 10%) |
|---|---|
| DTT, 1M | 3.3 µl |
| Buffer DLB | 36.3 µl |
| Component | Volume for 12 reactions (plus 10%) |
|---|---|
| DTT, 1M | 3.3 µl |
| H2O | 29.7 µl |
6.Place 4 µl cell material or purified DNA (in PBS) into a microcentrifuge tube. If using <4 µl of cell material or DNA, add PBS sc to bring the volume to 4 µl.
7.Add 3 µl Buffer D2.Mix carefully by gently flicking the tube and centrifuge briefly.
8.Place tube in the thermal cycler and run protocol “cell lysis & DNA denaturation” or “denaturation of purified DNA” (Table 4) depending on the starting material.
| Cell lysis and DNA denaturation | Denaturation of purified DNA | ||
|---|---|---|---|
| Time | Temperature | Time | Temperature |
| 10 min | 65°C | 3 min | 25°C |
| 1 min | 4°C | 1 min | 4°C |
9.After cell lysis and DNA denaturation is complete, add 3 µl Stop Solution. Mix carefully by flicking the tube and centrifuge briefly. Immediately place on ice.
10.Thaw REPLI-g sc DNA Polymerase on ice. Thaw all other components at room temperature, vortex, and then centrifuge briefly. The REPLI-g sc Dilution Buffer may form a precipitate after thawing. The precipitate will dissolve by vortexing for ∼10 s.
11.Prepare a master mix by adding components in the order listed in Table 5.Then mix and centrifuge briefly. Briefly vortex and centrifuge the mixture of buffer, H2O sc, DTT, and oligos before adding REPLI-g sc DNA polymerase. The master mix should be kept on ice and used immediately upon addition of REPLI-g sc DNA polymerase.
| Component | Volume per reaction | Volume for 12 reactions (plus 10%) |
|---|---|---|
| H2O sc | 6.5 µl | 85.8 µl |
| REPLI-g sc Dilution Buffer | 14.5 µl | 191.4 µl |
| REPLI-g sc universal oligo or REPLI-g sc advanced oligo | 14.5 µl | 191.4 µl |
| DTT (1:10) | 2.5 µl | 33 µl |
| REPLI-g sc DNA polymerase | 2 µl | 26.4 µl |
12.For each reaction, add 40 µl master mix to 10 µl denatured DNA (from step 10).
13.Place tube in the thermal cycler and run the protocol “Cycling conditions of MDA” listed in Table 1.
Support Protocol 1: PicoGreen™ QUANTIFICATION OF MDA AMPLIFIED DNA
This protocol is designed for quantification of double stranded REPLI-g amplified DNA using PicoGreen reagent. PicoGreen quantification is recommended if a high number of samples must be quantified at once. For single probes, use the Qubit dsDNA BR assay kits.
NOTE : Alternatively, Qubit quantification can also be performed according to manufacturer's protocol using Qubit dsDNA HS and BR assay kits depending on the expected concentration range (Thermo Fisher Scientific, cat. no. Q33230, Q32850, or Q33265).
CAUTION : Always wear a suitable lab coat, disposable gloves, and protective goggles when working with hazardous chemicals, and consult the appropriate material SDSs, available from the product supplier, before starting with the protocol.
Materials
-
Quant-iT™ PicoGreen™ dsDNA reagent (Invitrogen; Thermo Fisher Scientific, cat. no. P7581)
-
TE buffer (Thermo Fisher Scientific, cat. no. 12090015)
-
Human genomic DNA (e.g., Promega, cat. no. G3041)
-
MDA amplified DNA (from Basic Protocol 1)
-
2-ml microcentrifuge tube
-
96-well plates (suitable for use in a fluorescence microplate reader, e.g., Costar no. 3915 96-Well Black Polystyrene Plate)
-
Fluorescence microplate reader (e.g., TECAN® Ultra)
1.Make a 1/150 dilution of PicoGreen stock solution in TE buffer, in a 2-ml microcentrifuge tube. Each quantification reaction requires 20 µl. Cover the microcentrifuge tube in aluminum foil or place it in the dark to avoid photodegradation of the PicoGreen reagent. Prepare the PicoGreen/TE solution in a plastic container as the PicoGreen reagent may adsorb onto glass surfaces.
2.Prepare a 16 µg/ml stock solution of genomic DNA in TE buffer.
3.Make 200 µl of 1.6, 0.8, 0.4, 0.2, and 0.1 µg/ml DNA standards by further diluting the 16 µg/ml genomic DNA with TE buffer.
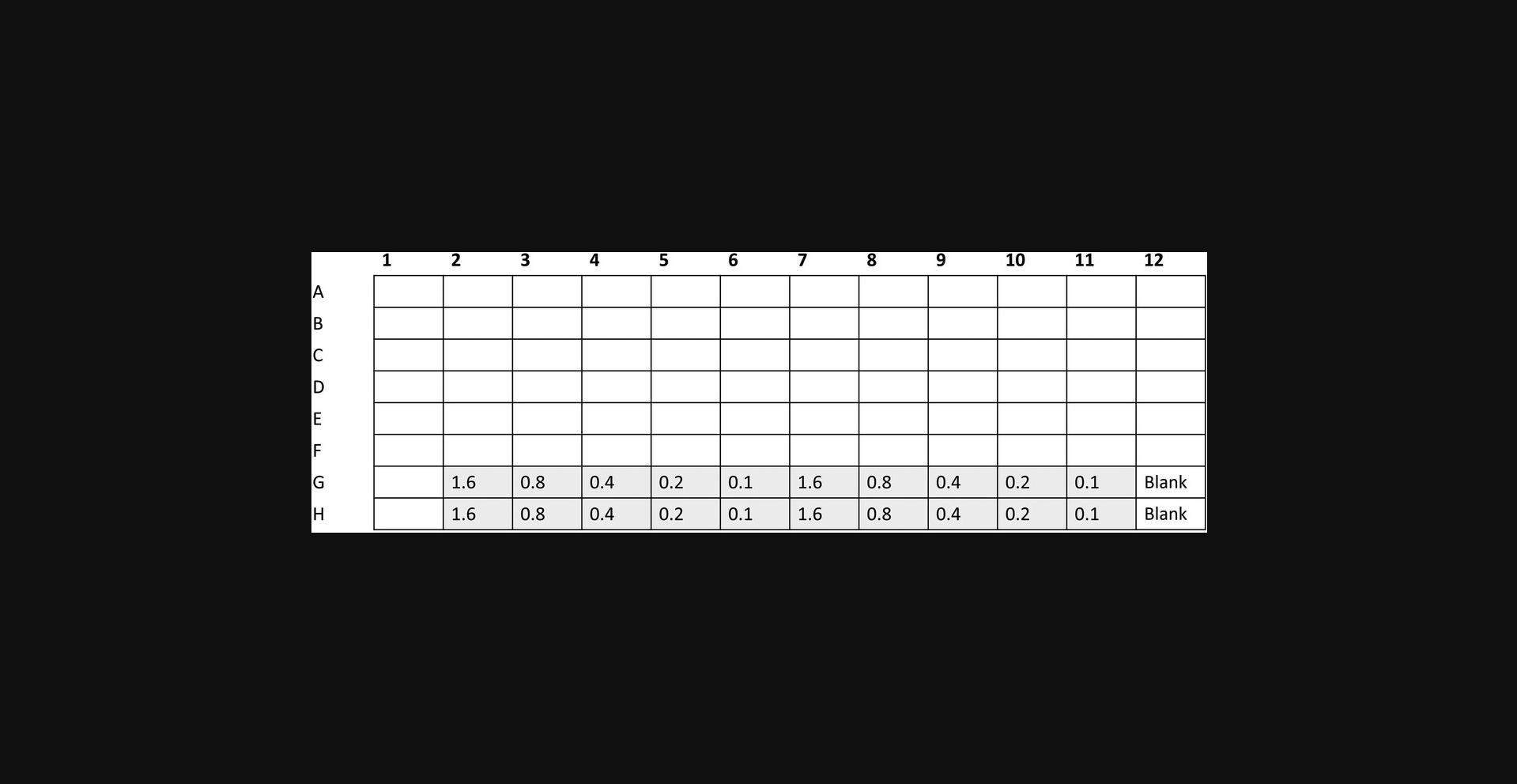
4.Transfer 20 µl of each DNA standard in duplicate into a 96-well plate labeled A as indicated in Figure 2.
5.Place 2 µl of each MDA amplified DNA sample (Basic Protocol 1, step 14) for quantification into a new 96-well plate and add 198 µl TE buffer to make a 1/100 dilution. Store the remaining MDA amplified DNA at –20°C. The 1/100 dilutions can also be stored at –20°C and used for future downstream sample analysis.
6.Place 2 µl diluted MDA DNA (from step 5) into an unused well of 96-well plate A and add 18 µl TE to make a 1/1000 dilution.
7.Add 20 µl PicoGreen working solution (from step 1) to each sample (amplified DNA and DNA standards) in 96-well plate A. Gently shake the plate on the bench top to mix the samples and reagent.
8.Centrifuge the 96-well plate 10 s to collect residual liquid from the walls of the wells.
9.Measure sample fluorescence using a fluorescence microplate reader and standard fluorescence filters (excitation 480 nm; emission ∼520 nm).
10.To calculate DNA concentration, generate a standard curve by plotting the concentration of DNA standards (µg/ml; x axis) against the fluorescence reading generated by the microplate reader (y axis). Plot an average of the fluorescence recorded for each DNA standard of the same concentration.
11.Use the standard curve to determine the concentration (µg/ml) of the diluted REPLI-g amplified DNA sample. This is achieved by plotting the fluorescence reading of the sample against the standard curve and reading the DNA concentration on the x axis.
12.The calculation of DNA concentration depends on the standard curve and the determination of the slope. For accurate results, the standard curve should be a straight line. Any deviation from this may cause inaccuracies in the measurement of REPLI-g amplified DNA concentrations.
13.Multiply the value determined in step 11 by 1,000 to show the concentration of undiluted sample DNA (because the sample DNA measured by PicoGreen fluorescence had been diluted 1 in 1000).
14.To determine the total amount of DNA in the sample, multiply the concentration of undiluted sample DNA (µg/ml; determined in step 13) by the reaction volume in milliliters (i.e., for a 50-µl reaction, multiply by 0.05).
Support Protocol 2: PURIFICATION OF AMPLIFIED DNA AFTER MDA
This protocol describes the procedure of cleanup and concentration of the MDA amplified DNA or cDNA using QIAseq beads. The input and elution volume may vary to meet the required concentration level.
Materials
-
MDA amplified DNA (from Basic Protocol 1)
-
QIAseq Single Cell DNA Library Kit UDI (QIAGEN, cat. no. 181703, 181705, or 181707):
- QIAseq Beads
- H2O sc
-
80% ethanol
-
Magnetic rack for 2-ml microcentrifuge tubes or PCR plates (Thermo Fisher Scientific)
-
DynaMag™-2 Magnet (Thermo Fisher Scientific, cat. no. 12321D) or DynaMag™-96 Magnet (Thermo Fisher Scientific, cat. no. 12027)
1.Dilute MDA amplified nucleic acid 1:2 with H2O sc.
2.Add 1× resuspended QIAseq beads slurry, e.g., 50 µl QIAseq beads and 50 µl diluted WGA sample, and mix well by pipetting.
3.Incubate mixture 5 min at room temperature to bind the DNA on the beads.
4.Immobilize beads on a magnetic stand for 2-5 min and carefully discard supernatant.
5.Wash beads by adding 200 µl fresh 80% ethanol to each pellet. Pellet beads on the magnetic stand for 2-5 min and then carefully discard supernatant.
6.Wash beads for a second time by repeating the wash step 5 once.
7.Incubate on the magnetic stand 5-10 min or until the beads are dry. Avoid over drying, which may result in lower DNA recovery. Remove from the magnetic stand.
8.Elute by resuspending in 20 µl H2O sc. Pellet beads on the magnetic stand. Carefully transfer 17 µl supernatant to a new PCR plate. Alternatively, store purified DNA at –20°C until further processing.
Basic Protocol 2: WHOLE TRANSCRIPTOME AMPLIFICATION FROM SINGLE CELLS
This protocol describes the amplification of polyadenylated mRNA starting from single cells. Figure 3 illustrates the steps involved in the whole transcriptome amplification. These are cell lysis, RNA denaturation, genomic DNA removal, followed by reverse transcription of mRNA or total RNA, cDNA ligation, and finally isothermal multiple displacement amplification of the ligated cDNA. The procedure results in sufficient yields of amplified cDNA and is suited for multiple applications, such as next-generation sequencing (RNA-Seq), real-time PCR, and microarray analysis. In a single reaction it can process 1 to 1,000 cells from all vertebrate species (e.g., human, mouse, rat, micromanipulated and sorted cells, or tissue culture cells). The protocol cannot be used for any cells with a cell wall, e.g., bacterial cells or plant cells. The protocol cannot process fixed cells that are treated with formalin or other cross-linking agents. Avoid any DNA or RNA contamination of reagents by using separate laboratory equipment (e.g., pipets, filter pipet tips, reaction vials) and set up the reaction in a location that minimizes the possibility of exogenous DNA contamination.
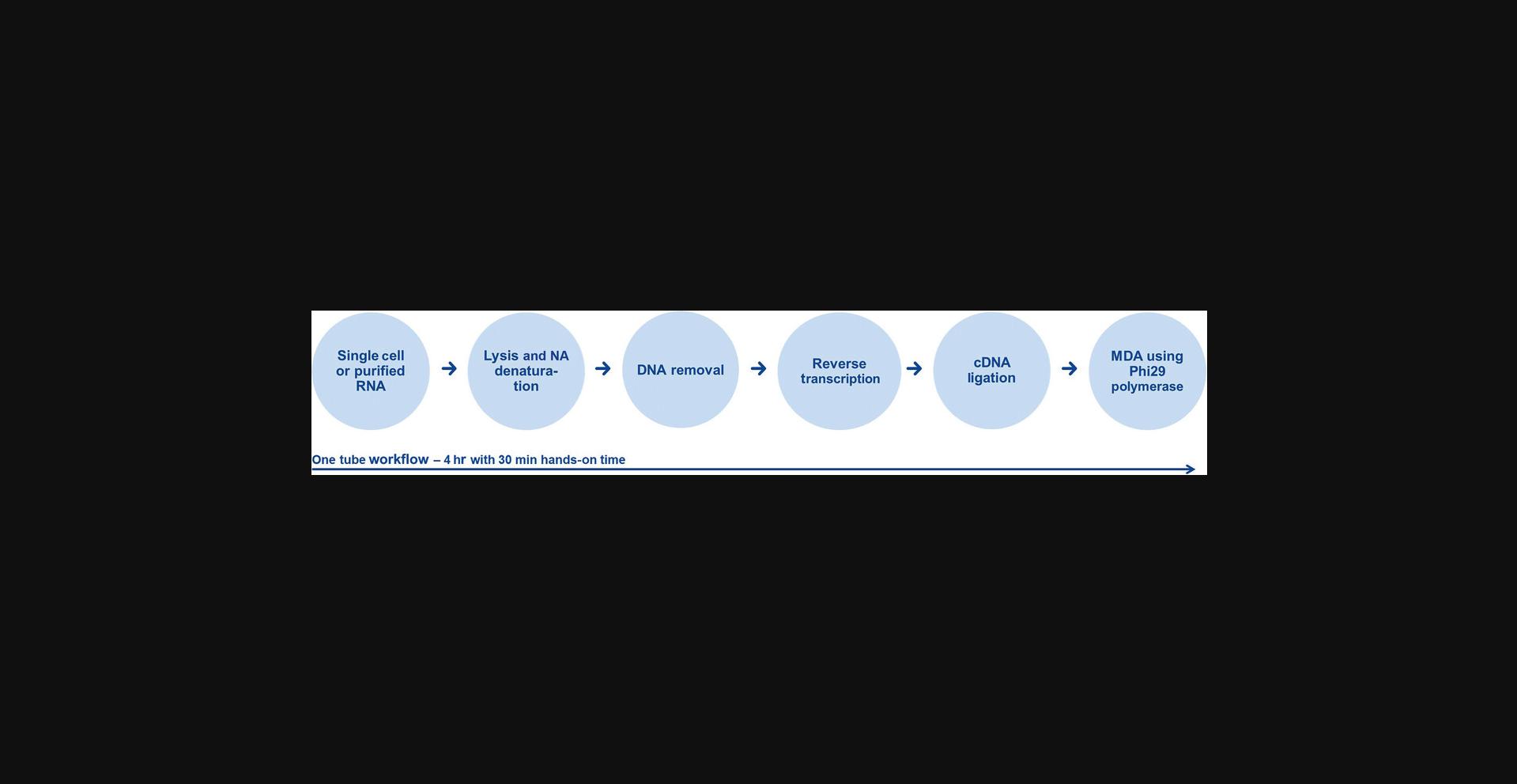
The presented procedure of isothermal amplification works optimally with long fragments. To increase the efficiency of this amplification, the generated cDNAs are ligated to each other prior to MDA. Very short RNA molecules, such as tRNAs or micro RNAs (miRNAs), cannot be amplified using this WTA.
Materials
-
QIAseq Single Cell RNA Library Kit UDI (QIAGEN, cat. no. 180703, 1801705, or 180707):
- Lysis buffer
- gDNA Wipeout buffer, WTA
- RT/Polymerase buffer
- Random primer
- Oligo dT primer
- Quantiscript® RT enzyme mix
- Ligase mix
- Ligase buffer
- REPLI-g sc dilution buffer
- REPLI-g sc advanced oligo for human cells or RNA or REPLI-g sc universal oligo
- REPLI-g SensiPhi® DNA polymerase (yellow lid)
-
H2O sc
-
Isolated cells in <7 µl PBS sc (PBS single cell)
-
Microcentrifuge tubes or PCR strips or plates
-
Thermal cycler
-
Microcentrifuge
-
Vortexer
-
Pipets: 1-10 µl, 20-200 µl, and 100-1,000 µl and pipet tips
NOTE : When working with such limited material, it is important to create and maintain an RNase-free environment by following proper microbiological and aseptic techniques. Any RNase contamination will lead to loss of material and insufficient, non-uniform amplification. The use of disposable plastic tubes and pipet tips from freshly opened boxes or bags is strongly recommended. Forty units of Recombinant RNasin® ribonuclease inhibitor (Promega, cat. no. N251A; 40 u/μl) may be added in the step of DNA wipe out. RNasin® ribonuclease inhibitor cannot be added during lysis because the used conditions will denature the enzyme. It is not recommended that RNasin® ribonuclease inhibitor is repeatedly added into the next reaction steps because this will alter the buffer composition and will inhibit the subsequent reactions.
1.Thaw all buffers at room temperature (15°C to 25°C) and vortex before use to ensure thorough mixing, spin down, and place on ice.
2.Thaw all enzymes and enzyme mixes (Quantiscript RT enzyme mix, ligase mix, and REPLI-g SensiPhi DNA polymerase) on ice. Mix by gently flicking the tube. Avoid any vigorous vortexing of enzymes and enzyme mixes because this may affect their activity.
3.Program a thermal cycler (see Table 6) for all incubation steps of the protocol.
| Step | Time | Temperature |
|---|---|---|
| Cell lysis |
5 min 3 min Hold |
24°C 95°C 4°C |
| gDNA removal |
10 min Hold |
42°C 4°C |
| Reverse transcription |
60 min 3 min Hold |
42°C 95°C 4°C |
| Ligation |
30 min 5 min Hold |
24°C 95°C 4°C |
| Whole transcriptome amplification |
2 hr 5 min Hold |
30°C 65°C 4°C |
- a
Set the heating lid to 50°C for all steps.
4.Place 7 μl cell material (in PBS) into a microcentrifuge tube. If using <7 μl of cell material, add H2O sc to bring the volume up to 7 μl and proceed immediately with step 5.
5.Add 4 µl lysis buffer. Mix carefully by gently flicking the tube and centrifuge briefly.
6.Incubate at 24°C for 5 min followed by 95°C for 3 min. Cool to 4°C.
7.Add 2 µl gDNA wipeout buffer, mix by vortexing, and centrifuge briefly.
8.Incubate at 42°C for 10 min. If more time is needed to prepare the next step, place on ice.
9.Prepare Quantiscript RT mix as outlined in Table 7.
| Component | Volume per reaction | Volume for 12 reactions (plus 10%) |
|---|---|---|
| RT/polymerase buffer | 4 µl | 52.8 µl |
| Random primer | 1 µl | 13.2 µl |
| Oligo-dT primer | 1 µl | 13.2 µl |
| Quantiscript RT enzyme mix | 1 µl | 13.2 µl |
10.Add 6 µl Quantiscript RT mix to the sample from step 8, mix by vortexing, and centrifuge briefly.
11.Incubate at 42°C for 60 min to allow reverse transcription of RNA. Stop reaction by incubating at 95°C for 3 min, then cool on ice.
12.Prepare ligation mix by adding the components in the order shown in Table 8.
| Component | Volume per reaction | Volume for 12 reactions (plus 10%) |
|---|---|---|
| Ligase buffer | 8 µl | 105.6 µl |
| Ligase mix | 2 µl | 26.4 µl |
13.Add 10 µl ligation mix to the RT reaction from step 11.Mix by vortexing and centrifuge briefly.
14.Incubate at 24°C for 30 min. Stop reaction by incubating at 95°C for 5 min, then cool on ice.
15.Prepare REPLI-g SensiPhi amplification mix (see Table 9). Mix by vortexing and centrifuge briefly.
| Component | Volume per reaction | Volume for 12 reactions (plus 10%) |
|---|---|---|
| REPLI-g sc dilution buffer | 14.5 µl | 191.4 µl |
| REPLI-g sc universal oligo or REPLI-g sc advanced oligo | 14.5 µl | 191.4 µl |
| REPLI-g SensiPhi DNA polymerase | 1 µl | 13.2 µl |
16.Add 30 µl REPLI-g SensiPhi amplification mix to the ligation reaction from step 14.
17.Incubate at 30°C for 2 hr. Stop reaction by incubating at 65°C for 5 min, then cool on ice.
Alternate Protocol: WHOLE TRANSCRIPTOME AMPLIFICATION FROM PURIFIED RNA
This protocol is for amplification of polyadenylated mRNA using purified total RNA or mRNA. This protocol is identical with the Basic Protocol 2 with the deviation of RNA denaturation and neutralization because cell lysis is obsolete for this workflow.
Also, quality control of mRNA integrity prior to amplification using capillary electrophoretic devices is highly recommended.
rRNA depletion will eliminate the rRNA contamination, which is more prominent when working with purified total RNA, and will increase the enrichment of other important biotypes to study transcriptomics such as mRNA and large non-coding RNA (lncRNA).
NOTE : This protocol may also be used with purified RNA from CTCs, enriched using AdnaTest (QIAGEN, cat. No. 395092), followed by nucleic acid purification using AllPrep DNA/mRNA Nano kit (QIAGEN, cat. no. 80272) and the protocol Simultaneous Purification of Genomic DNA and mRNA from Low-Biomass Samples in the AllPrep DNA/mRNA Nano kit Handbook, using the option for mRNA isolation.
Additional Materials (also see Basic Protocol 2)
- Purified total RNA or mRNA, >500 pg
- QIAseq FastSelect −rRNA HMR Kit, for 8 samples (QIAGEN, cat. no. 334385, 334386, 334387, 334388)
- Additional material provided by the QIAseq Single Cell RNA Library UDI Kit (QIAGEN, cat. no. 180703)
NOTE : It is important to create and maintain a nucleic acid and RNase-free environment by following proper microbiological and aseptic techniques. RNase contamination will lead to reduced MDA yields, as well as low number of detected genes and a high number of chimeric reads after sequencing. Nucleic acid contamination will result in non-mapped reads and inaccurate transcript counts. The use of disposable plastic tubes and pipet tips from freshly opened boxes or bags is strongly recommended.
1.Pipet 8 μl purified RNA (>500 pg) into a microcentrifuge tube. If using <8 μl of purified RNA, add H2O sc to make up to 8 μl total volume.
2.Add 3 μl NA denaturation buffer, mix by vortexing, and centrifuge briefly.
3.Incubate at 95°C for 3 min; then cool to 4°C.
4.Add 2 µl gDNA wipeout buffer, mix by vortexing, and centrifuge briefly.
5.Incubate at 42°C for 10 min to degrade gDNA. Place on ice.
6.Add 1 μl QIAseq FastSelect −rRNA HMR to 4 μl RT polymerase buffer.
Mix and vortex.
| Component | Volume per reaction | Volume for 12 reactions (plus 10%) |
|---|---|---|
| RT polymerase buffer | 4 µl | 48 µl |
| QIAseq FastSelect −rRNA HMR | 1 µl | 12 µl |
7.Add 5 μl master mix to the RNA sample from step 5 and incubate as described in Table 11 to allow blocking of ribosomal RNA.
| Input RNA | Step | Cycling conditions |
|---|---|---|
| Purified RNA | 1 | 2 min at 75°C |
| 2 | 2 min at 70°C | |
| 3 | 2 min at 65°C | |
| 4 | 2 min at 60°C | |
| 5 | 2 min at 55°C | |
| 6 | 2 min at 37°C | |
| 7 | 2 min at 25°C | |
| 8 | Hold at 4°C |
8.Add 1 μl of each Primer mix (oligo-dT and random primer) and 1 μl Quantiscript RT enzyme mix to the 18 μl RNA sample from the previous step. Proceed with RT reaction and incubation step as described in the Basic Protocol 2, step 11.
Basic Protocol 3: ENRICHMENT OF COMPLETE SMALL GENOMES USING TARGET-SPECIFIC PRIMERS IN MDA
This protocol is for enrichment of complete small genomes, e.g., mitochondrial genome or viral DNA in a high genomic DNA background. The protocol replaces the random oligonucleotides by a few target-specific primers in the MDA amplification to specifically enrich viral DNA and other small genomes in the presence of a high host DNA background. This protocol allows enrichment of mitochondrial genome or viral DNA starting from 105 copies out of 500 pg to 10 ng of purified genomic eukaryotic DNA template.
Specific primers for the enrichment of small genomes can be designed using open-source online primer design tools, e.g., Primer3 (https://bioinfo.ut.ee/primer3-0.4.0/) or Primer-BLAST (https://www.ncbi.nlm.nih.gov/tools/primer-blast/). Primer-Blast includes the additional feature of specificity checking. With this option enabled, the program will align the primers against a selected database. Based on their matches to putative non-specific targets and their orientations, it will determine whether a primer pair can generate an amplification product on any unwanted targets in this database. The program will return a collection of primers that do not generate a valid PCR product on unintended sequences and are therefore specific to the intended template.
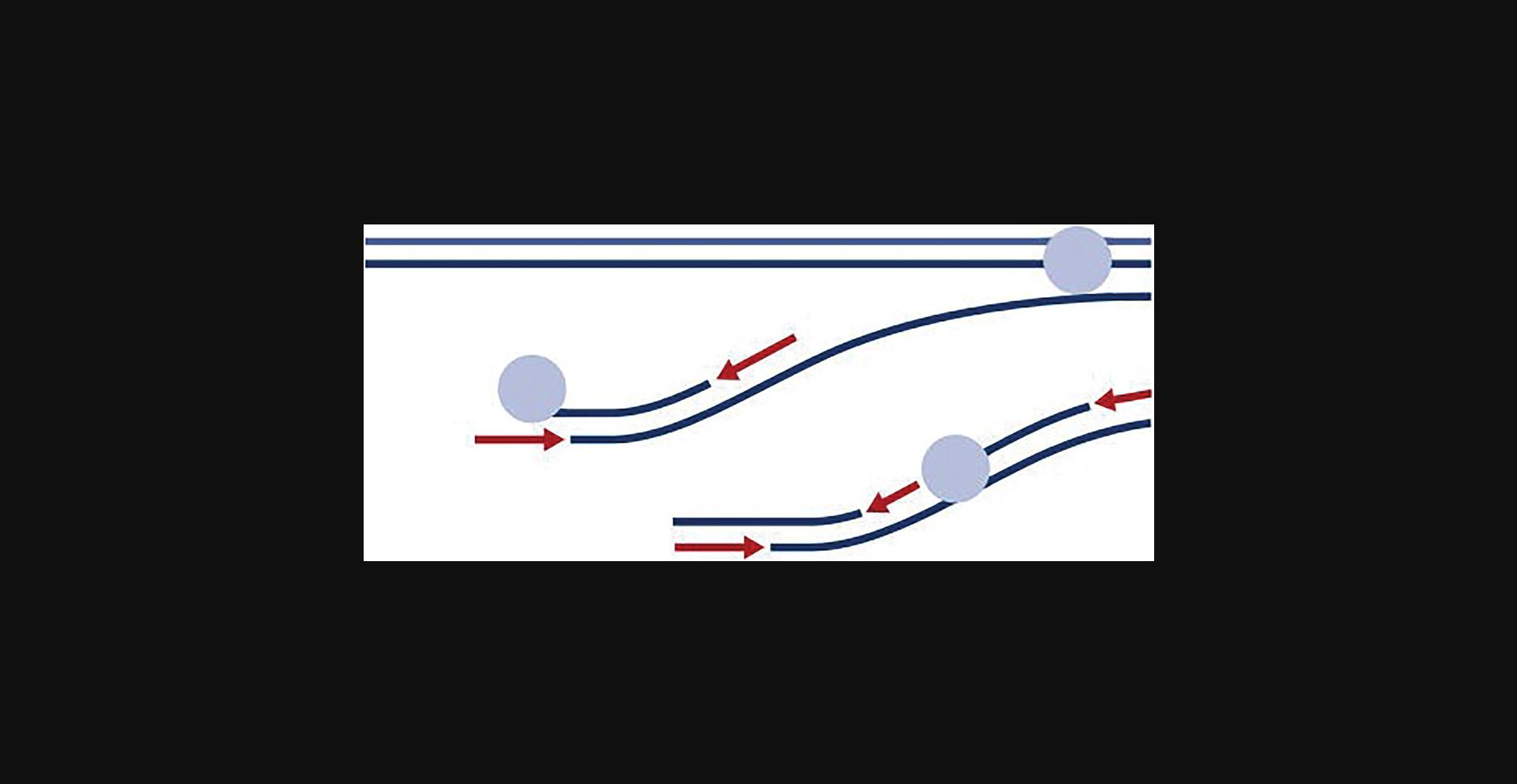
For design parameters, it is recommended that T m of primers is set to 34°C to 40°C and length to between 17 and 21 bp. Amplicon length should be 300 to 4,000 bp. We recommend choosing alternating primers on both strands every 2,000 to 3,000 bp to capture the total length of the genome intended to be amplified as shown in Figure 4.

Primers should be stabilized on the 3′-primer end with phosphorothioate modifications on the last two 3′ bases as shown in the example 5′-NNNNNNNNNNNN-3′. This will protect degradation of the primer during the MDA. The primer mix should include primers at a concentration of 10 µM each for the MDA reaction.
Materials
-
Purified DNA
-
QIAseq Single Cell DNA Library Kit UDI (QIAGEN, cat. no. 181703, 181705, or 181707):
- REPLI-g sc DNA Polymerase
- REPLI-g sc Dilution Buffer
- Buffer DLB
- Stop Solution
- PBS sc, 1×
- DTT, 1 M
- H2O sc
- QIAseq Human Control DNA
-
Target-specific oligonucleotide mix, 10 µM each
-
Microcentrifuge tubes or PCR strips or plates
-
Thermal cycler
-
Microcentrifuge
-
Vortexer
-
Pipets: 1-10 µl, 20-200 µl, and 100-1000 µl and pipet tips
1.Thaw H2O sc and REPLI-g sc dilution buffer at room temperature, vortex, and then centrifuge briefly.
2.Prepare sufficient Buffer D1 (denaturation buffer) for the total number of whole genome amplification reactions (Table 12).
| Component | Volume for 12 reactions (plus 10%) |
|---|---|
| DTT, 1M | 7.3 µl |
| Buffer DLB (reconstituted in 500 µl H2O sc) | 25.7 µl |
3.Prepare sufficient Buffer N1 (neutralization buffer) for the total number of whole genome amplification reactions (Table 13).
| Component | Volume for 12 reactions (plus 10%) |
|---|---|
| Stop solution | 9.9 µl |
| H2O sc | 56.1 µl |
4.Place 2.5 µl DNA into each well of a 96-well plate or microcentrifuge tube.
5.Add 2.5 µl Buffer D1.Mix carefully by gently flicking the tube and centrifuge briefly.
6.Incubate 3 min at room temperature (25°C).
7.Add 5 µl Stop Solution. Mix carefully by flicking the tube, then centrifuge briefly and place immediately on ice. Proceed immediately by adding the master mix.
8.Prepare a master mix according to Table 14.Mix and centrifuge briefly. Scale up accordingly if performing several reactions at once by preparing a master mix sufficient for the total number of reactions.
| Component | Volume per reaction | Volume for 12 reactions (plus 10%) |
|---|---|---|
| H2O sc | 7.5 µl | 99 µl |
| REPLI-g sc dilution buffer | 29 µl | 382.8 µl |
| Target-specific oligonucleotide mix (10 µM each) | 1.5 µl | 19.8 µl |
9.Add 38 µl master mix to the 10 µl DNA from step 7.Mix by flicking the tube and spin down to collect liquid at the bottom of the tube. Place in a cycler or preheated heating block and incubate at 75°C for 5 min, then cool down to room temperature for 5 min to allow annealing of the primers on the template DNA.
10.Thaw REPLI-g sc DNA polymerase on ice. Tip the tube gently to mix and centrifuge briefly.
11.For each amplification reaction, add 2 µl REPLI-g sc DNA polymerase to the 48 µl DNA mix (from step 9). Mix by flicking the tube and then centrifuge briefly.
12.Incubate at 33°C for 8 hr.
13.Inactivate REPLI-g sc DNA polymerase at 65°C for 3 min.
14.If the amplified DNA will not be used immediately, store it either at 4°C (for short-term storage, e.g., up to 1 week) or at –20°C (for long-term storage, e.g., up to 5 years).
Basic Protocol 4: COMPLETE VIRAL RNA AMPLIFICATION USING TARGET-SPECIFIC PRIMERS IN MDA
This protocol enables enrichment of viral RNA genomes from total-RNA preparations, by replacing the random oligos and oligo-dT in the RT and MDA amplification of Basic Protocol 2 by target-specific primers. It is optimized to enrich RNA from as little as 1 pg in the presence of up to 5 ng contaminating RNA.
Specific primers for the enrichment of small genomes can be designed using open-source online primer design tools, e.g., Primer3 (https://bioinfo.ut.ee/primer3-0.4.0/) or Primer-BLAST (https://www.ncbi.nlm.nih.gov/tools/primer-blast/). Primer-Blast includes the additional feature of specificity checking. With this option enabled, the program will align the primers against a selected database. Based on their matches to putative non-specific targets and their orientations, it will determine whether a primer pair can generate an amplification product on any unwanted targets in this database. The program will return a collection of primers that do not generate a valid PCR product on unintended sequences and are therefore specific to the intended template.
For design parameters, it is recommended that the T m of primers is set to 34°C to 40°C and the length between 17 and 21 bp. Amplicon length should be 300 to 4,000 bp. We recommend choosing alternating primers on both strands every 2,000 to 3,000 bp to capture the total length of the genome intended to be amplified as shown in Figure 4.
Primers should be stabilized on 3′-primer end with phosphorothioate modifications on the last two 3′ bases as shown in the example 5′-NNNNNNNNNNNN-3′. This will protect degradation of the primer during the MDA. The primer mix should include primers in a concentration of 25 µM each for the RT reaction and 10 µM each for the MDA reaction.
NOTE : In cases of an extremely low number of copies (<5,000 copies) of viral RNA in high total RNA background (>1 ng total RNA), enrichment efficiency will be enhanced by rRNA depletion, using the QIAseq FastSelect –rRNA HMR Kit. Blocking of ribosomal RNA occurs prior to the Quantiscript RT as described in the Alternate Protocol.
Additional Materials (also see Basic Protocol 2)
-
QIAseq Single Cell RNA Library Kit UDI (QIAGEN, cat. no. 180703, 1801705, or 180707):
- NA Denaturation Buffer
- gDNA Wipeout Buffer, WTA
- RT/Polymerase Buffer
- Quantiscript® RT Enzyme Mix
- Ligase Mix
- Ligase Buffer
- REPLI-g sc Dilution Buffer
- REPLI-g SensiPhi® DNA Polymerase (yellow lid)
-
H2O sc
-
Purified RNA
-
Target-specific RT-primer mix, 25 µM each
-
Target-specific oligonucleotide mix primer, 10 µM each
-
Microcentrifuge tubes or PCR strips or plates
-
Thermal cycler
-
Microcentrifuge
-
Vortexer
-
Pipets: 1-10 µl, 20-200 µl, and 100-1,000 µl and pipet tips
1.Prepare all reagents as described in Basic Protocol 2.Thaw all buffers at room temperature (15°C to 25°C) and vortex before use to ensure thorough mixing, spin down, and place on ice. Thaw all enzymes and enzyme mixes (Quantiscript RT enzyme mix, ligase mix, and REPLI-g SensiPhi DNA polymerase) on ice and mix by gently flicking the tube. Avoid any vigorous vortexing of enzymes and enzyme mixes because this may affect their activity. Program a thermal cycler (see Table 6) for all incubation steps of the protocol.
2.Place 8 μl purified RNA (>500 pg) into a microcentrifuge tube. If using <8 μl of purified RNA, add H2O sc to make up volume to 8 μl.
3.Add 3 μl NA denaturation buffer, mix by vortexing, and centrifuge briefly.
4.Incubate at 95°C for 3 min; then cool to 4°C.
5.Add 2 µl gDNA wipeout buffer, mix by vortexing, and centrifuge briefly.
6.Incubate at 42°C for 10 min. If more time is needed to prepare the next step, place on ice.
7.Prepare the Quantiscript RT mix (see Table 15).
| Component | Volume per reaction | Volume for 12 reactions (plus 10%) |
|---|---|---|
| RT/polymerase buffer | 4 µl | 52.8 µl |
| Target-specific RT primer (25 µM each) | 1 µl | 13.2 µl |
| Quantiscript RT enzyme mix | 1 µl | 13.2 µl |
8.Add 6 µl Quantiscript RT mix to the lysed cell sample, mix by vortexing, and centrifuge briefly.
9.Incubate at 42°C for 60 min. Stop reaction by incubating at 95°C for 3 min, then cool on ice.
10.Prepare ligation mix by adding the components in the order shown in Table 16.
| Component | Volume per reaction | Volume for 12 reactions (plus 10%) |
|---|---|---|
| Ligase Buffer | 8 µl | 105.6 µl |
| Ligase Mix | 2 µl | 26.4 µl |
11.Add 10 µl ligation mix to the RT reaction from step 9.Mix by vortexing and centrifuge briefly.
12.Incubate at 24°C for 30 min. Stop reaction by incubating at 95°C for 5 min, then cool on ice.
13.Prepare REPLI-g SensiPhi amplification mix (see Table 17). Mix by vortexing and centrifuge briefly.
| Component | Volume per reaction | Volume for 12 reactions (plus 10%) |
|---|---|---|
| REPLI-g sc dilution buffer | 28 µl | 369.6 µl |
| Target-specific oligonucleotide mix (10 µM each) | 1 µl | 13.2 µl |
| REPLI-g SensiPhi DNA polymerase | 1 µl | 13.2 µl |
14.Add 30 µl REPLI-g SensiPhi amplification mix to the ligation reaction from step 12.
15.Incubate at 30°C for 3 hr. Stop reaction by incubating at 65°C for 5 min, then cool on ice.
Basic Protocol 5: ENZYMATIC FRAGMENTATION AND ADAPTER LIGATION OF MDA AMPLIFIED MATERIAL
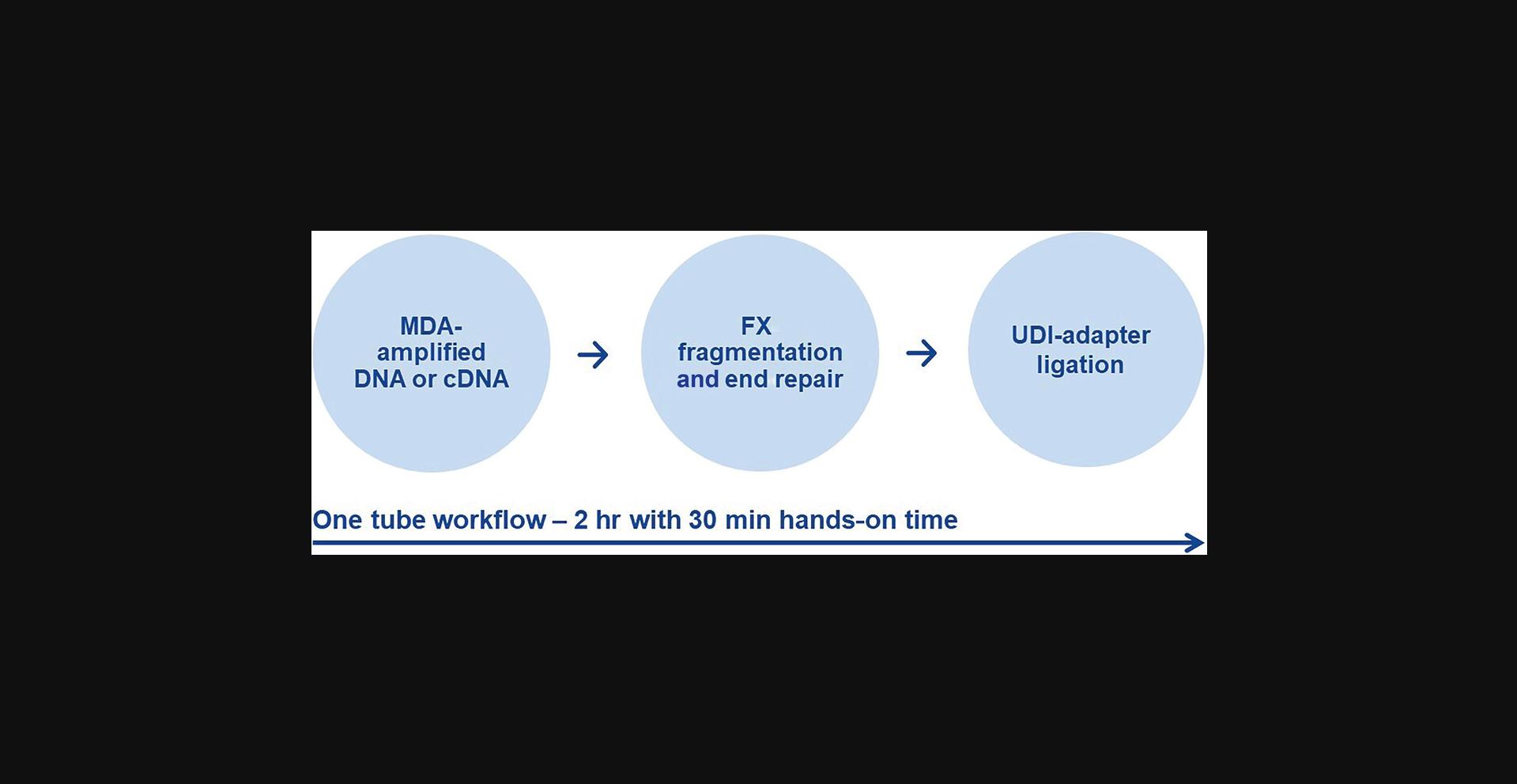
This one tube workflow describes the random enzymatic fragmentation based on the QIAseq FX reagents, followed by end-repair, A-addition, and adapter ligation. Excessive adapters and adapter-dimers are removed using QIAseq beads and are ready for quantification and use in NGS on NGS instruments from Illumina. The workflow of the library generation is described in Figure 5.
Materials
-
Amplified DNA (see Basic Protocol 1, Support Protocols 1 and 2) or amplified cDNA (see Basic Protocols 2, 3, and 4, and Alternate Protocol 1)
-
QIAseq Single Cell DNA Library Kit UDI (QIAGEN, cat. no. 181703, 181705, or 181707) or QIAseq Single Cell RNA Library Kit UDI (QIAGEN, cat. no. 180703, 1801705, or 180707):
- FX Enzyme Mix
- FX Buffer
- FX Enhancer
- DNA Ligase
- DNA Ligase Buffer
- Plate with UDI adapters compatible with Illumina sequencing platforms
- QIAseq Beads
- H2O sc
-
80% ethanol
-
Microcentrifuge tubes or PCR strips or plates
-
Thermal cycler
-
Microcentrifuge
-
Vortexer
-
Pipets: 1-10 µl, 20-200 µl, and 100-1,000 µl and pipet tips
-
Magnetic rack for 2-ml microcentrifuge tubes or PCR plates (Thermo Fisher Scientific)
-
DynaMag™-2 Magnet, (Thermo Fisher Scientific, cat. no. 12321D) or DynaMag™-96 Magnet (Thermo Fisher Scientific, cat. no. 12027)
1.Thaw all kit components on ice. Once reagents are thawed, mix buffers thoroughly by briefly vortexing tubes to avoid any localized concentrations. Briefly centrifuge reagents before use.
2.Equilibrate QIAseq beads to room temperature for 20-30 min before use and mix them thoroughly.
3.Program a thermal cycler according to Table 18.Start cycler and when the temperature has reached 4°C pause the program.
| Incubation time for generation of fragments with average size 400-450 bp | ||||
|---|---|---|---|---|
| Step | Temperature | gDNA (WGA) | cDNA (WTA) | Small genomes |
| 1 | 4°C | 1 min | 1 min | 1 min |
| 2 | 32°C | 15 min | 30 min | 15 min |
| 3 | 65°C | 30 min | 30 min | 30 min |
| 4 | 4°C | Hold | Hold | Hold |
4.Dilute 500 ng of amplified material in 10 µl H2O sc (50 ng/µl). If the amplified DNA is below this concentration, use the QIAseq beads to concentrate nucleic acids as described in the Support Protocol 2.
5.Pipet 10 µl diluted DNA into PCR tubes or strips and place them on ice or in a cooling block.
6.Prepare the FX reaction mix on ice according to Table 19 and mix by pipetting. Add the components of the FX reaction mix in the same order as stated in the table. Before adding the FX enzyme mix, pipet up and down the buffer mix. You can scale up the FX reaction mix according to the number of samples required.
| Component | Volume per reaction | Volume for 12 reactions (plus 10%) |
|---|---|---|
| FX buffer, 10× | 5 µl | 66 µl |
| H2O sc | 20 µl | 264 µl |
| FX enhancer | 5 µl | 66 µl |
| FX enzyme mix | 10 µl | 132 µl |
7.Add 40 μl FX reaction mix to each diluted amplified gDNA sample on ice and gently vortex to mix.
8.Briefly spin down the PCR plate/tubes, immediately transfer to the prechilled thermocycler (4°C), and resume the program. Once the fragmentation program is complete, transfer samples to ice and proceed immediately with adapter ligation.
9.Vortex and spin down the UDI adapter plate. Remove the protective adapter plate lid, carefully pierce the foil seal, and transfer 5 µl from one DNA adapter well to each 50-µl sample from the previous protocol. Track the barcodes used for each sample.
10.Replace the adapter plate lid and freeze unused adapters.
11.Prepare ligation master mix (per DNA sample) in a separate tube on ice according to Table 20.Mix well by gently vortexing at low rpm.
| Component | Volume per reaction | Volume for 12 reactions (plus 10%) |
|---|---|---|
| DNA ligase buffer, 5× | 20 µl | 264 µl |
| H2O sc | 15 µl | 198 µl |
| DNA ligase | 10 µl | 132 µl |
12.Add 45 µl ligation master mix to each sample from step 9.Close tubes or seal plate, mix well by short pulse vortexing, spin down, and incubate at 20°C for 15 min.
13.Perform purification by adding 60 µl (0.6×) of resuspended QIAseq beads slurry to each ligated sample and mix well by pipetting or gently vortexing.
14.Incubate mixture 5 min at room temperature.
15.Spin down to collect remaining droplets and pellet beads on a magnetic stand 2 min, then carefully discard supernatant.
16.Wash beads by adding 200 µl fresh 80% ethanol to each pellet. Pellet beads on the magnetic stand 2 min, then carefully discard supernatant.
17.Repeat wash step 16 once for a total of two ethanol washes.
18.Incubate on the magnetic stand 5-10 min or until the beads are dry. Avoid over-drying, which may result in lower DNA recovery. Remove from the magnetic stand.
19.Elute by resuspending in 52.5 µl H2O sc. Pellet beads on the magnetic stand. Carefully transfer 50 µl supernatant to a new PCR plate/tube.
20.Perform a second 1.1× QIAseq beads purification using 55 µl resuspended QIAseq beads to each sample and mix.
21.Repeat steps 14-18 to rebind the DNA on the beads, wash, and dry beads.
22.Finally elute the library by resuspending in 26 µl H2O sc. Pellet beads on the magnetic stand. Carefully transfer 23.5 µl supernatant into a new PCR plate. Store purified libraries at –20°C until ready for sequencing.
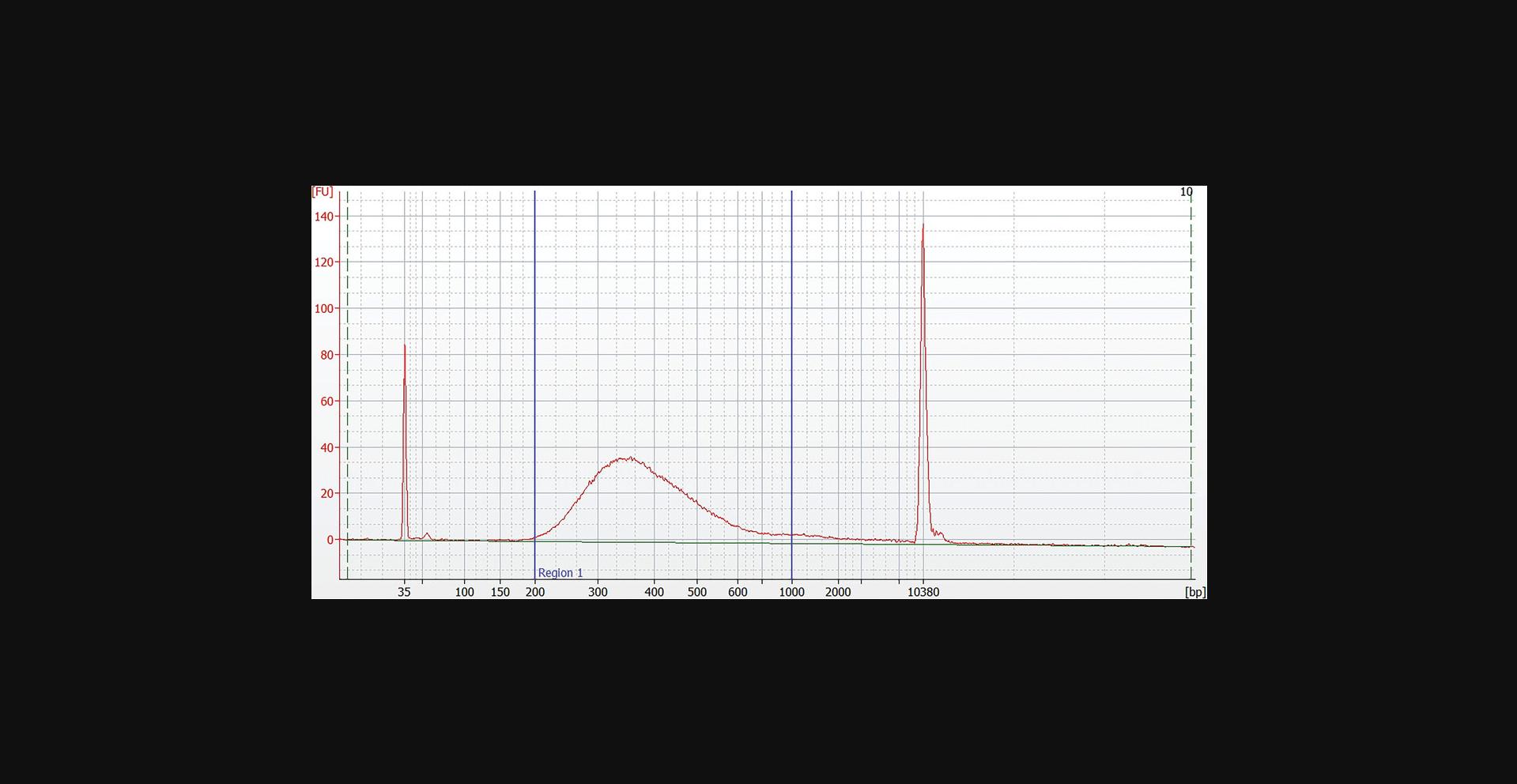
23.Assess the quality of the library using a capillary electrophoresis device or comparable method. Check for the correct size distribution of library fragments and for the absence of adapters or adapter dimers (Fig. 6).
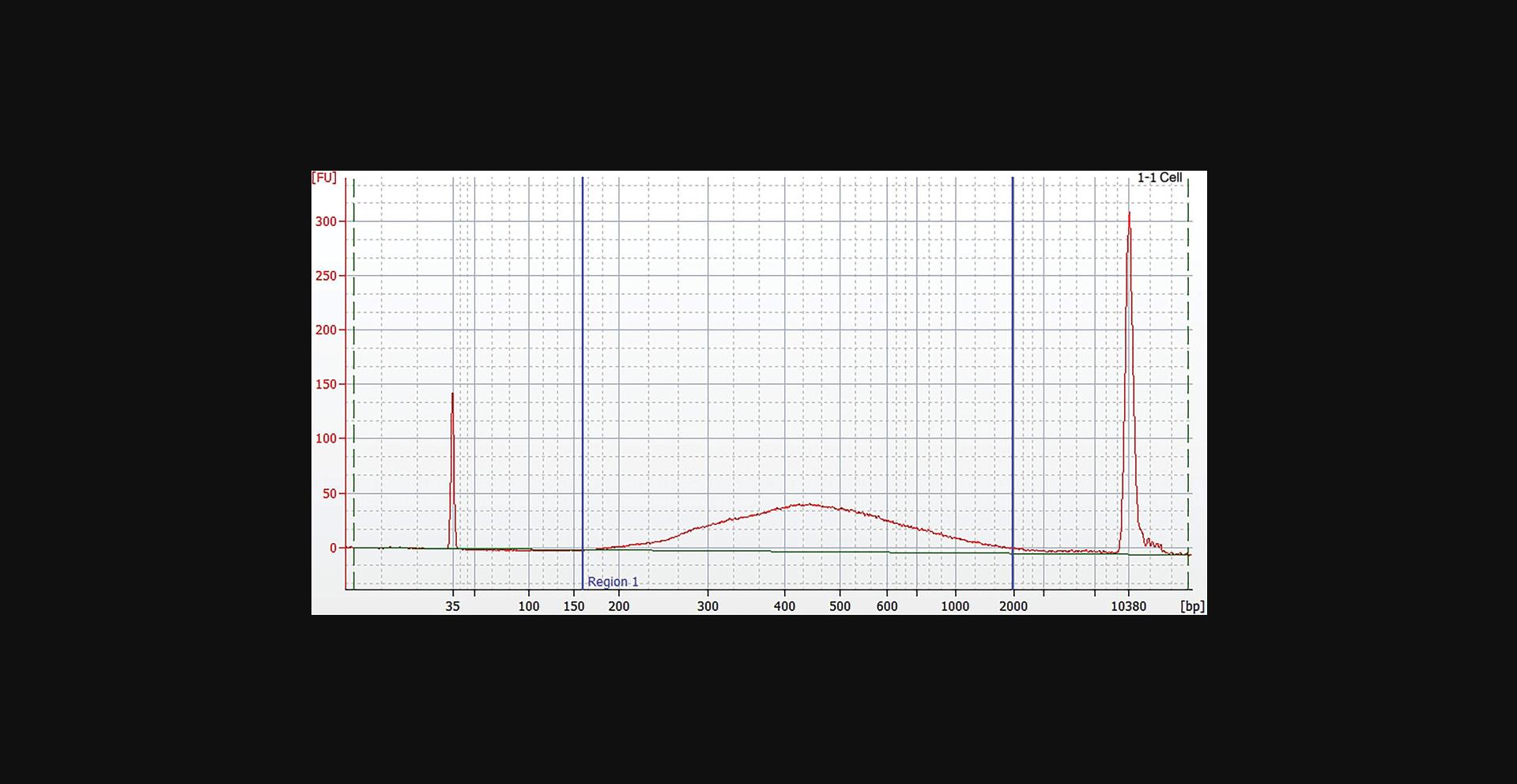
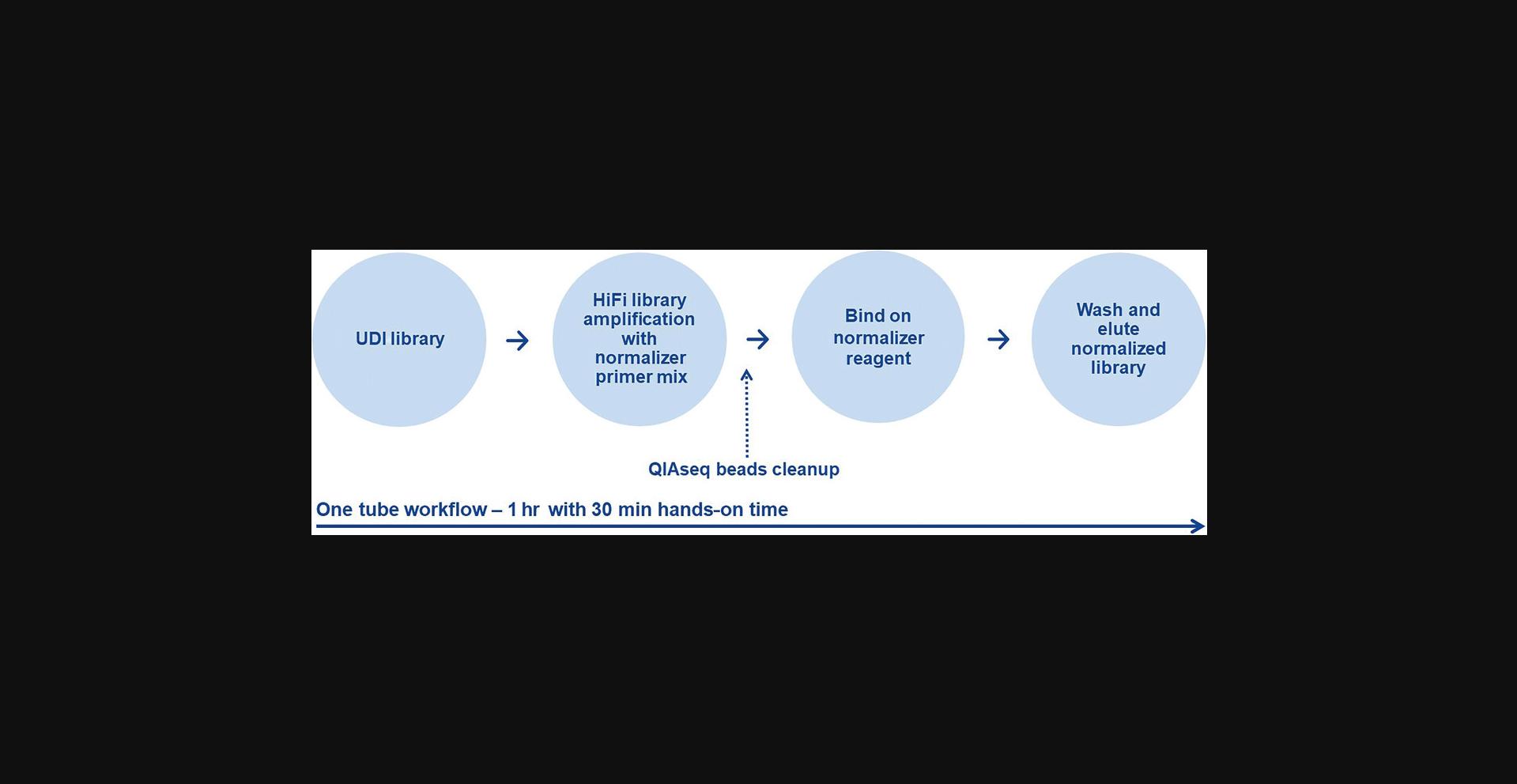
Basic Protocol 6: NORMALIZATION OF LIBRARY CONCENTRATION USING MAGNETIC BEADS
This protocol is for the normalization of NGS libraries compatible with Illumina platforms using the QIAseq Library Normalizer Kit and is optional. The protocol eliminates the need of quantifying libraries individually. When processing multiple libraries in parallel, it accelerates the procedure of pooling the libraries, without the need of a reagent and time consuming quantification. The used magnetic beads bind and release a specific amount of library in a highly controlled manner. After elution, all normalized libraries have a concentration of 4 nM and can be easily pooled together for sequencing. The workflow is described in Figure 7.
Materials
-
QIAseq Library Normalizer Kit (QIAGEN, cat. no. 180703, 180705):
- Normalizer primer mix
- Normalizer reagent
- Normalizer wash buffer
- Normalizer elution buffer
-
QIAseq Single Cell DNA Library Kit UDI (QIAGEN, cat. no. 181703, 181705, or 181707), or QIAseq Single Cell RNA Library Kit UDI (QIAGEN, cat. no. 180703, 1801705, or 180707):
- HiFi PCR MMix
- QIAseq beads
-
80% ethanol
-
H2O sc
-
Tris·HCl, 10 mM, pH 8.0
-
Thermal cycler
-
Magnetic stand
Library amplification prior to normalization
1.Thaw all reagents on ice. Once reagents are thawed, mix them thoroughly by vortexing to avoid any localized concentrations.
2.Program a thermal cycler with the conditions outlined in Table 21.
| Time | Temperature | Number of cycles |
|---|---|---|
| 2 min | 98°C | 1 |
|
20 s 30 s 30 s |
98°C 60°C 72°C |
4 for WGA and WTA libraries, or 8 for small genome and viral RNA libraries |
| 1 min | 72°C | 1 |
- a
Abbreviations: WGA, whole genome amplification; WTA, whole transcriptome amplification.
3.Prepare library amplification reaction mix according to Table 22.
| Component | Volume per reaction | Volume for 12 reactions (plus 10%) |
|---|---|---|
| HiFi PCR master mix, 2× | 25 µl | 330 µl |
| Normalizer primer mix | 1.5 µl | 19.8 µl |
4.Add 26.5 µl library amplification reaction mix to 23.5 µl of the generated library from Basic Protocol 5, step 22.Mix by gently vortexing and spin down.
5.Place mixes in the cycler and start the program (Table 21).
6.Once amplification is completed, proceed with library cleanup using 1× QIAseq beads (50 µl).
7.Add 50 µl resuspended QIAseq beads slurry to each ligated sample and mix well by pipetting or gently vortexing.
8.Incubate mixture 5 min at room temperature.
9.Pellet beads on a magnetic stand for 2 min and carefully discard supernatant.
10.Wash beads by adding 200 µl fresh 80% ethanol to each pellet. Pellet beads on the magnetic stand 2 min, then carefully discard supernatant.
11.Repeat wash step 9 once for a total of two ethanol washes.
12.Incubate on the magnetic stand for 5-10 min or until the beads are dry. Remove from the magnetic stand.
13.Elute by resuspending in 32.5 µl H2O sc. Pellet beads on the magnetic stand. Carefully transfer 30 µl supernatant to a new LoBind tube.
QIAseq library normalization
14.Preheat normalizer wash buffer in a heat block or water bath to 55°C. If needed, thoroughly mix wash buffer and prepare aliquots for preheating. Prepare 450 μl of wash buffer per normalization reaction.
15.Pipet 15 μl of each modified library into a 1.5-ml tube or well of a PCR plate. Leave the tube/plate at room temperature.
16.Vortex normalizer reagent at least 60 s to achieve optimal homogenization.
17.Add 5 μl of the homogenized normalization reagent to each 15 μl of prepared library and mix well.
18.Incubate 10 min at room temperature.
19.Add 200 μl pre-warmed normalizer wash buffer (55°C) to each tube.
20.Pellet beads on a magnetic stand and wait until the solution is clear, then carefully discard supernatant without disturbing the pellet.
21.Add 200 μl pre-warmed normalizer wash buffer (55°C) to each pellet. Switch the tube/plate position on the magnet to wash the beads, then wait until the solution is clear and carefully discard supernatant without disturbing the pellet.
22.Remove all remaining liquid using a 10-μl pipet tip, then proceed to the next step immediately.
23.Add 26 μl normalizer elution buffer to each pellet and mix well by vortexing. Make sure all beads are submerged and all liquid is at the bottom of the tube.
24.Incubate 5 min at 55°C in a heating block or thermal cycler.
25.Pulse spin to collect all liquid, then pellet beads on a magnetic stand and wait until the solution is clear. Carefully transfer 25 μl of the supernatant to a new tube/plate.
26.Proceed to library pooling or store the normalized libraries at − 20°C.
COMMENTARY
Background Information
The introduced method of multiple displacement amplification (MDA; see Fig. 10) is an isothermal whole genome amplification, which utilizes polymerases with high processivity and strand-displacement activity that extend from a randomly primed site and can replicate up to 100 kb without dissociating from the genomic DNA template (de Bourcy et al., 2014). MDA technology is used in the presence of exonuclease-resistant primers to achieve high yields of DNA products from all kinds of mammalian and bacterial tissues (Dean et al., 2002; Raghunathan et al., 2005).
Compared to PCR-based whole genome amplification methods, MDA can be easily miniaturized, is less sensitive to failure due to contamination, and allows NGS applications with low bias and high genome coverage (de Bourcy et al., 2014).
The introduced whole transcriptome amplification (WTA) method uses a novel variant of the Phi 29 polymerase, the high sensitivity and high fidelity REPLI-g SensiPhi polymerase for MDA and an optimized set of buffers and reagents for efficient cell lysis, effective removal of genomic DNA (gDNA), sensitive reverse transcription of the RNA, and accurate amplification of cDNA. Depending on the input material, cells, or purified RNA it can be combined with rRNA depletion for maximal recovery of mRNA.
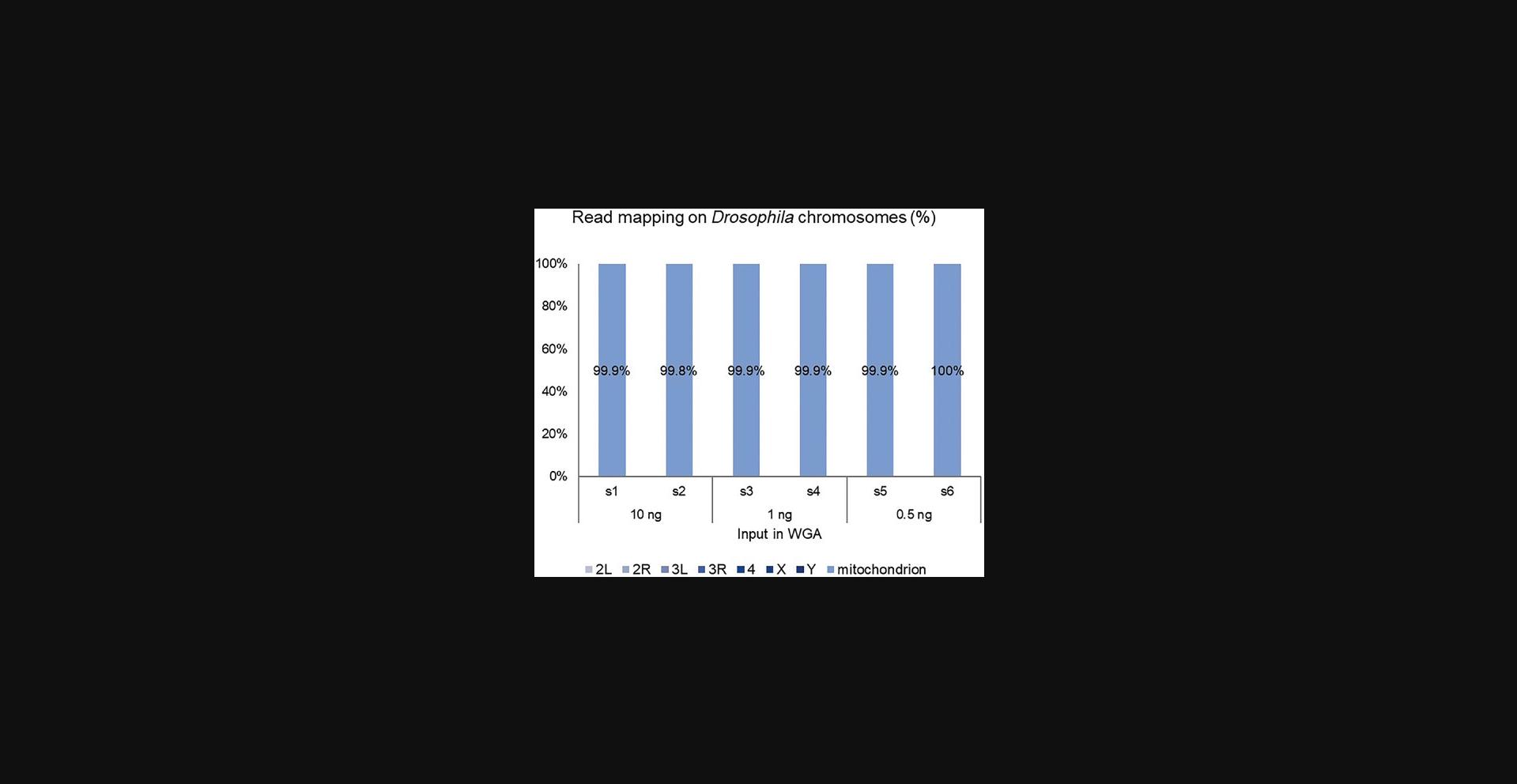
Both methods have been modified to accept target-specific primer to allow PCR-free amplification of complete small genomes or DNA and RNA viruses in a very high nucleic acid background as demonstrated for Drosophila mitochondrial-DNA enrichment in Figure 11. The figure demonstrates the highly efficient enrichment of a mitochondrial genome with complete read mapping to mitochondrial DNA (mtDNA), and almost no reads were mapped to other Drosophila chromosomes. The features of Phi29 polymerase to process long fragments were used to amplify complete genomes from ultra-low input by only using a few primers, which were designed every 2,000 to 3,000 bp on the reference genome of the species which needed to be enriched. This represents a new art of enrichment, which is more cost efficient than PCR-based methods, which significantly need a higher density of primers to cover a complete genome. The HiFi feature of the enzyme ensures accurate variant analysis of these small genomes.
The library preparation using FX technology incorporates an all-enzymatic DNA fragmentation into a streamlined, optimized protocol that does not require additional sample cleanup before adapter ligation. A simple, three-reaction protocol that takes place in the same tube enables straightforward automation of library preparation on various liquid-handling platforms, reducing hands-on time, and run-to-run variability. This method can accommodate a wide range of DNA input and represents a more efficient and flexible method, compared, for example, to transposon-based methods, which are optimized for a certain amount of DNA input. Further this technology uses sequence-independent enzymes for fragmentation and generates libraries with minimal bias, delivering results comparable to mechanical shearing (Tourlousse et al., 2021).
The last method represents a tool for efficient normalization of libraries, without the need of time and cost consuming qPCR quantification and subsequent normalization by dilution. This enables standardized high throughput library generation and pooling. It uses modified library amplification primers and magnetic beads to efficiently bind and release the library fragments in a strictly controlled mode and allows optimal pooling conditions and optimal clustering on Illumina flow cells.
All workflows can be combined with each other to address a variety of applications from single-cell genomics and transcriptomics, metagenomics as well as viral DNA and RNA sequencing.
When starting with intact cells, another important variable that will influence the outcome of the described methods is the handling used to isolate and collect the single cells. The chosen method should be precise, gentle, and deliver intact, viable cells. One of the best methods to target and collect single cells is micromanipulation (Frohlich & Konig, 2000). This method is suitable only for low throughput experiments because it is highly laborious. To increase the numbers of single cells, automated solutions are needed. Especially when studying heterogeneity in tissues, these must be dissociated to produce cell suspensions, and the released cells can then be enriched in cell sorters according to specific markers, which are expressed on their surface. Instruments that can select and enrich rare cells based on cell surface markers are also used to isolate tumor cells that circulate in blood (Rushton et al., 2021). In addition, immunomagnetic beads (AdnaTest) have also been employed to enrich circulating tumor cells from blood (Hanssen et al., 2016). Flow cytometry using fluorescence-activated cell sorting (FACS) is by far the most efficient method, and is widely used for isolating large numbers of single cells or nuclei from liquid suspensions (Frohlich & Konig, 2000; Navin & Hicks, 2011; Nawy, 2014). New developments of cell sorters and dispensers, based on microfluidic cartridges, have accelerated and simplified cell sorting to allow isolation of high-quality viable cells, suitable for NGS applications (Yumoto et al., 2020).
Critical Parameters
If using only one cell, the art of isolating the cell and the condition of the cell is a critical point. For the MDA reaction, integrity of DNA plays an important role. High molecular weight DNA will lead to uniform amplification, while fragmented DNA will lead to less uniform amplification with increased drop offs or complete drop out of regions. The used method for the cell isolation must be optimized for each cell type in order to ensure the viability of the cells and consequently the high integrity of DNA. In addition to DNA integrity, the accessibility of DNA will affect the quality of amplification, too. For example, the WGA protocol (Basic Protocol 1) will not work efficiently with bacterial cells if these begin sporulation, because lysis and DNA release will be inefficient (Piggot & Coote, 1976).
The complete lysis of the cell is the requirement for successful WGA/WTA. It is particularly important to completely mix the cell with the cell lysis buffer without contacting the cell material with a pipet tip because this will lead to loss of material. Gently flick the tube several times and centrifuge to collect the sample.
MDA using a random primer is, as is every random amplification procedure, extremely sensitive to contaminating nucleic acids because they will be co-amplified and in the worst case, they will outcompete the amplification of the test sample. It is important to maintain a nucleic acid free working environment. If possible, set up reaction mixes and reactions in a clean PCR-hood.
Troubleshooting
Table 23 shows a list of issues, which may occur during the execution of the described protocol and explains the possible causes and corrective actions.
| Problem | Possible cause | Solution |
|---|---|---|
| MDA reduced yields | Cells are not suitable for WGA or WTA | Use viable cell; cell viability must be >90% |
| MDA reduced yields | gDNA or RNA not suitable for whole genome amplification | Qualify integrity by capillary electrophoresis |
| MDA reduced yields | Reaction inhibition by salts or alcohols present in the DNA preparation | Clean up or dilute purified genomic DNA to reduce concentration of inhibitory substances and reamplify |
| MDA reduced yields | Reaction time is too short | Recommended MDA time in the corresponding protocols is sufficient to amplify nucleic acid from said material. In some cases, sufficient yield is possible even with shorter incubation, but this should be confirmed experimentally. |
| MDA reduced yields | Reaction temperature is too high | Check the thermal cycler for the correct lid temperature (70°C) and the correct reaction temperature (30°C) during the whole genome amplification reaction. If thermal cycler is not calibrated run a control amplification reaction at a lower temperature (e.g., 25°C to 28°C), which should give the appropriate yield. |
| Reduced/no locus representation | Cells are not viable and DNA/RNA is degraded or lysis is insufficient | Use high quality starting material, avoid contact of pipet tips with the cell material, and perform mixing by gently flicking the tube |
| Reduced/no locus representation | Micro-dissected cells do not contain the whole nucleus | When isolating cells using microdissection, ensure that the section thickness allows the capture of the whole nucleus and that the nucleus is not damaged |
| Reduced/no locus representation | Sample is contaminated; contaminating DNA may outcompete the amplification of single cell genome | Maintain clean nuclease and DNA and RNA free lab environment. If possible, work under a PCR hood. |
| Low library yields | MDA yields and input in FX reaction were lower than expected |
Quantify yield of WGA using PicoGreen reagent. Typically, 100 ng of WGA DNA generates enough Illumina-compatible library to use directly for sequencing without any additional amplification. If the final library yield is not sufficient for the expected number of sequencing runs, a library amplification step can be performed following the adapter ligation step. |
| Low library yields | Low quality DNA or FX reaction inhibition | Do not exceed 5 µl MDA-DNA input in the FX reaction or perform cleanup of MDA-DNA |
| Low library yields | Normalizer reagent was not mixed efficiently | Follow the exact recommendations in the protocol |
| Presence of shorter fragments between 60 and 120 bp in library | These peaks are adapters and adapter-dimers that are generated when ligation is inhibited, or adapter depletion after library preparation is not efficient | Check for correct temperature during ligation. If library yields are sufficient, perform an additional library cleanup using the 0.8× QIAseq beads |
- a Abbreviations: MDA, multiple displacement amplification; WGA, whole genome amplification; WTA, whole transcriptome amplification.
Understanding Results
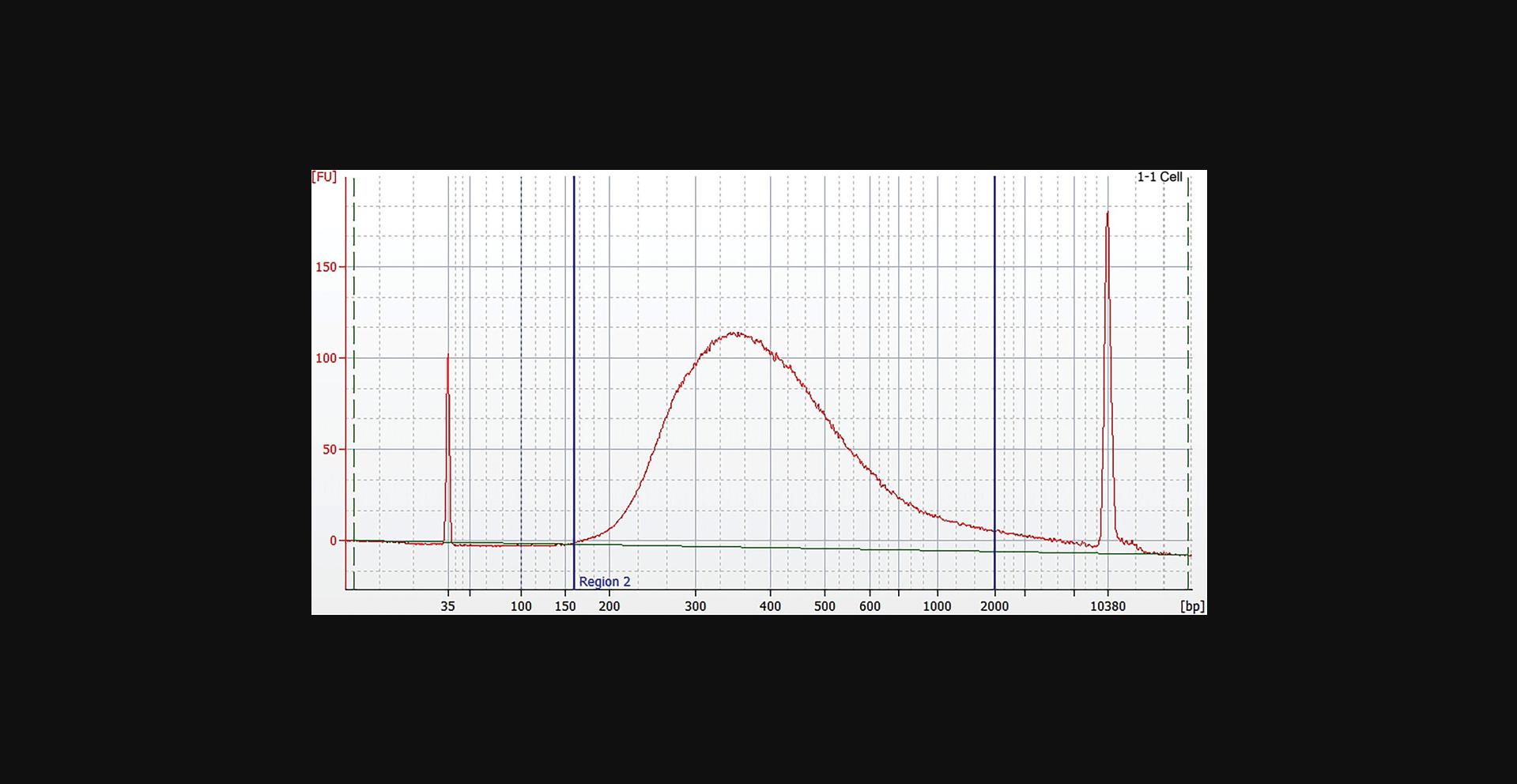
The average yield of the WGA is 20 to 40 µg DNA and of the WTA 10 to 20 µg of cDNA depending on the amount and the quality of the starting material. The procedure of enrichment of small genomes using target-specific primers results in 1 to 10 µg amplified material. The library concentration after normalization is 4 nM with an average fragment size of 400 to 450 bp (Fig. 9).
Time Considerations
The WGA from single cells will take 2.5 hr to complete with 15 min hands-on time. WGA of small genomes will take 4.5 to 8.5 hr to complete with 15 min hands-on time.
The WTA will take ∼4 hr while PicoGreen quantification will take 30 min to 1 hr, depending on the number of samples. Enrichment of viral RNA using the MDA procedure with target-specific primers will take 5 to 7 hr.
Library preparation using the FX procedure will take ∼2 hr with 30 min hands-on time. The library normalization will take ∼1 hr with 30 min hands-on time. Control of fragment size, e.g., with the Agilent Bioanalyzer will need an additional 30 min with only 15 min hands-on time.
Acknowledgments
This work is not supported by any funding.
Author Contributions
Ioanna Andreou : Conceptualization, investigation, methodology, resources, writing original draft; Markus Storbeck : Investigation, resources, writing review and editing; Peter Hahn : Methodology, writing review and editing; Samuel Rulli : Conceptualization, writing review and editing; Eric Lader : Conceptualization, supervision, writing review and editing.
Conflict of Interest
Authors are employees of QIAGEN, the supplier of the reagents used in this work. They have no other conflicting interests.
Open Research
Data Availability Statement
The data that support the findings of this study, which are not included in this article, are available from the corresponding author upon reasonable request.
Literature Cited
- de Bourcy, C. F., De Vlaminck, I., Kanbar, J. N., Wang, J., Gawad, C., & Quake, S. R. (2014). A quantitative comparison of single-cell whole genome amplification methods. PLoS One , 9, e105585. https://doi.org/10.1371/journal.pone.0105585
- Dean, F. B., Hosono, S., Fang, L., Wu, X., Faruqi, A. F., Bray-Ward, P., Sun, Z., Zong, Q., Du, Y., Du, J., Driscoll, M., Song, W., Kingsmore, S. F., Egholm, M., & Lasken, R. S. (2002). Comprehensive human genome amplification using multiple displacement amplification. Proceedings of the National Academy of Sciences of the United States of America , 99, 5261–5266. https://doi.org/10.1073/pnas.082089499
- Frohlich, J., & Konig, H. (2000). New techniques for isolation of single prokaryotic cells. FEMS Microbiology Reviews , 24, 567–572. https://doi.org/10.1016/S0168-6445(00)00045-0
- Hanssen, A., Wagner, J., Gorges, T., Taenzer, A., Uzunoglu, F. G., Driemel, C., Stoecklein, N. H., Knoefel, W. T., Angenendt, S., Hauch, S., Atanackovic, D., Loges, S., Riethdorf, S., Pantel, K., & Wikman, H. (2016). Characterization of different CTC subpopulations in non-small cell lung cancer. Scientific Reports , 6, 28010. https://doi.org/10.1038/srep28010
- Navin, N., & Hicks, J. (2011). Future medical applications of single-cell sequencing in cancer. Genome Medicine , 3, 31. https://doi.org/10.1186/gm247
- Nawy, T. (2014). Single-cell sequencing. Nature Methods , 11, 18. https://doi.org/10.1038/nmeth.2771
- Piggot, P. J., & Coote, J. G. (1976). Genetic aspects of bacterial endospore formation. Bacteriological Reviews , 40, 908–962. https://doi.org/10.1128/br.40.4.908-962.1976
- Raghunathan, A., Ferguson, H. R. Jr., Bornarth, C. J., Song, W., Driscoll, M., & Lasken, R. S. (2005). Genomic DNA amplification from a single bacterium. Applied and Environmental Microbiology , 71, 3342–3347. https://doi.org/10.1128/AEM.71.6.3342-3347.2005
- Rushton, A. J., Nteliopoulos, G., Shaw, J. A., & Coombes, R. C. (2021). A review of circulating tumour cell enrichment technologies. Cancers , 13(5), 970. https://doi.org/10.3390/cancers13050970
- Tourlousse, D. M., Narita, K., Miura, T., Sakamoto, M., Ohashi, A., Shiina, K., Matsuda, M., Miura, D., Shimamura, M., Ohyama, Y., Yamazoe, A., Uchino, Y., Kameyama, K., Arioka, S., Kataoka, J., Hisada, T., Fujii, K., Takahashi, S., Kuroiwa, M., … Terauchi, J. (2021). Validation and standardization of DNA extraction and library construction methods for metagenomics-based human fecal microbiome measurements. Microbiome , 9(1), 95. https://doi.org/10.1186/s40168-021-01048-3
- Yumoto, M., Hemmi, N., Sato, N., Kawashima, Y., Arikawa, K., Ide, K., Hosokawa, M., Seo, M., & Takeyama, H. (2020). Evaluation of the effects of cell-dispensing using an inkjet-based bioprinter on cell integrity by RNA-seq analysis. Scientific Reports , 10, 7158. https://doi.org/10.1038/s41598-020-64193-z
Internet Resources
CTC and AdnaTest Flyer.
QIAseq kits for MDA-based WGA, which include resources such as performance data, ordering information, handbooks, Material Safety Data Sheets (MSDSs), and frequently asked questions.
QIAseq kits for MDA-based WTA, which include resources such as performance data, ordering information, handbooks, MSDSs, and frequently asked questions.
QIAseq FastSelect kits for rRNA depletion, which include resources such as performance data, ordering information, handbooks, MSDSs, and frequently asked questions.
Open source primer design tool.
Open source primer design tool.
Products for Qubit-based DNA quantification, which include resources such as performance data, ordering information, handbooks, MSDSs, and frequently asked questions.
NGS supporting services and resources.
Citing Literature
Number of times cited according to CrossRef: 1
- Ioanna Andreou, Markus Storbeck, Peter Hahn, Samuel Rulli, Eric Lader, Exome Sequencing Starting from Single Cells, Current Protocols, 10.1002/cpz1.70017, 4 , 9, (2024).

