ORFanID Web-based Search Engine to Identify Orphan and Taxonomically Restricted Genes
Thushara Galbadage, Vinodh Gunasekera, Emanuel Tundrea, Richard S. Gunasekera
Abstract
ORFanID is a web-based software engine designed to identify ORFan genes from genomes of interest; from a given list of DNA or protein sequences within the NCBI databases. The selection of the taxonomy level of interest can define the scope of the search for orphan genes. Detectable homologous sequences are found by the software for candidate genes in the NCBI databases. Based on these findings, the ORFanID engine identifies and depicts orphan genes. Results may be viewed and analyzed graphically for scientific research and inquiry. As the enigma of orphan genes unravels, we believe ORFanID will provide critical insights into the origin, function, and prevalence of ORFan genes in genomes.
The last step contains a supplemental video with extra context and tips, as part of the protocols.io Spotlight series, featuring conversations with protocol authors.
Steps
Accessing ORFanID
Navigate to ORFanID's webpage at http://www.orfangenes.com/
Sample Searches
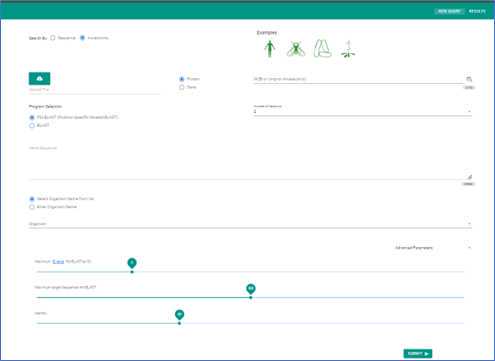
If you don't have a specific gene sequence or an accession number, use the sample options provided. Four sample icons are located at the bottom left of the search screen. Clicking on any of these will pre-fill the input field with either a gene sequence or an accession number. Click "Search" to proceed.
Input Methods
Choose between searching for a gene sequence or using an accession number. For instance, to search a Homo sapiens sample, use the toggle switch to specify your preference.
ORFanID offers three search methods:
a. Uploading a FASTA file.
b. Inputting an accession number or numbers. (e.g., the E. coli sample has three given accession numbers)
c. Directly submitting gene sequences.
Search Guidelines
When searching multiple gene sequences, separate each with a new line or space.
You'll need to specify the organism for your search. If the desired organism isn't listed, refer to the NCBI taxonomy database to obtain its full scientific name and taxonomy ID. Enter this information, ensuring the taxonomy ID is in parentheses.
Choose between searching by protein or gene using the provided radio selection boxes.
Additional Options
There's an optional Nickname field on the upper left. Use it to label and identify your search results for future reference.
Adjust the three advanced parameters if needed. By default, the maximum e-value for the BLAST algorithm is set to three, and the maximum target sequences are set at 550. Hover over the pop-up for detailed info.
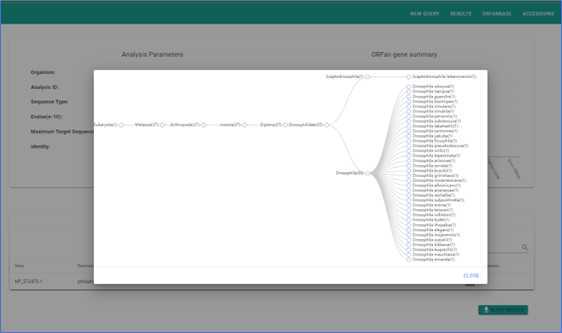
Submission and Results
After confirming your search criteria, the organism, and any other fields, click "Submit". A pop-up will prompt you to review your search. If satisfied, proceed to submit. Depending on the complexity of your query, results may take between 3 to 15 minutes.
Spotlight video
#尊敬的用户,由于网络监管政策的限制,部分内容暂时无法在本网站直接浏览。我们已经为您准备了相关原始数据和链接,感谢您的理解与支持。
<iframe title="YouTube video player" src="https://www.youtube.com/embed/iiHV8-pe3m8?si=QLOiWcmelSSUUNsq" height="315" width="560"></iframe>