NCBI's Conserved Domain Database and Tools for Protein Domain Analysis
Mingzhang Yang, Mingzhang Yang, Myra K. Derbyshire, Myra K. Derbyshire, Roxanne A. Yamashita, Roxanne A. Yamashita, Aron Marchler-Bauer, Aron Marchler-Bauer
Conserved Domain Database
domain architecture
protein annotation
protein classification
protein domains
protein function
protein naming
Abstract
The Conserved Domain Database (CDD) is a freely available resource for the annotation of sequences with the locations of conserved protein domain footprints, as well as functional sites and motifs inferred from these footprints. It includes protein domain and protein family models curated in house by CDD staff, as well as imported from a variety of other sources. The latest CDD release (v3.17, April 2019) contains more than 57,000 domain models, of which almost 15,000 were curated by CDD staff. The CDD curation effort increases coverage and provides finer-grained classifications of common and widely distributed protein domain families, for which a wealth of functional and structural data have become available. The CDD maintains both live search capabilities and an archive of pre-computed domain annotations for a selected subset of sequences tracked by the NCBI's Entrez protein database. These can be retrieved or computed for a single sequence using CD-Search or in bulk using Batch CD-Search, or computed via standalone RPS-BLAST plus the rpsbproc software package. The CDD can be accessed via https://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml. The three protocols listed here describe how to perform a CD-Search (Basic Protocol 1), a Batch CD-Search (Basic Protocol 2), and a Standalone RPS-BLAST and rpsbproc (Basic Protocol 3). © 2019 The Authors.
Basic Protocol 1 : CD-search
Basic Protocol 2 : Batch CD-search
Basic Protocol 3 : Standalone RPS-BLAST and rpsbproc
INTRODUCTION
The Conserved Domain Database (CDD) of the National Center for Biotechnology Information (NCBI) is a collection of protein family and protein domain models. A domain is defined as a compact, discrete unit of 3D structure, typically in the range of 50 to 200 amino acids in size, and as a unit of molecular evolution that can be utilized to establish evolutionary classifications; a domain is usually associated with discrete aspects of protein function, such as enzyme activity, membrane transport, or nucleic-acid binding, to name a few. Domain models in the CDD include many fine-grained hierarchical classifications for selected domain families established with the help of phylogenetic analyses and manually curated by CDD staff, as well as sets of domain models imported from external high-quality and comprehensive resources, collected as annotated multiple sequence alignments and converted into position-specific score matrices. The current CDD collection (version 3.17) contains 57,242 total models: 14,908 models from the CDD curation effort, 35 NCBIfams (Haft et al., 2018), 1012 models from SMART v6.0 (Letunic, Doerks, & Bork, 2014), 16,709 models from Pfam v31 (Finn et al., 2016), 4873 COGs v1.0 (Tatusov et al., 2001), 10,885 NCBI Protein Clusters (Klimke et al., 2009), and 4488 models from TIGRFAM v15 (Haft et al., 2013).
The conserved domain summary pages give access to a wealth of data associated with each domain family, including hierarchical classifications, taxonomic information, sequence alignments, structural interaction data, domain architectures, functional site annotations, and literature. Figure 1 diagrams some of the variety of information available to the user in navigating the CDD. In an effort to take advantage of these multiple types of information, the CDD uses Reverse Position-Specific BLAST (RPS-BLAST), also known as CD-Search (Conserved Domain Search), in its interactive web-based implementation to match protein sequences with domain and family models, providing a live search service for protein and nucleotide queries, as well as pre-computed (at a pre-set E -value) domain and site annotations for the majority of protein sequences in the NCBI's Entrez system. The CDD has been integrated with several resources at the NCBI, including BLAST, Protein, and Gene, and with external collections such as InterPro (Apweiler et al., 2000; Mitchell et al., 2019; https://www.ebi.ac.uk/interpro), in order to provide a comprehensive workflow that will fit most user's needs.
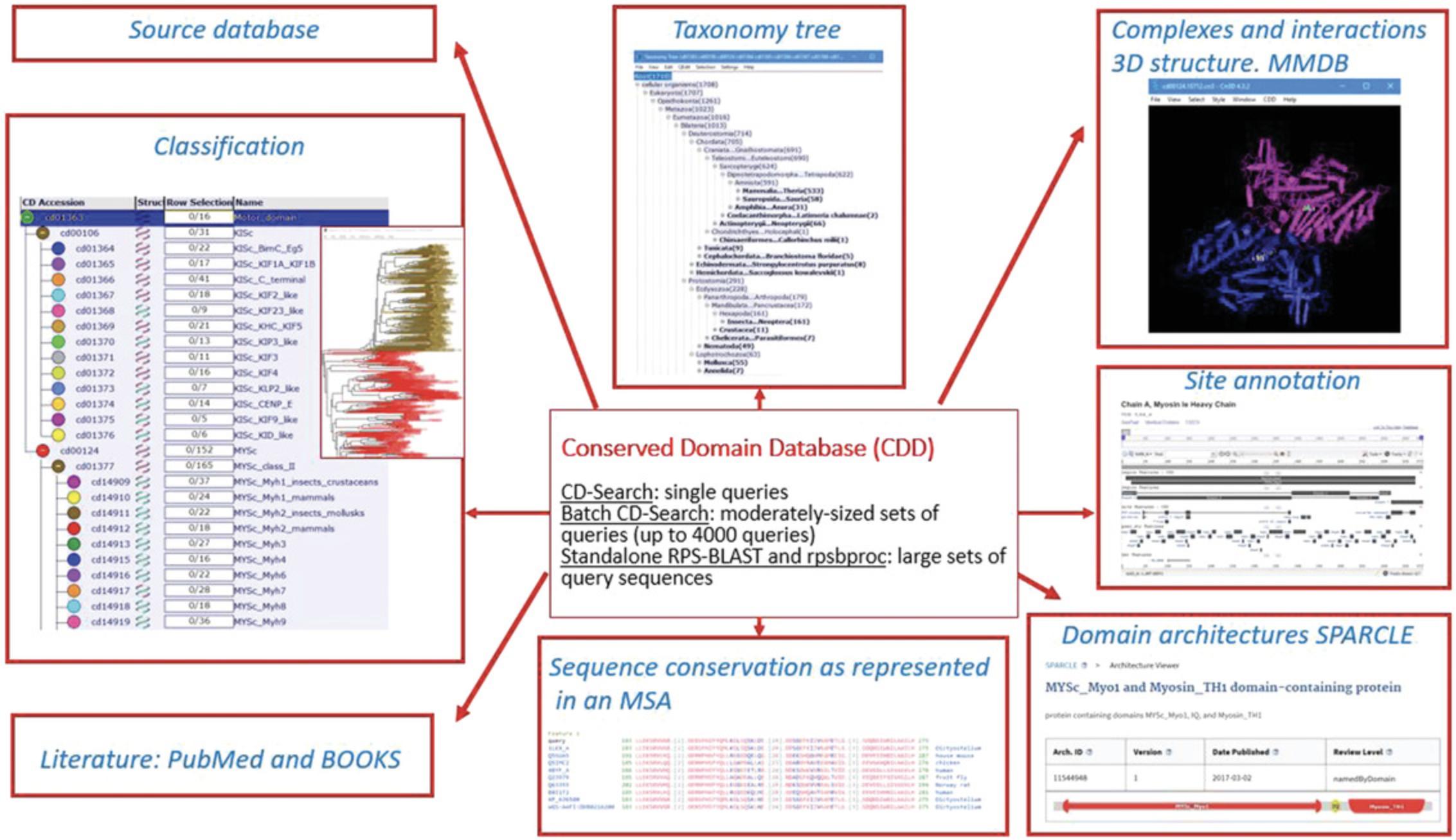
You can access the CDD resource by using CD-Search for a single nucleotide or protein sequence query, Batch CD-Search for up to 4000 queries at a time, or standalone RPS-BLAST plus rpsbproc running searches on your local infrastructure. You can also query Entrez (https://www.ncbi.nlm.nih.gov/cdd/) to access the CDD's domain information in the CDD resource. In Basic Protocols 1 to 3, we describe how to use each of these services so that you can customize the settings, and we outline commonly used workflows. In addition, we provide links to Help documentation (Table 1) to aid you as you navigate these pages.
Basic Protocol 1: CD-SEARCH
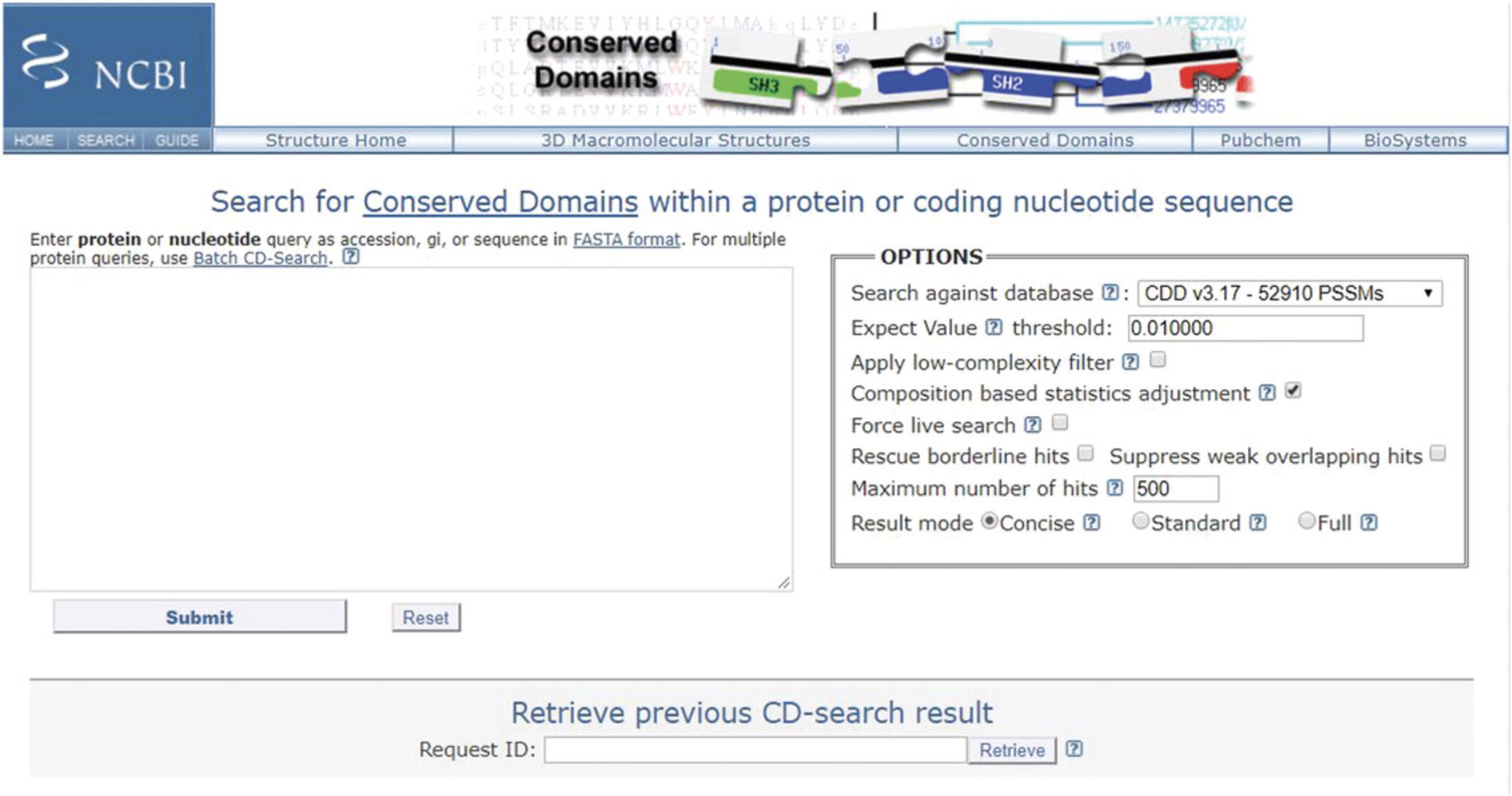
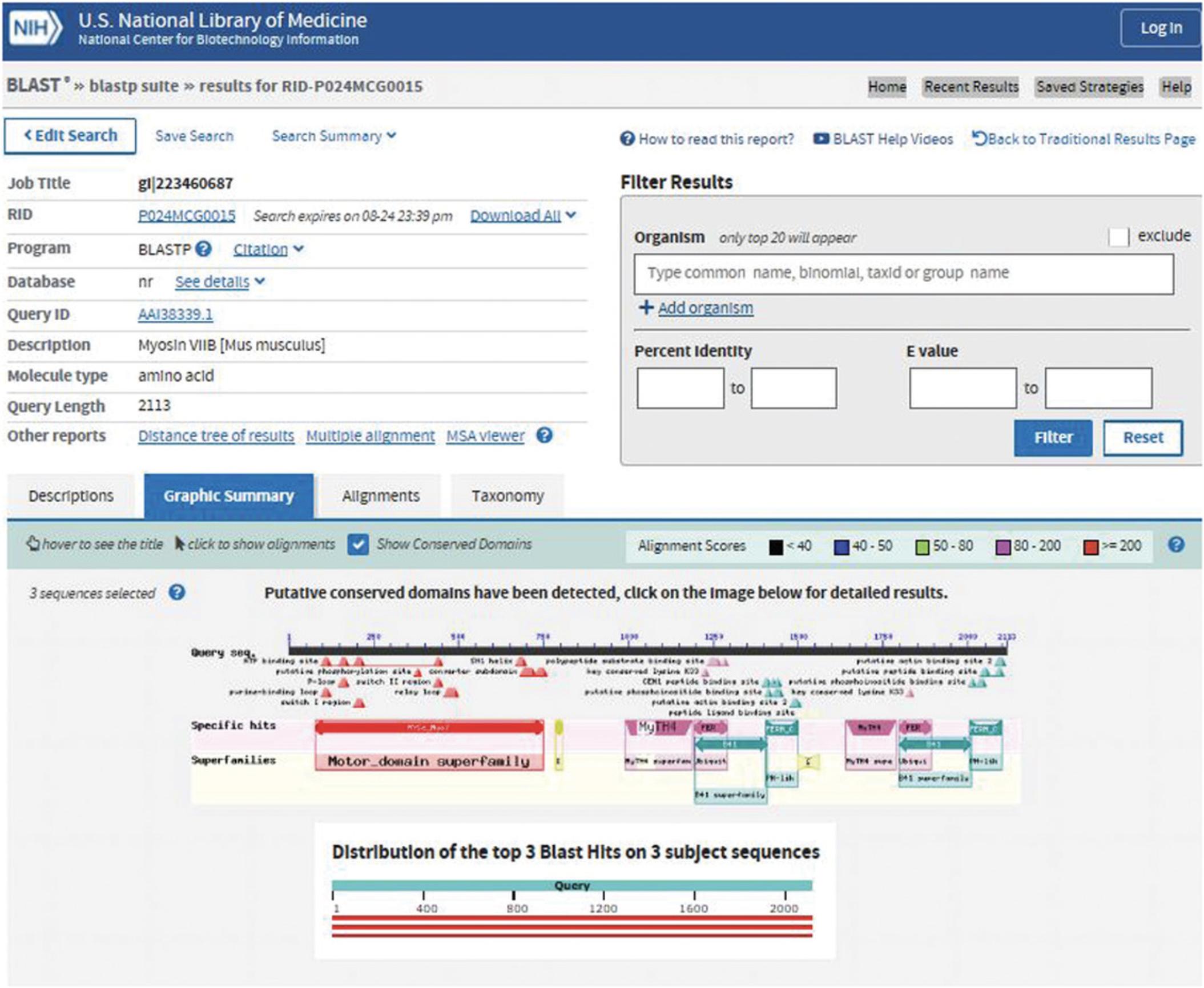
The NCBI's CD-Search service (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi; Figure 2) allows users to query a nucleotide or protein sequence against the CDD database via a sequence identifier or by pasting in the sequence in FASTA or raw text format. For the majority of queries provided as valid sequence identifiers, the default CD-Search settings display results of pre-computed RPS-BLAST searches (storing up to 500 hits each) that were run against the entire CDD database—including CDs curated by CDD staff along with additional sources from Pfam (Finn et al., 2016), SMART (Letunic et al., 2014), KOG (Tatusov et al., 2003), COG (Tatusov et al., 2001), PRotein K(c)lusters (PRK; Klimke et al., 2009) and TIGRFAMs (Haft et al., 2013)—at an E -value threshold of 0.01.The results are displayed by default in a concise format that shows the best-scoring domain model for each region of the query sequence plus the associated domain superfamily. If a region is annotated by a model that does not score well enough to be classified as a “specific hit,” only the superfamily annotation is shown. Default CD-Search parameters employ a score adjustment to address compositional bias, which largely abolishes the need to mask out low-complexity regions. Basic Protocol 1 demonstrates how to identify protein domains for a single nucleotide or protein sequence.
Necessary Resources
Hardware
Workstation with Internet access
Software
Web browser
Files
Protein sequence in FASTA format, accession number, or gi (GeneInfo) number
1.Open the protein sequence search page: https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi (see Figure 2).
2.In the text box, type the accession or gi number, or paste in the sequence of your protein or nucleotide of interest, in FASTA format.
3.To run the search with the default settings, press the Submit button.
4.View the results as they appear in HTML format (see Figure 3).
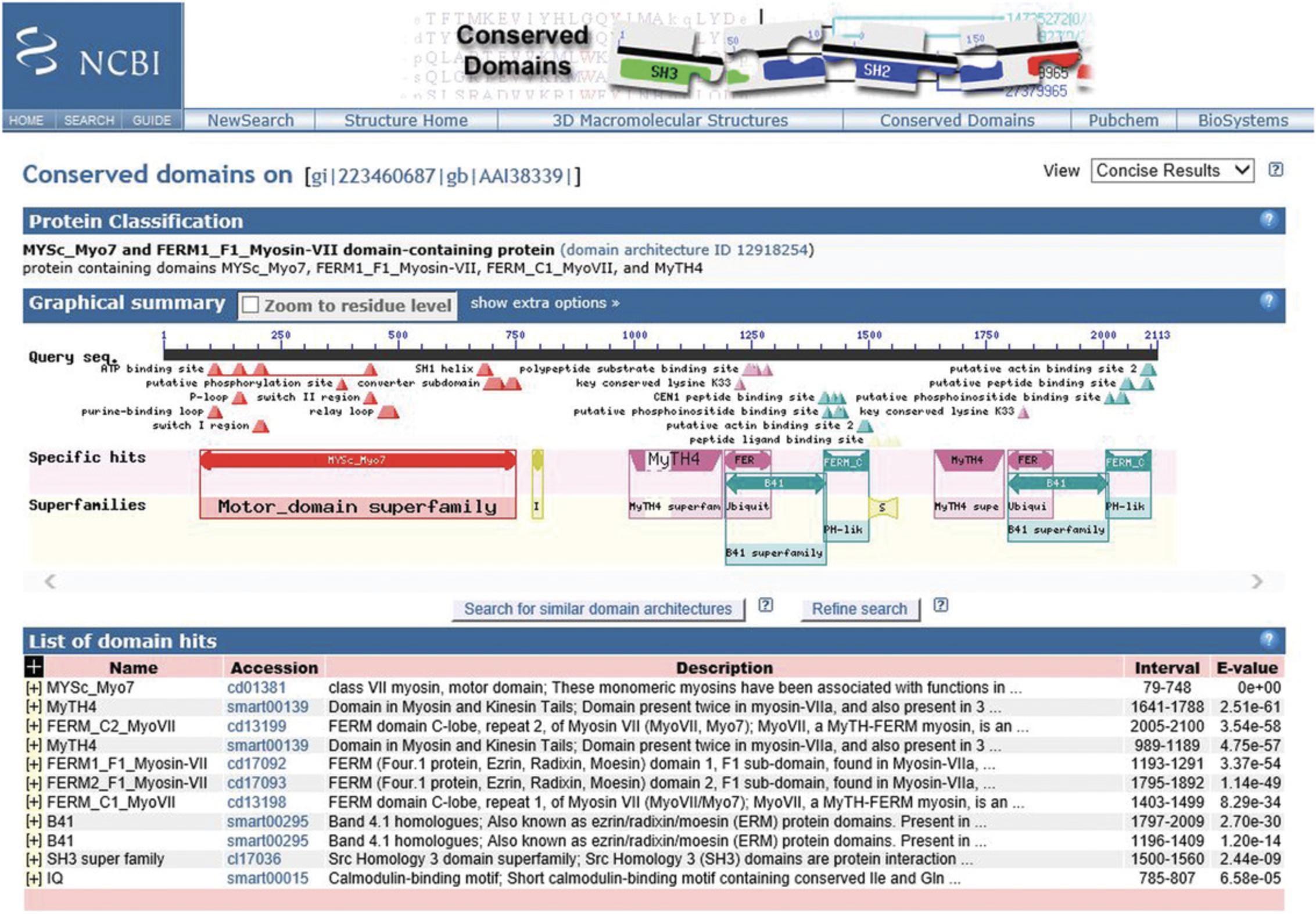
5.Select the scope of your graphical summary display by going to the top right-hand corner of the display and using the View pulldown menu to select either Concise Results , Standard Results , or Full Results.
6.Scroll over the annotations marked by triangles under the Query sequence in the Graphical Summary to reveal a pop-up window with information about a functional feature mapped to the query sequence via a domain hit. The pop-up window links to a CD summary page, which shows the multiple sequence alignment of protein sequences used to curate the model, annotated with hash marks denoting the location of the conserved feature residues, and providing the option to examine evidence supporting the feature.
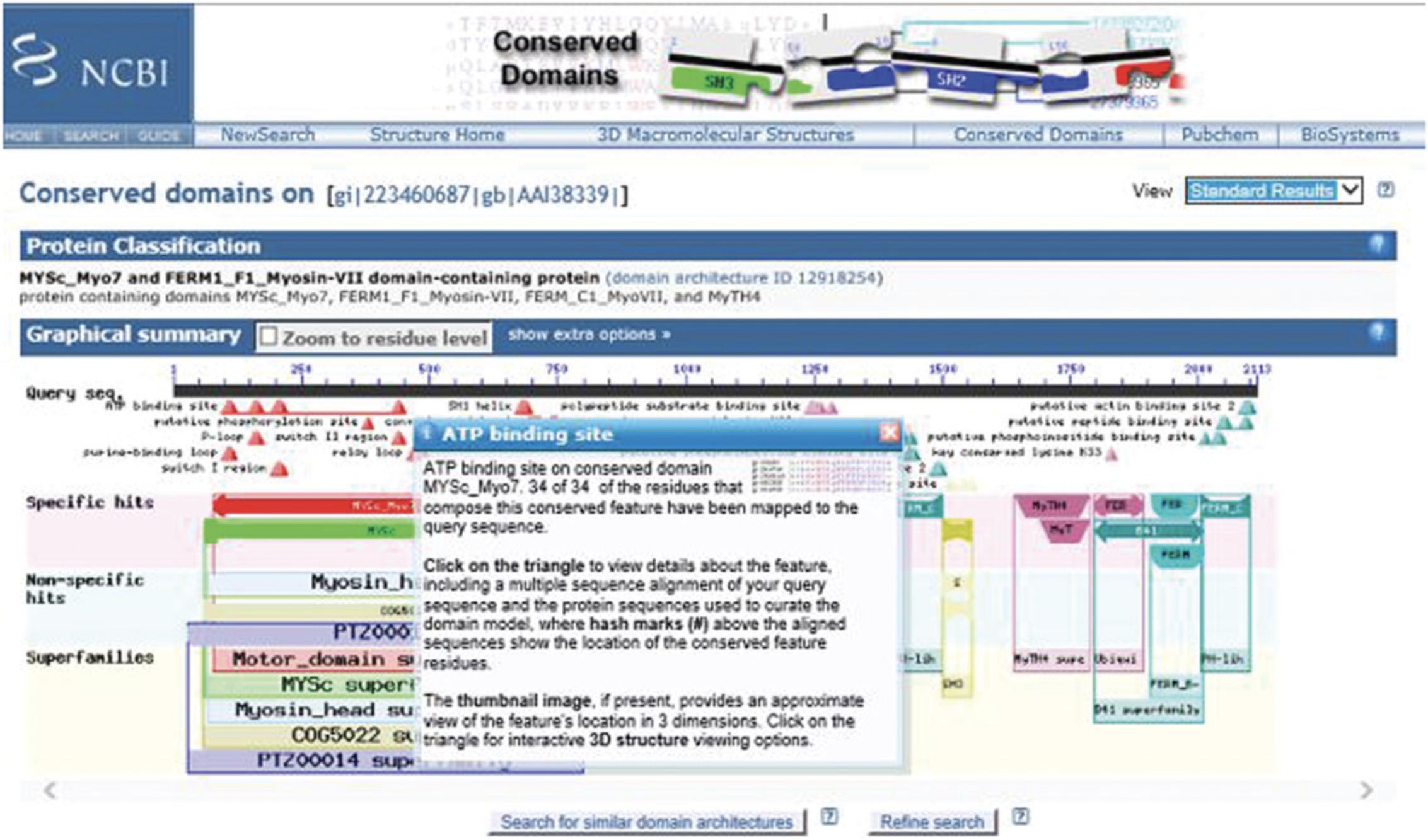
7.Scroll over the cartoon of the CD domain to reveal a pop-up panel showing the E -value, accession ID, name, and description. This also highlights the corresponding domain hit (shown in green) in the List of domain hits.
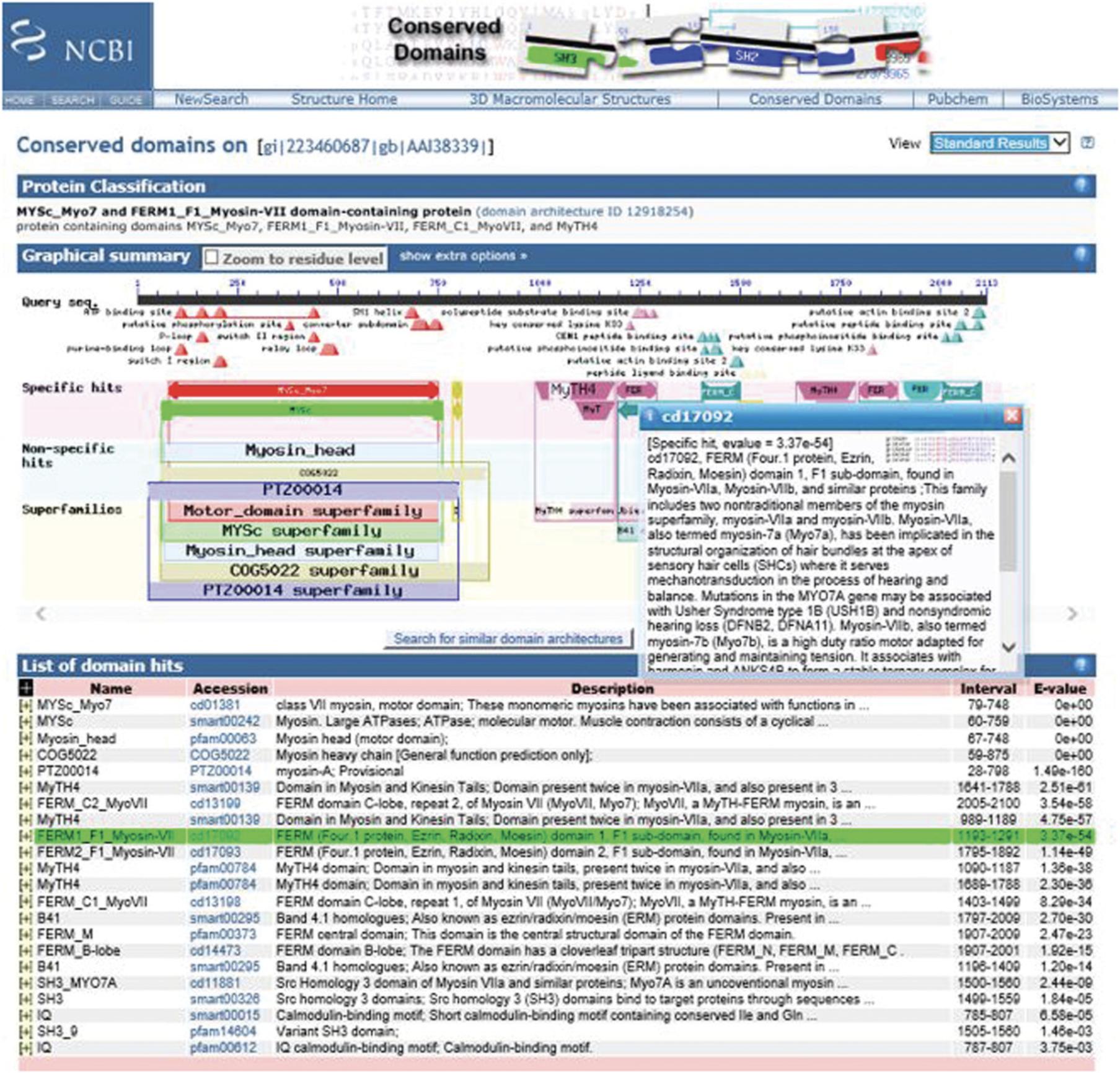
8.Click on the plus [+] in the List of domain hits to see how your query is aligned with the domain model.

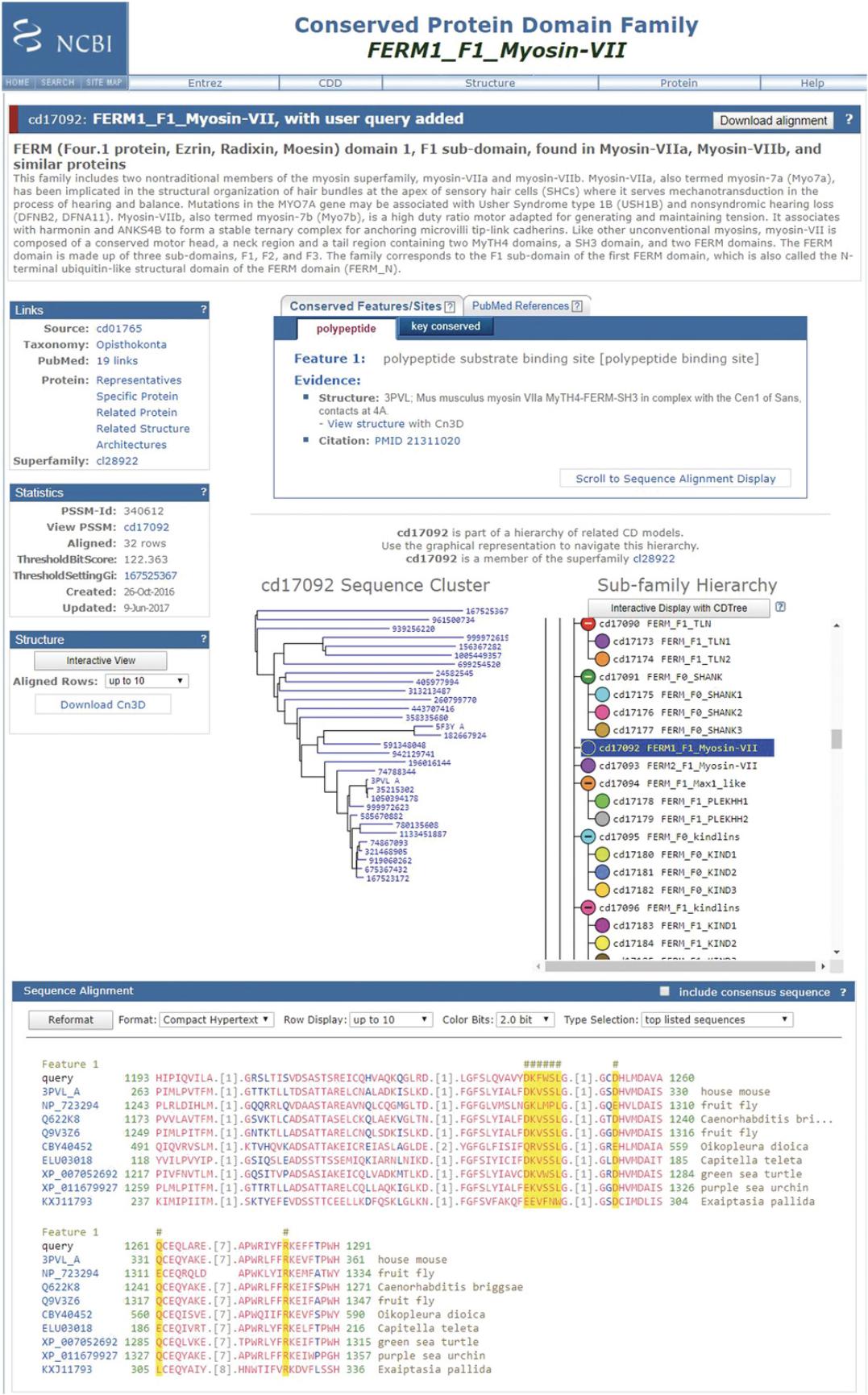
9.To launch and view the CD summary page on your domain of interest, click on the CD link in the List of Domain Hits, and click on the cartoon “bubble” of the CD of interest or on the symbols (triangles) indicating the location of feature annotations. Invoking the CD summary pages via links from the Graphical Summary will result in your query imbedded into the sequence alignment on the CD summary page.
Basic Protocol 2: BATCH CD-SEARCH
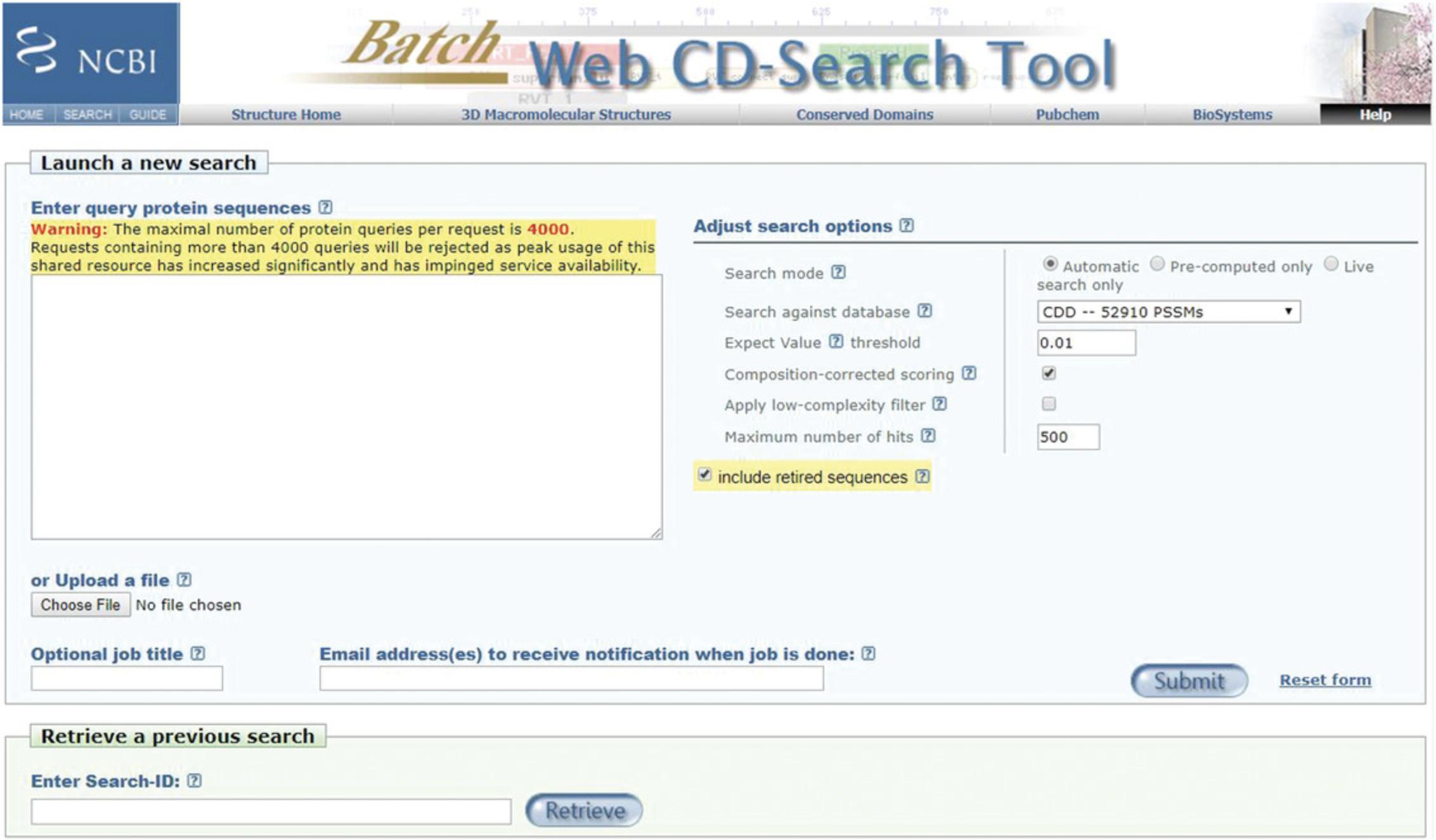
Use Batch CD-Search (https://www.ncbi.nlm.nih.gov/Structure/bwrpsb/bwrpsb.cgi) to compute and retrieve domain annotations for a batch of protein queries. Basic Protocol 2 demonstrates how to identify protein domains for a batch of protein queries up to 4000 sequences. The limits may be adapted in the future due to the high peak usage of this shared resource.
Necessary Resources
Hardware
Workstation with Internet access
Software
Web browser
Files
- A list of protein sequences in FASTA format, raw text format (lines of sequence data, without the FASTA definition line), accession number, or gi (GeneInfo) number, and separated by line breaks; different query types can be mixed in a single Batch CD-Search (for more detailed information on input format, consult the Batch CD-Search Help documentation, at https://www.ncbi.nlm.nih.gov/Structure/cdd/cdd_help.shtml#BatchRPSBInput; we provide a test set of 1348 sequences named “MYCs Myosin motor domain cd00124 sequences” in the Supplementary Materials)
- The Batch CD-Search page (https://www.ncbi.nlm.nih.gov/Structure/bwrpsb/bwrpsb.cgi; Figure 9)
Run the search
1.Enter your list of query proteins directly into the text box, or upload the list as a text file.
2.Add a title to your job in the Optional job title text box.
3.Input your email address(s) in the Email address(es) text box, so that you will be notified when the job is complete.
4.Run Batch CD-Search by pressing the “Submit ” button or hitting Enter.
5.View the Preliminary Results. If the search has been successful, a preliminary web page will be returned displaying the message “Search completed successfully” and with Sample data.
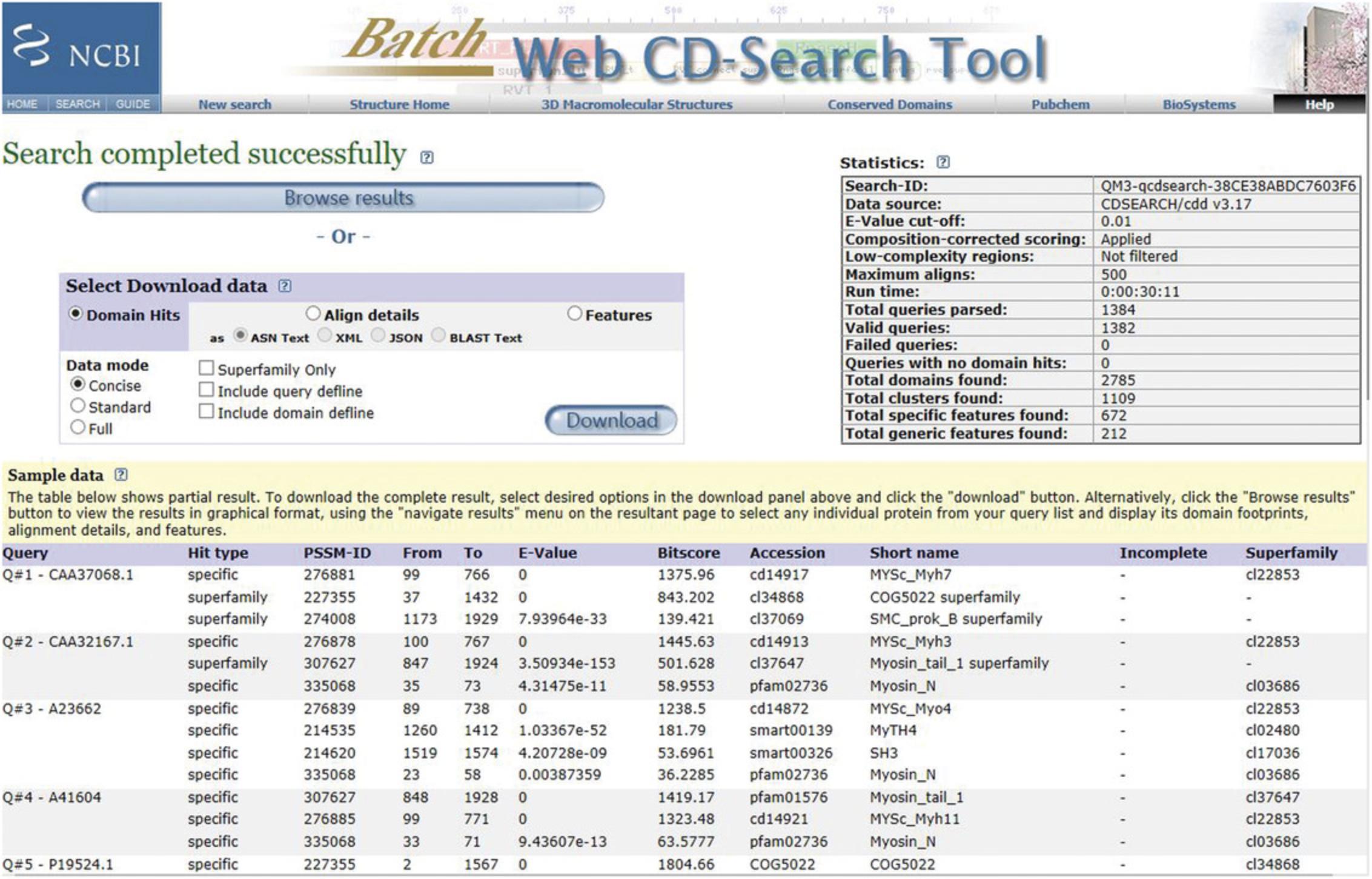
6.Save the Search-ID : Save the complete Search ID string found at the top of the Statistics box to access the complete results (master data structure) for up to 2 days after the search is first run.
7.Browse the complete results (master data structure).
Browse results
8.Press the Browse results button on the Preliminary Results web page.
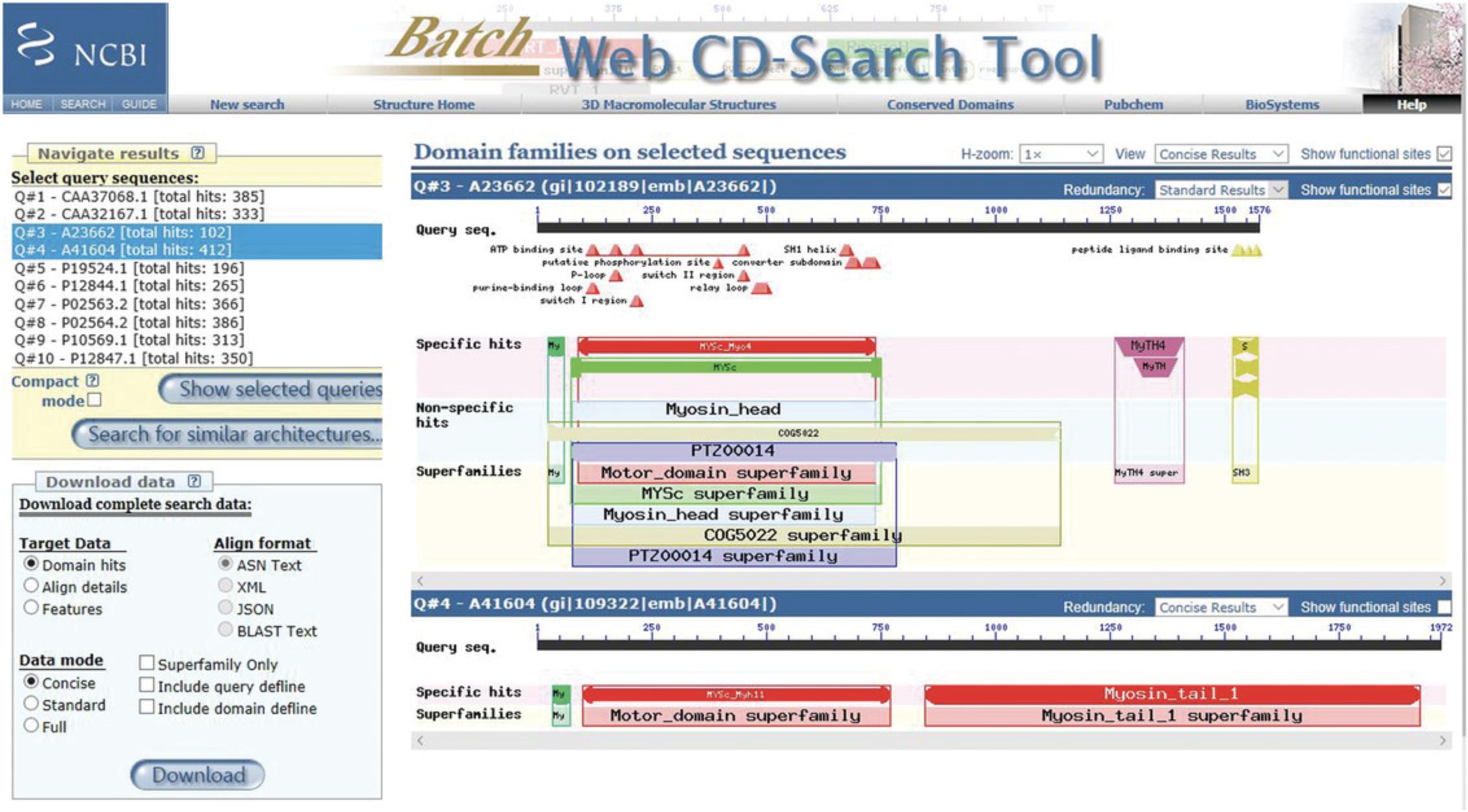
9.Browse and compare multiple results. In the Navigate Results panel, select multiple query sequences by holding down the keyboard Ctrl key and using the keyboard arrow keys to scroll through the query list. Then press the Show selected queries button to display your selections.
10.To view results in Compact mode, in the Navigate Results panel, check the Compact Mode box, and then press the Show selected queries button.
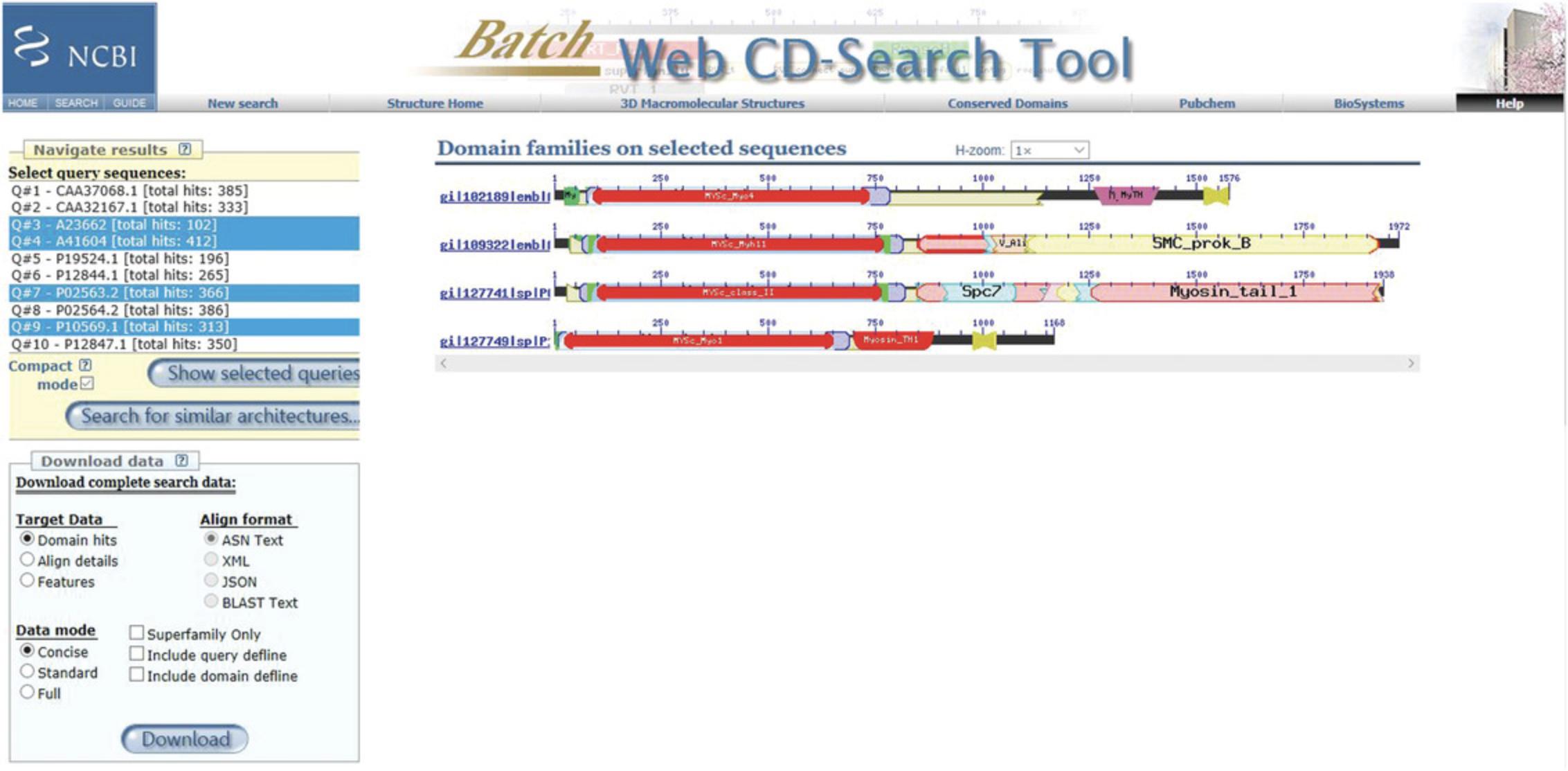
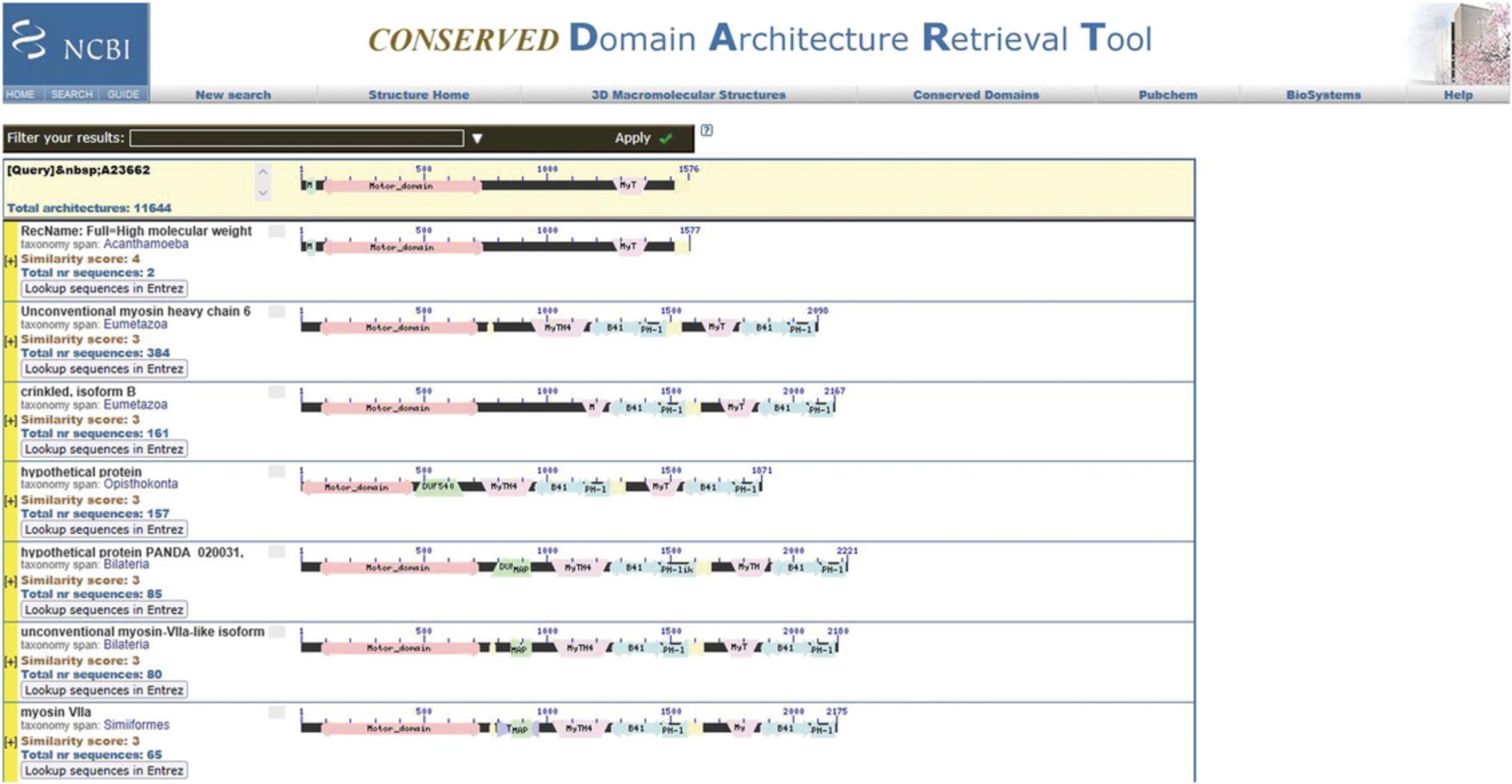
11.To search for similar architectures, in the Navigate Results panel, select a query sequence, and then press the Search for similar architectures button.
Download the data
12.From the Browse results page , you can download three categories of Target Data : Domain hits, Align details, and Features.
13.To download Domain Hit Data, on the Browse results page , select the Download data panel, with the default setting (Target data : Domain Hits and Data mode : Concise), and press the Download button.
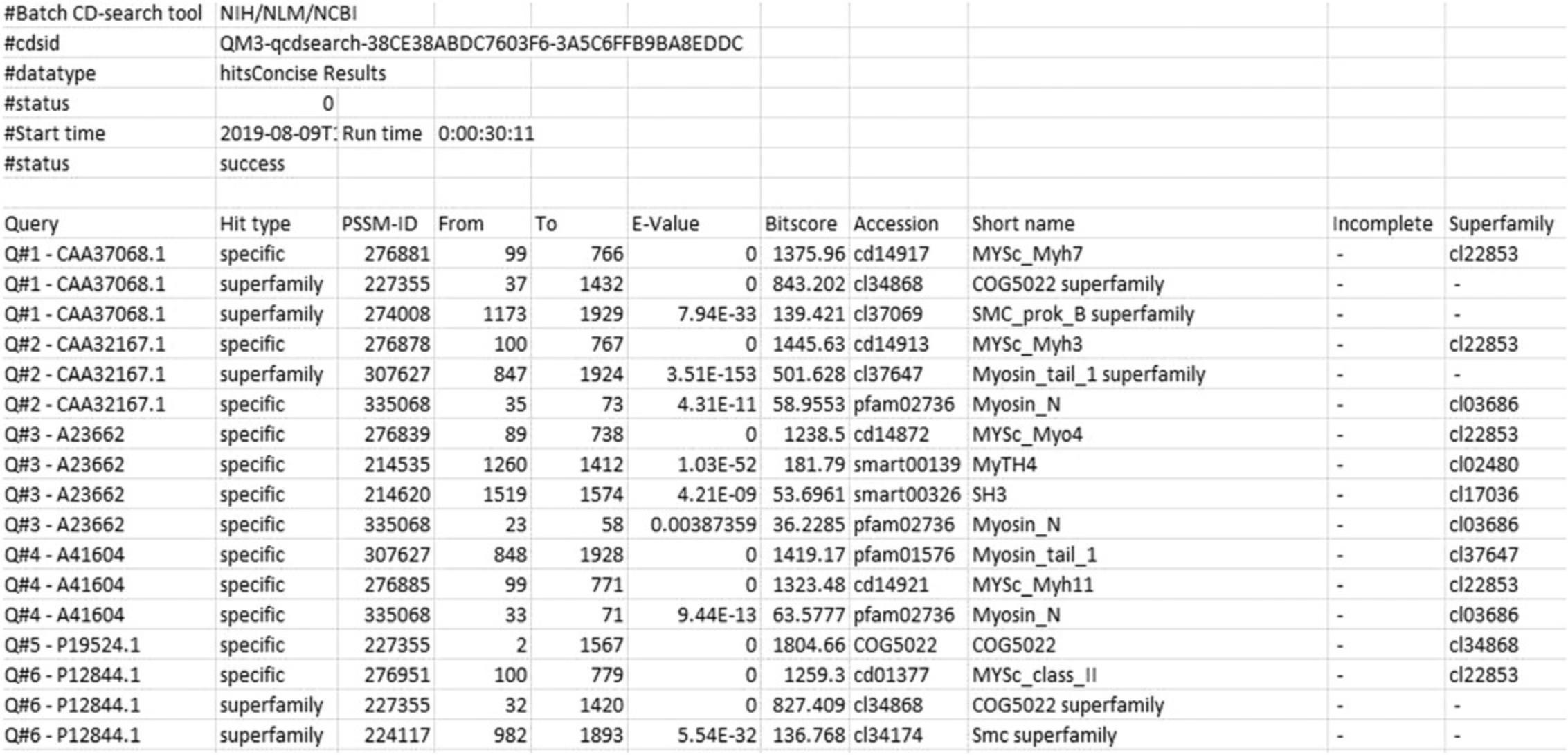
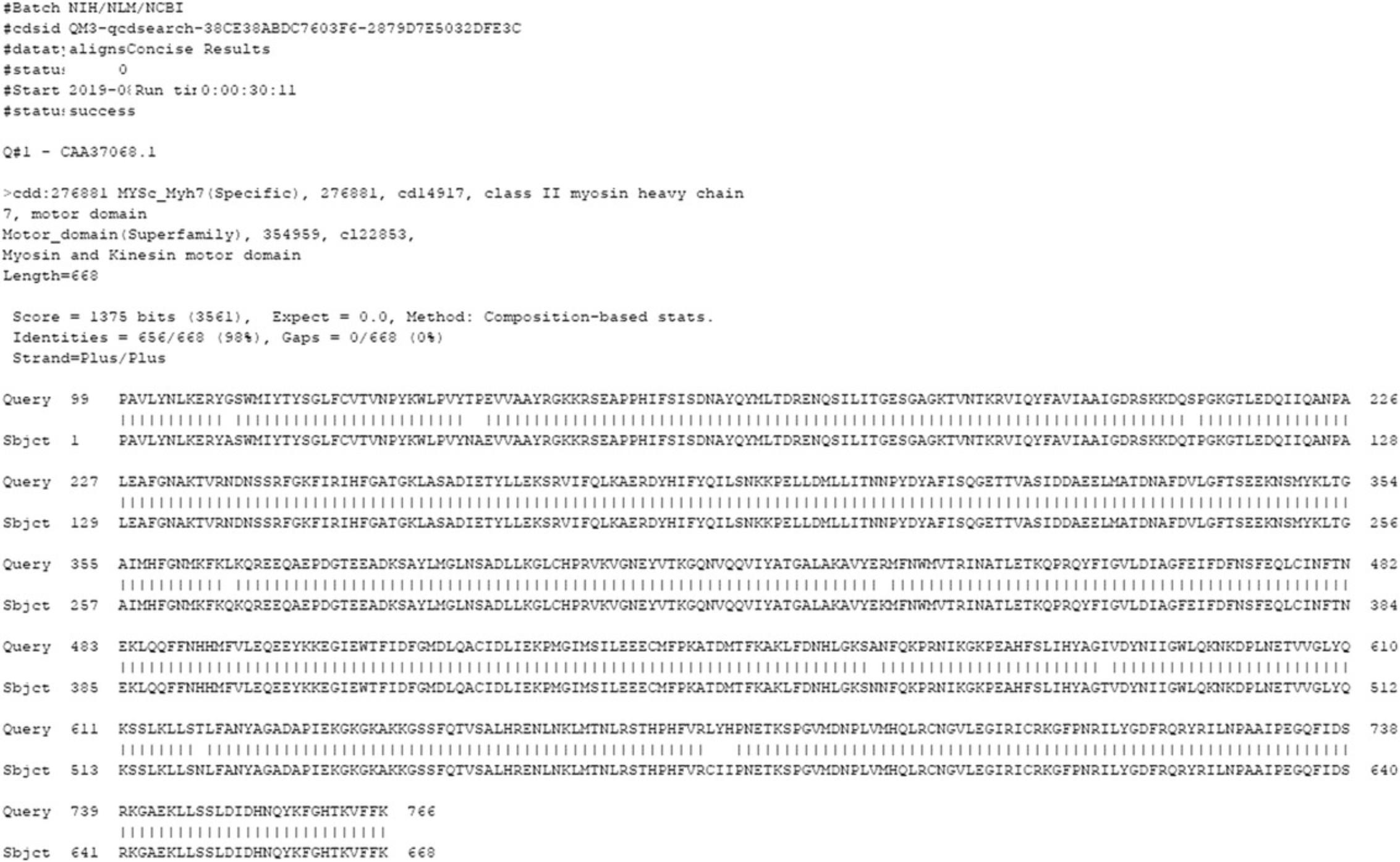
14.To download Alignment details data, from the Browse results page , in the Download data panel, with appropriate settings (Target Data : Align details; Align format : BLAST text; and Data mode : Concise), press the Download button.
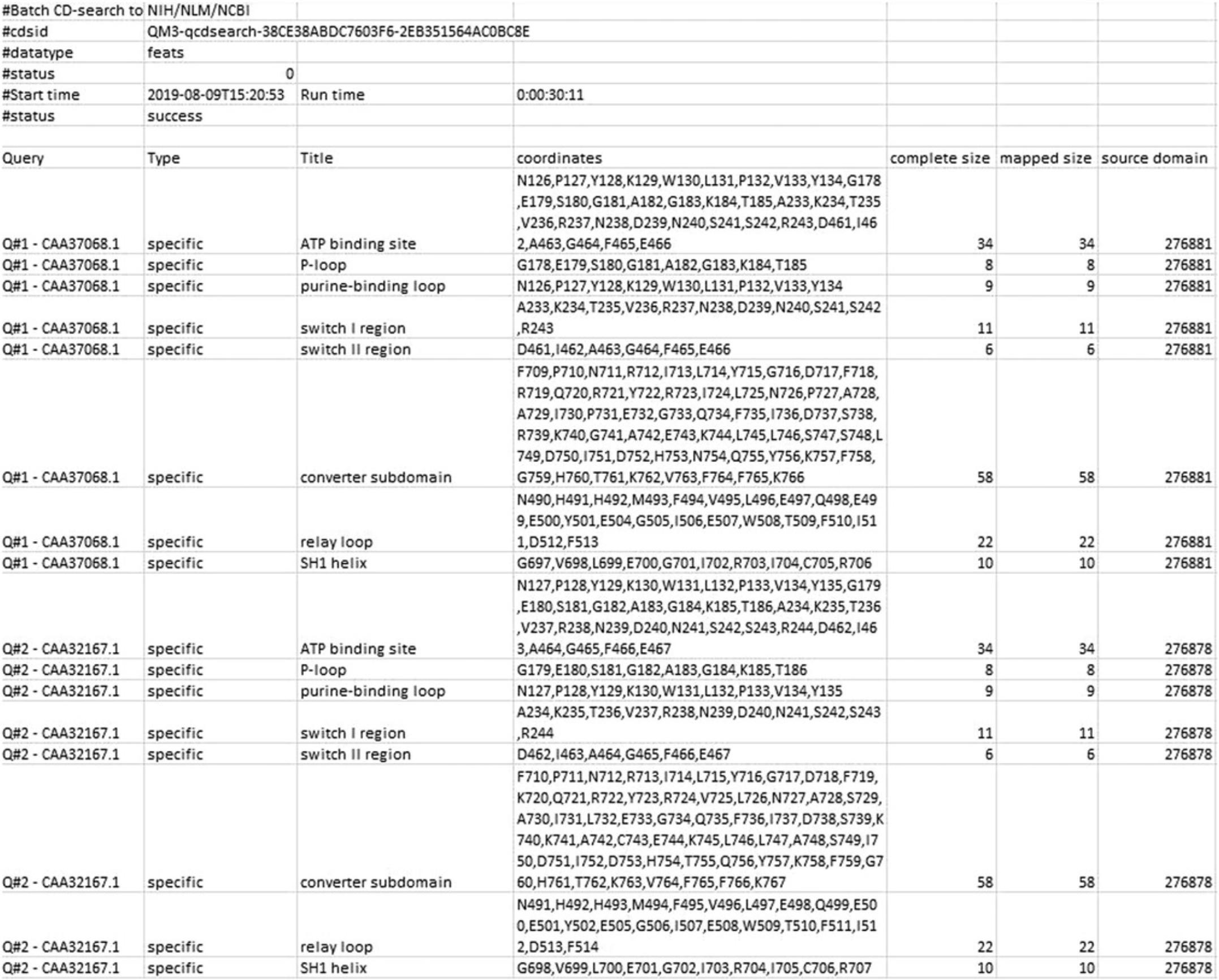
To download Features Data, open the Download data panel in the Browse results page , with appropriate settings (Target data : Features, Align format : ASN Text, and Data mode : Concise), and press the Download button.
Basic Protocol 3: STANDALONE RPS-BLAST AND rpsbproc
Use Standalone RPS-BLAST and rpsbproc (https://ftp.ncbi.nih.gov/pub/mmdb/cdd/rpsbproc/e) to compute and retrieve domain annotation programmatically. Basic Protocol 3 demonstrates how to identify protein domains for a batch of protein queries of greater than 4000.
Necessary Resources
Hardware
An internet-connected Linux, Windows, or Mac workstation
Software
- Web browser, for downloading files from FTP site
- The tar utility, to extract files from compressed archive files: A built-in utility for the Linux, Windows, and Mac platforms, found in Shell (Linux), Windows Command Processor (Windows), and Terminal (Mac), respectively
- The gzip utility, required to decompress files: For the Linux and Mac platforms, commonly a built-in utility by default; for the Windows platform, the specified software, including 7-Zip, WinZip, and others, can be used
- The curl utility, for downloading files from FTP site (optional): For the Linux platform, commonly installed by default; for Windows and Mac platforms, can be downloaded from https://curl.haxx.se/download.html and installed manually
- Specific FTP software, for downloading files from FTP site more efficiently (optional): e.g., FileZilla
Files
Input queries in FASTA format: i.e., protein or nucleotide sequences
Preliminary Steps
Detailed instructions on how to retrieve the RPS-BLAST executable and rpsbproc utility and run them locally can be found in the rpsbproc README file at the CDD FTP site (https://ftp.ncbi.nih.gov/pub/mmdb/cdd/rpsbproc/README).
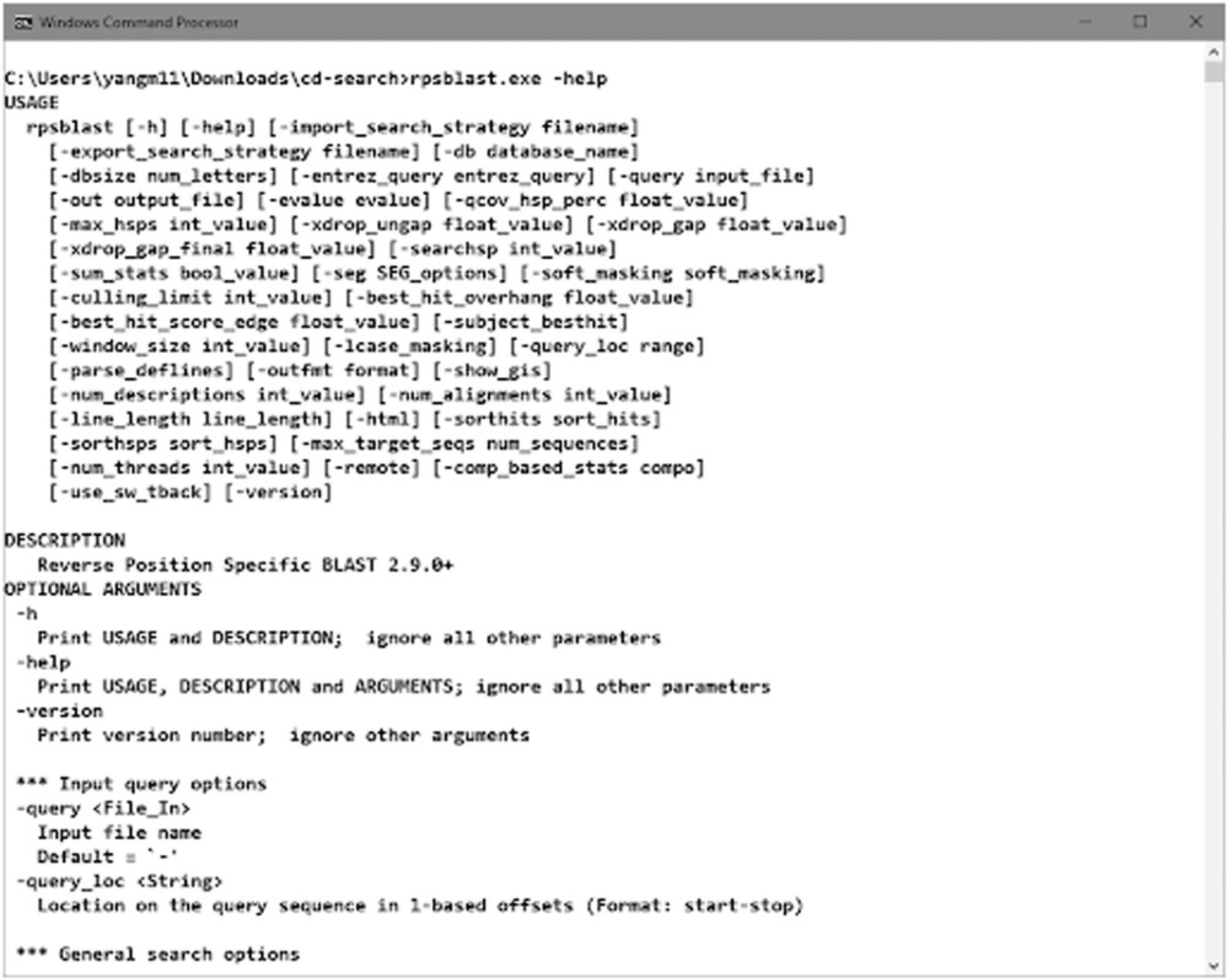
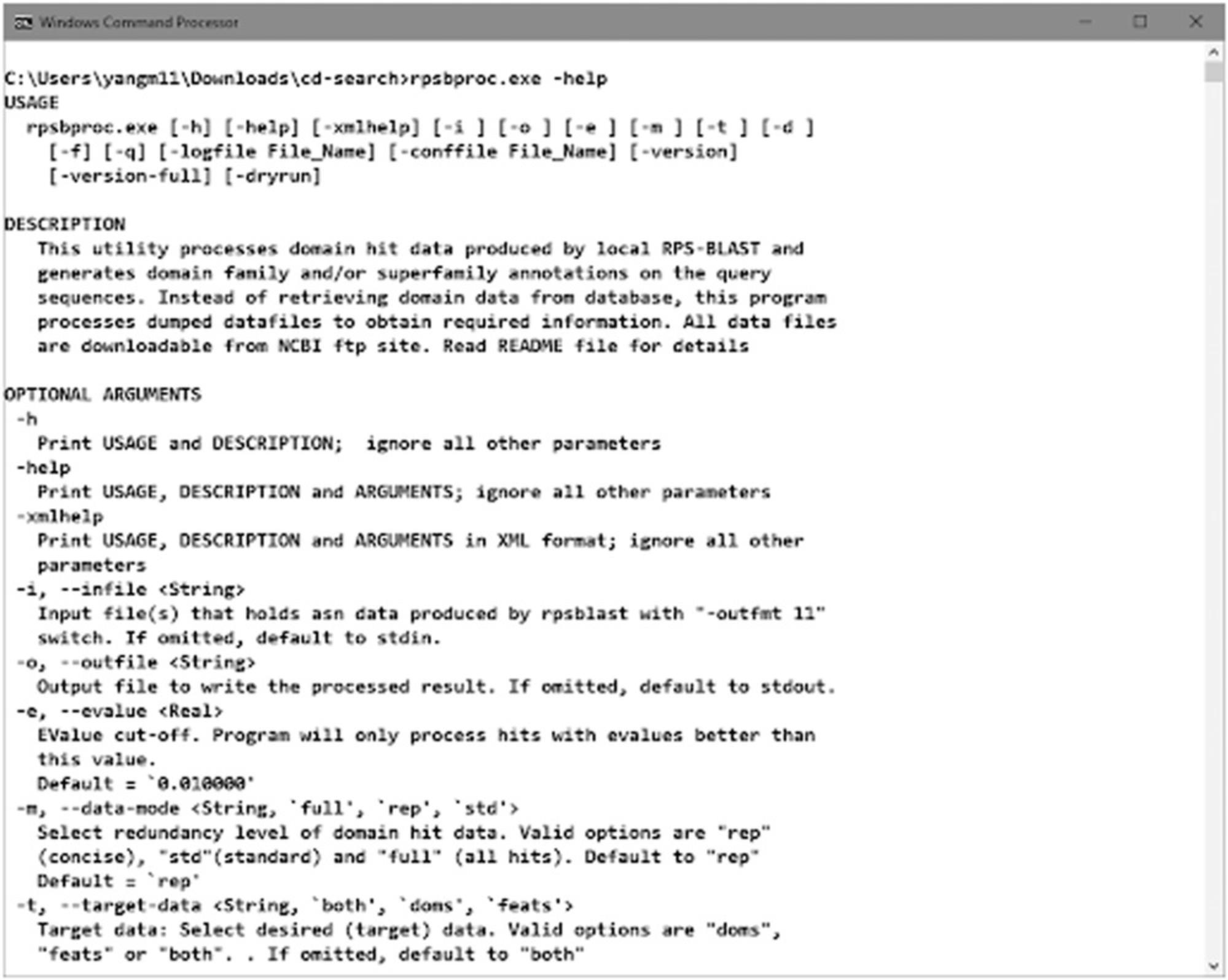
The standalone RPS-BLAST packaged with the pre-built BLAST executables (“rpsblast” for protein queries and “rpstblastn” for nucleotide queries) is available at the NCBI BLAST FTP site and as part of the NCBI C++ toolkit distribution. Detailed documentation for BLAST at NCBI, including RPS-BLAST, can be found in BLAST® Command Line Applications User Manual (https://www.ncbi.nlm.nih.gov/books/NBK279690/). Run the command rpsblast with argument “-help” to check the usage information (Figure 17).
For each query sequence, standalone RPS-BLAST lists the conserved domain models that scored below a certain E -value threshold (by default set to 10), sorted by E -value. For each hit, information such as the conserved domain's PSSMID, a set of scores (E -value, BitScore, etc.), and the sequence alignment between the conserved domain and the query sequence can be returned. In order to run the rpsbproc utility, the output file generated by RPS-BLAST executables needs to be stored in ASN.1 format, using “.asn” as the filename extension.
The rpsbproc command line utility is an addition to the standalone version of RPS-BLAST. It post-processes the RPS-BLAST output to give a compact and nonredundant view of the search results (such as would be returned by the Batch CD-Search). rpsbproc reads the output of rpsblast/rpstblastn and fills in domain superfamily and functional site information, as well as structural motifs, for each region of the sequence. It then re-sorts the hits and calculates a set of nonredundant representative hits. The result is presented in a tab-delimited flat file and can be looked at either programmatically or manually. Run rpsbproc command with argument “-help” to check the usage information (Figure 18).
To run RPS-BLAST locally and use rpsbproc to process the output, you must first collect the applications needed. You can download the pre-built rpsblast, rpstblastn, and rpsbproc binaries from the NCBI FTP site, which are directly executable on Windows and Linux platforms, with no complex installation required. For those who need (or desire) to build these utilities locally, you can download the source code tarballs from the NCBI FTP site. Please note that these programs are NCBI C++ toolkit applications and require the NCBI C++ toolkit to build. Please follow the README file to build these utilities locally. For Linux and Mac users, please refer to the rpsbproc README file for detailed instruction to run standalone RPS-BLAST and rpsbproc utility. Below are step-by-step instructions for running these executables on a Windows platform.
Procedure
1.Download the rpsbproc README file (https://ftp.ncbi.nih.gov/pub/mmdb/cdd/rpsbproc/README) to your project folder for reference in the following steps.
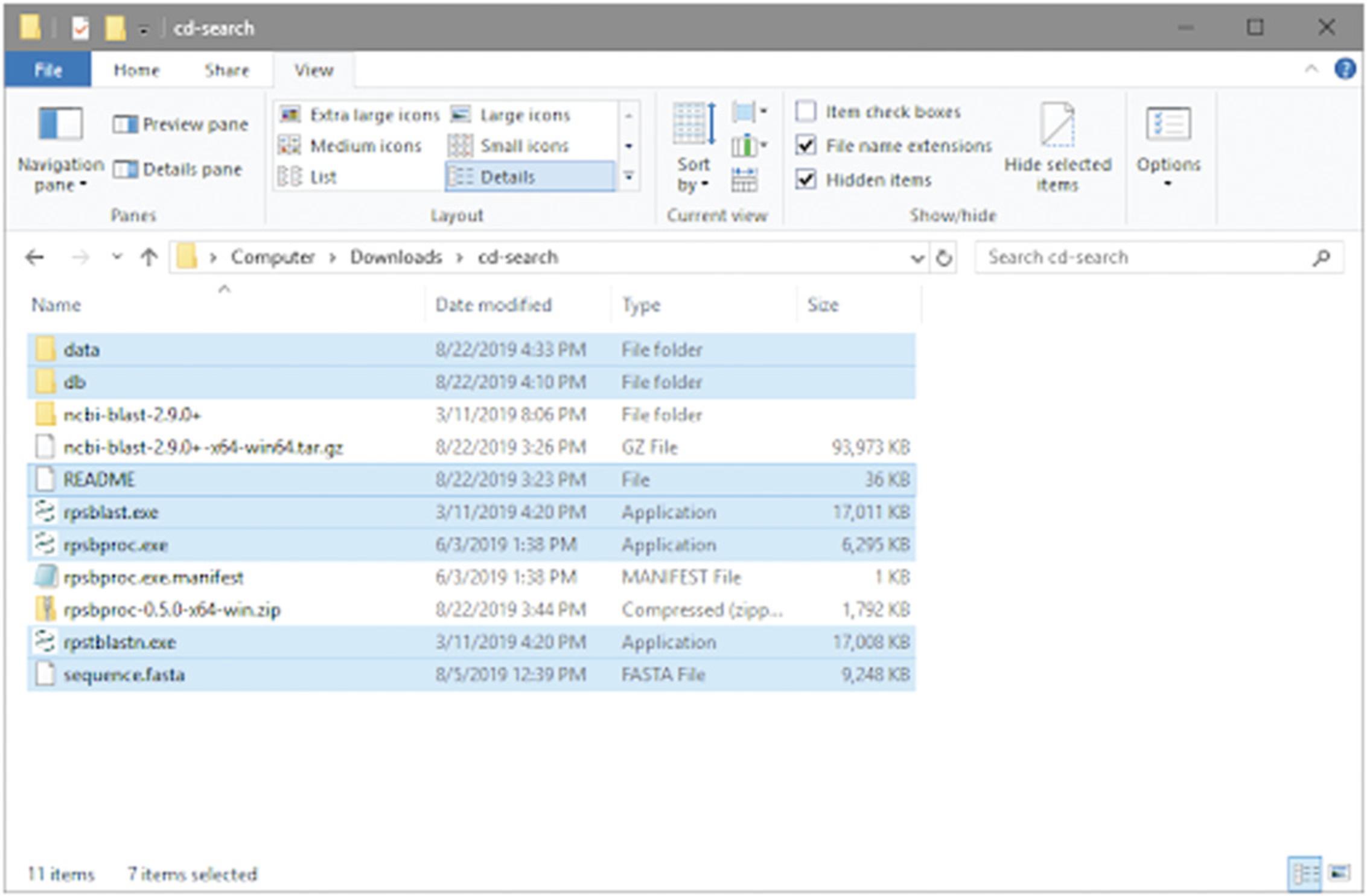
2.Retrieve the RPS-BLAST executable by downloading the RPS-BLAST executable (ncbi-blast-2.9.0+-x64-win64.tar.gz) to the project folder from the NCBI BLAST FTP site (https://ftp.ncbi.nih.gov/blast/executables/LATEST/). Then, open the Windows Command Processor (cmd.exe) and navigate to the project folder to run the command below to uncompress the downloaded file, which creates a folder named ncbi-blast-2.9.0+ in the project folder.
- tar -zxf "ncbi-blast-2.9.0+-x64-win64.tar.gz"
Navigate to the bin sun-folder in ncbi-blast-2.9.0+, and copy the executables rpsblast.exe and rpstblastn.exe to the project folder.
3.Retrieve the rpsbproc executable by downloading the executable (rpsbproc-0.5.0-x64-win.zip) to the project folder from the NCBI CDD FTP site (https://ftp.ncbi.nih.gov/pub/mmdb/cdd/rpsbproc/). In the Windows Command Processor, navigate to the project folder and run the command below to uncompress the zip-file downloaded.
- tar -zxf rpsbproc-0.5.0-x64-win.zip
Now you have rpsbproc.exe and rpsbproc.exe.manifest files in the project folder.
4.Create the search database for RPS-BLAST by downloading the preformatted search database (files) from the CDD FTP site (https://ftp.ncbi.nih.gov/pub/mmdb/cdd/little_endian/) to the folder named db under the project folder. Uncompress the files separately in the current db directory using the commands below:
- tar -zxf Cdd_LE.tar.gz
- tar -zxf Cdd_NCBI_LE.tar.gz
- tar -zxf Cog_LE.tar.gz
- tar -zxf Kog_LE.tar.gz
- tar -zxf Pfam_LE.tar.gz
- tar -zxf Prk_LE.tar.gz
- tar -zxf Smart_LE.tar.gz
- tar -zxf Tigr_LE.tar.gz
5.Create the data folder by downloading the domain-annotation files (listed below) from the CDD FTP site (https://ftp.ncbi.nih.gov/pub/mmdb/cdd/) to the data folder.
- bitscore_specific.txt
- cddannot.dat.gz
- cddannot_generic.dat.gz
- cddid.tbl.gz
- cdtrack.txt
- family_superfamily_links
6.Put your FASTA file containing query sequences into the project folder. sequence.fasta was used in this example.
7.Run RPS-BLAST by opening the Windows Command Processor (cmd.exe). Navigate to the project folder and run RPS-BLAST using the command below. Backslashes are used because this command is run on a Windows command processor.
- rpsblast.exe -query sequence.fasta -db .\db\Cdd -evalue 0.01 -outfmt 11 -out sequence.asn
8.Run the rpsbproc executable using the command below to annotate the results generated by RPS-BLAST.
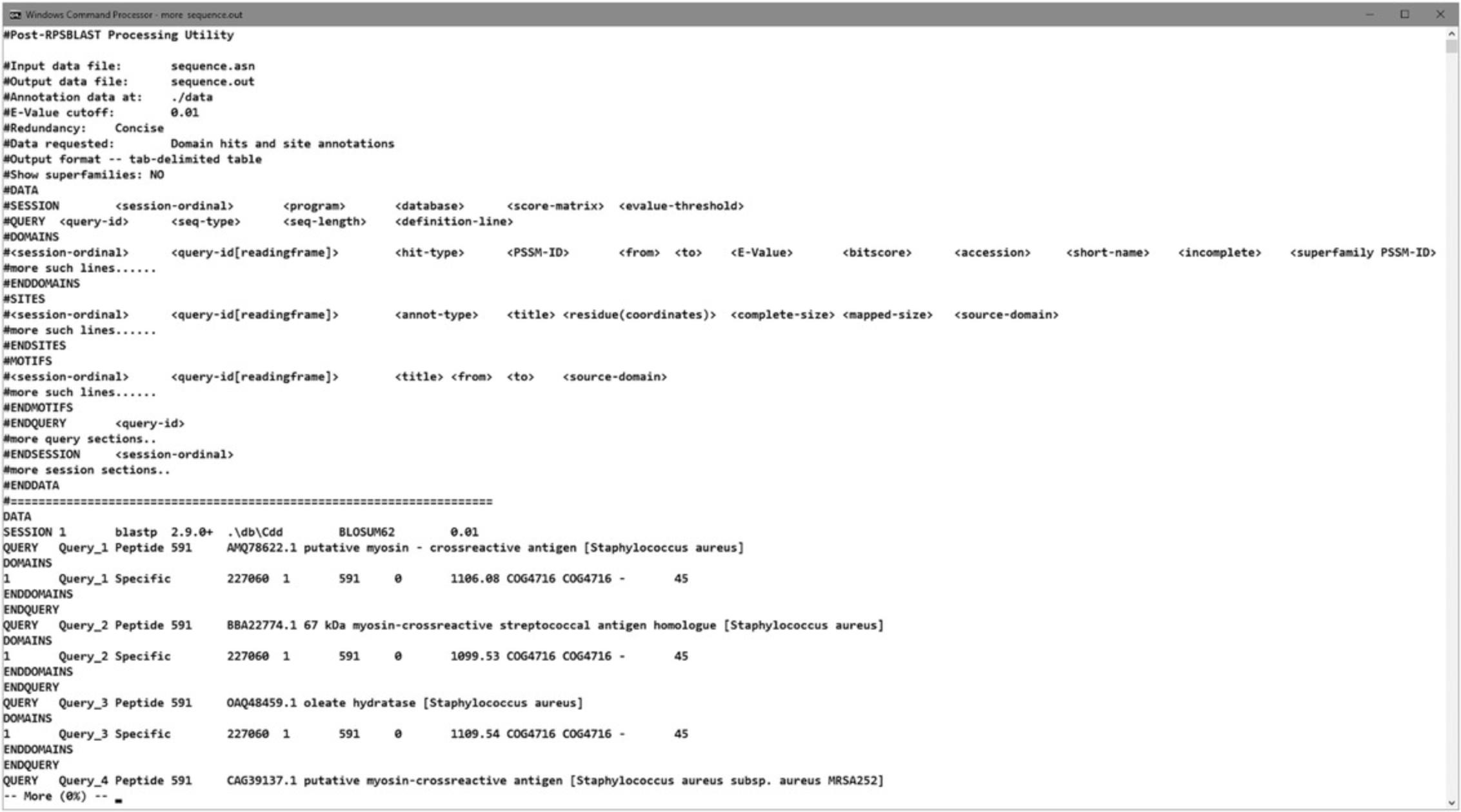
- rpsbproc.exe -i sequence.asn -o sequence.out -e 0.01 -m re
9.View the results. The output file has a tab-delimited format and can be opened with WordPad, Excel, or similar editors.
GUIDELINES FOR UNDERSTANDING RESULTS
Basic Protocol 1
CD-Search allows users to query a nucleotide or protein sequence against the CDD database via its accession number or gi number, or by pasting in the sequence in FASTA or raw text format using RPS-BLAST. The CDD database includes CDs curated in house by the NCBI along with additional sources from Pfam, SMART, KOG, COG, PRK, and TIGRFAM. The results are displayed by default in a concise format that shows the best-scoring domain model for each region of the query together with the corresponding domain superfamily, and the superfamily annotation only if the hit was not strong enough to be classified as specific (high confidence).
The resulting CD-Search results display contains three sections: Protein classification, Graphical summary, and List of domain hits for the query. At the top, above the protein classification section, it shows the query as well as the view that is currently being used (Concise Results, Standard Results, or Full Results). The CD Summary page can be launched from either the Graphical summary or the List of domain hits and contains detailed information about your domain of interest.
Section 1: Protein Classification
The Protein classification section displays a suggested name for the query protein, a label that may specify a suggested function, and a link to the SPARCLE (Subfamily Protein Architecture Labeling Engine; Marchler-Bauer et al., 2017) classification (Figure 3).
Section 2: Graphical Summary
The Graphical summary shows the domain hits and annotated features. Feature annotations are denoted by triangles colored the same as the domains they correspond to. The results mode display can be chosen in the CD Search panel or changed after the search is run by changing the selection in the View panel (see Figure 3). The standard display format shows the best-scoring domain model from each data source (best Pfam hit, best COGs hit, etc.). The full display format shows all matching domain models identified by RPS-BLAST for each region of the query sequence and can be very redundant. The display can be customized to hide the display of site annotation features by selecting the show extra options and deselecting the Show site features , as well as magnifying the display using the Horizontal zoom and the Zoom to residue level selections.
Hovering over the triangles in the site features triggers a pop-up window with information on the number of feature residues that map to the query sequence. Clicking on the triangle takes you to the CD summary page, where your query is embedded into the CD alignment with the residues involved in the site features highlighted and marked with hash marks (#).
Hovering over the domains triggers a pop-up window with a description of the domain and highlights the corresponding row in the List of domain hits panel. Clicking on the domain graphic also takes you to the CDD page with your query embedded into the CD alignment.
Section 3: List of Domain Hits
The List of domain hits lists the conserved domains identified on the query sequence. For each conserved domain identified, it displays its short name, its accession number, a description of the domain, the interval on the query that is covered by the domain footprint, and the E -value. Clicking on the (+) next to each name reveals the full description of the domain and shows the alignment of the query sequence to a representative (consensus) sequence of the domain model, together with the numerical domain model identifier (PSSM-ID) and the alignment bit score. Click on the domain model's accession number to view the multiple sequence alignments of the proteins used to develop the corresponding domain model. Note that your query sequence is not embedded in this version of the CD summary page.
If a live search was performed, the BLAST Request ID (RID) is shown at the bottom of the Standard and Full displays and allows you to retrieve the search results using the RID anytime within the 36 hr following the search, without having to re-execute it.
To change the search settings, click on the Refine Search button (which will retain your query) or select New Search from the selection bar immediately below the logo at the top of the page. Go to the OPTIONS panel. Use the Search against database option pulldown to select a specific database. Change the E -value to stricter or more permissive by changing the value in the Expect Value threshold option. If you would like to mask out compositionally biased regions, check Apply low-complexity filter (the graphical display of results will then highlight masked-out regions on the query). Composition based statistics adjustment , which is selected by default, abolishes the need to mask out compositionally biased regions in query sequences, for the most part. Keep both the Composition based statistics adjustment and the Apply low-complexity filter options on at the same time to filter out some false positives that may still slip through the cracks of the composition-correction, or click both of them off to find more distant relatives for compositionally biased queries. To perform a live search, check the Force live search box (it will be checked if you choose settings different from the CD-Search default). You can also Rescue borderline hits and Suppress weak overlapping hits by selecting the appropriate boxes (Derbyshire, Lanczycki, Bryant, & Marchler-Bauer, 2012).
CD Summary Page
At the top of the CD summary page, you will see the CD accession number and a description of the CD. Below this you may see a box with a tab for Conserved Features/Sites , which contains the name(s) of the annotated site(s), evidence of various types (structure evidence, PMID references to literature, and free-text comments), and a tab for PubMed References that lists relevant articles about the specific domain or protein family and more generic reviews of the wider superfamily. Annotation selections are highlighted in the Sequence Alignment panel and noted by hash marks at the very bottom of the page that show how the query sequence is aligned with respect to the CD sequences.
Below the Conserved Features/Sites and PubMed References panel, there is a Sequence Cluster tree of the CD that matched your query. If the domain is part of a hierarchical classification, you will also see a tree-like representation of that hierarchy, with the CD that matched your query highlighted with a dark blue background, as shown in Figure 7.
Click on the Interactive Display with the CDTree button after selecting to download the selected CD or the entire hierarchy for viewing and further analysis with the CDTree software package (https://www.ncbi.nlm.nih.gov/Structure/cdtree/cdtreeInstall.shtml).
The right hand side of the CD summary page contains information blocks titled Links (Source, Taxonomy, PubMed, Protein, and Superfamily), Statistics (PSSM-Id, View PSSM, Aligned, ThresholdBitScore, ThresholdSettingGi, Created and Update dates), and Structure information , where you can download Cn3d , a molecular structure and multiple sequence alignment viewer (https://www.ncbi.nlm.nih.gov/Structure/CN3D/cn3d.shtml) to visualize and manipulate the sequence and structure alignment for your query in the context of its CD hit.
Select the Interactive View after setting the number of aligned rows that you would like displayed in Cn3D. Upon launching, it displays three panels: a CDD Descriptive Items panel that shows some of the information found on the CD summary page (name, description, annotation, and references), a visualization window that shows the model's 3D structures if present, and a multiple sequence alignment window containing the query sequence embedded in the CD alignment.
Basic Protocol 2
Basic Protocol 2 can run CD-Search on a batch of up to 4000 proteins in a single request and accessed via a web service or programmatically. A single Batch CD-Search returns annotation data in a tabular form suitable for further processing, including domain hit from-to intervals, E -values and scores, domain model names and accessions, and the positions of functional sites such as catalytic residues, binding sites, and motifs. A wealth of information on your protein collection is returned in a single search.
When Browsing results, please find help for interpreting the graphical results for each individual protein in Basic Protocol 1 (single CD-Search): Guidelines for understanding the results: Graphical summary.
Basic Protocol 2 as described is run in the default mode. There are many options you may opt to modify.
The default search mode described is the automatic search , which either runs a live RPS-BLAST search or retrieves precalculated results for each single item on the list depending on its sequence format. For most query sequences specified via sequence identifiers, precalculated RPS-BLAST results are available and will be retrieved; if no results are available, a live search will be executed. For queries entered as FASTA or raw sequence, live searches will be run. You may opt for a Live search only mode, which runs a live RPS-BLAST search for every item on the query list, or a Pre-computed only mode, which only retrieves a precalculated RPS-BLAST result where available but will ignore other queries.
The default mode (automatic search) runs against the complete CDD database (i.e., includes the CDD in-house-curated models and those from external sources including Pfam, SMART, KOG, COG, PRK, and TIGRFAMs) at an E -value threshold of 0.01. The Search against database pull-down menu provides the option to limit your search to only the NCBI in-house-curated subset, or to any one of the other databases included in the CDD. The current version number for each database can be found at https://www.ncbi.nlm.nih.gov/Structure/cdd/docs/cdd_news.html. You also have the option to enter a different E -value threshold.
The default mode includes obsolete or preliminary sequences, and the output flags these as non-current. You may opt to exclude these by unchecking its box on the search page.
In the default mode, the Apply a low-complexity filter is turned off, but you may elect to turn it ON by checking its box on the search page to mask compositionally biased regions in the query protein sequences.
In the default mode, the Maximum number of hits returned is 500, as the number of expected domain hits is small for an average protein.
As the number of queries per Batch CD-Search run is limited, and as the maximum throughput of the resource is restricted by the number of servers available on the back end, you may opt to run searches locally on your own hardware. Basic Protocol 3 describes how to run standalone RPS-BLAST plus the rpsbproc command-line utility. It returns annotation in a tabular format similar to that of Batch CD-Search, suitable for further processing, and allows you to run RPS-BLAST with customized PSSM subsets.
Basic Protocol 3
Basic Protocol 3 runs standalone RPS-BLAST and rpsbproc to process a large amount of protein/nucleotide sequences, and returns annotation data similar to that of batch CD-Search (Basic Protocol 2), which include domain hits, site annotations, and structural motifs. Additionally, it allows you the option of running RPS-BLAST locally on your own machine and, optionally, with your own PSSM subsets.
The output file generated by the rpsbproc utility comprises two sections. The first section displays the program information, parameters used for data processing, and a “template” explaining the format and content of each column of the data table. All the lines in this section start with a “#” character so that programs can treat them as “comment” lines that can be safely ignored.
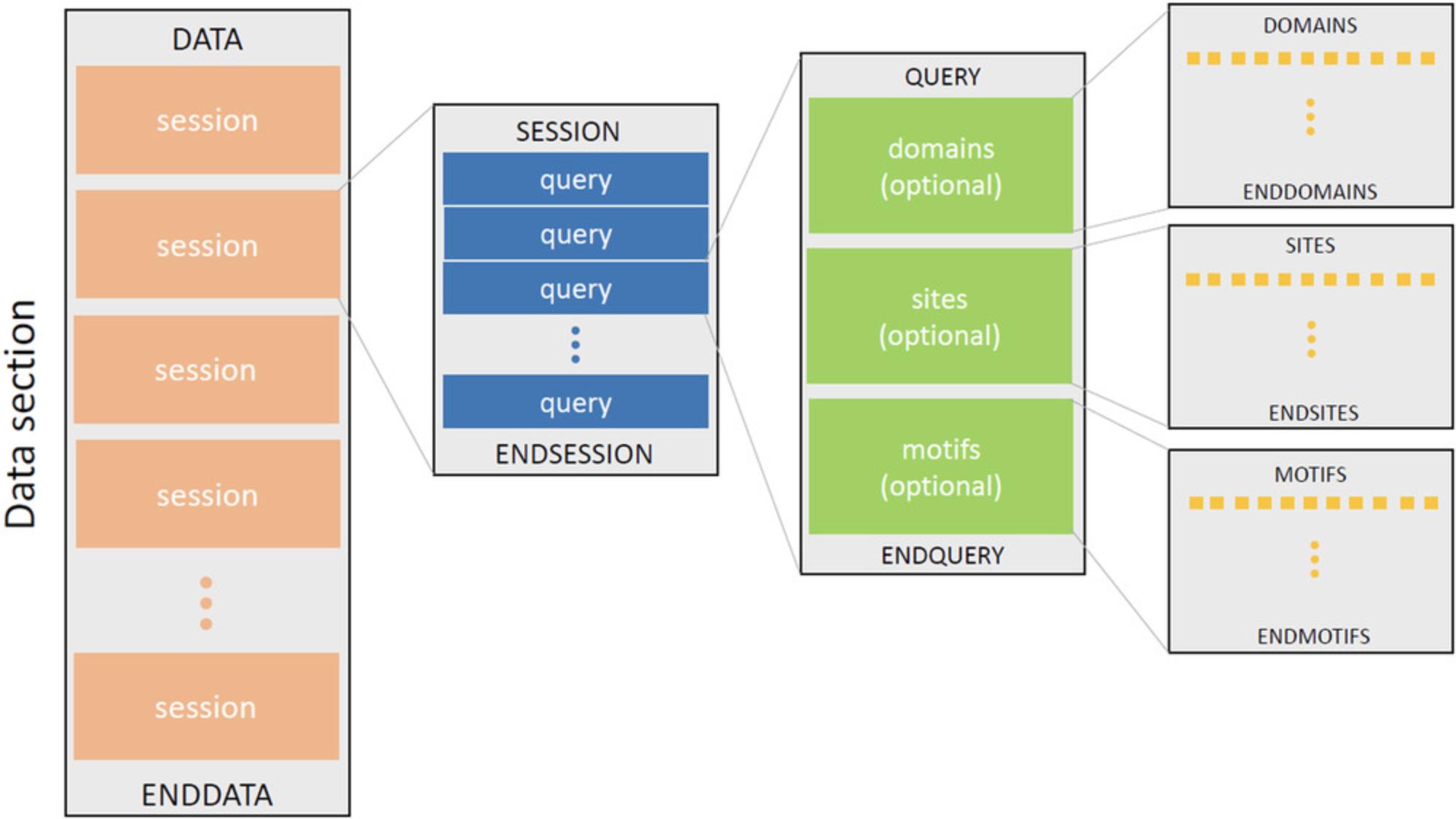
The second section, known as the data section, contains the real data intended to be programmatically processed. All columns are delimited with a tab character (“\t”). The data section always starts with a DATA token and ends with an ENDDATA token. In between, there can be several sessions, each of which start with a SESSION token and end with an ENDSESSION token. Each session is given an ordinal and unique number, which is known as the session ID. Each session is composed of queries, which are unit blocks of sessions. Every single query block contains three optional sections, namely domains, sites, and motifs. The full structure of the data section is illustrated in Figure 21. The domains, sites, and motifs sections contain rows of values, corresponding to the column names defined in the first section of the rpsbproc output file. In the domain section, for example, each row represents a domain hit, including the following information: session ID; query ID; hit type; PSSM ID; start position; end position; E -value; bit score, accession; short name; and whether the alignment is incomplete on the N terminus, C terminus, or both; and superfamily PSSM ID (similar to the data shown in Figure 14).
COMMENTARY
Background Information
A protein domain is typically associated with a function, such as enzyme catalysis or nucleic-acid binding, and is a unit of molecular evolution; via comparative sequence analysis, protein domain sequences can be organized into an evolutionary classification. The CDD's curated domain collections are often classified to a very fine-grained level with the help of available 3D structure to guide multiple sequence alignments, and are manually annotated with functional sites using evidence from 3D structure and other information, including the published literature. Having information about a protein's domain(s) can give you (the user) a wealth of information about your protein of interest. In the cases of unclassified or novel proteins, this domain information provides vital clues to protein function, and often domain annotation is the only available hint toward molecular and cellular function for novel uncharacterized proteins.
In addition to results from the in-house curation effort, the CDD contains domain models from external sources such as Pfam. Agreement between annotations from two or more resources provides users confidence about the domains identified, whereas disagreements between them—which may be as trivial as different domain boundary definitions, or more serious in the case where different functional domains are identified for the same region of a query—may indicate that results should be interpreted with caution.
The three CD-Search protocols described in this paper outline methods for users to submit queries of a single protein or in batches of very large numbers of proteins. The results from these searches—such as domain model identification and accessions, domain footprints (from-to intervals) on the query, E -values and scores, and the locations of functional sites and interactions—can for larger numbers of queries be returned in a tabular form suitable for further processing.
The CDD was first described in the literature in 2002 (Marchler-Bauer et al., 2002). Version v1.54 then contained 3693 models, including contributions from the CDD's in-house curation, Pfam, and SMART. CDD v3.17 (April 3, 2019) contains 57,242 total models from all Source databases, 14,908 of them from the CDD curation effort.
Critical Parameters
The current limitation of 4000 sequences for Batch CD-Search was imposed by the CDD due to high peak usage of this shared resource; you will be alerted to any future changes to this upper limit on the Batch CD-Search page.
To demonstrate the various CD-Searches for Basic Protocols 1 to 3, we have provided test sets. The Batch CD search test set was derived from an in-house-curated MYSc myosin motor domain intermediate model (cd00124) of the cd01353 Motor Domain hierarchy, which was released on February 5, 2015. The Standalone RPS-Blast and rpsbproc test set is a FASTA file that contains all protein records returned by searching NCBI Protein database with the search term myosin AND “Staphylococcus aureus” (https://www.ncbi.nlm.nih.gov/protein/?term=myosin+AND+"Staphylococcus+aureus") on August 5, 2019. The rspbproc utility available at the CDD FTP site was the version released June 29, 2015. The searches were carried out in August 2019 against CDD database version 3.17, released April 3, 2019. Please note that using updated versions of the CDD database, RPS-BLAST, and rpsbproc utility may result in slightly different results.
The CDD predicts domains on your protein(s) of interest and provides important clues about its function. To pursue options for further analysis, readers are encouraged to launch SPARCLE (the Subfamily Protein Architecture Labeling Engine; see Guidelines for Understanding Results section on Basic Protocol 1) from the domain architecture ID link, on the CD-Search Results page (Figure 5), to investigate further protein classification. SPARCLE is a CDD resource that allows comparative analyses of protein families on the basis of conserved domain architecture and for the functional characterization and labeling of protein sequences that have been grouped by their characteristic conserved domain architecture. SPARCLE can also be accessed directly from the SPARCLE home page (https://www.ncbi.nlm.nih.gov/sparcle). For example, you could search in SPARCLE/advanced search builder with "Myosin" in the name field. Detailed SPARCLE help is available by clicking the question mark box on the SPARCLE results page.
The three CD-Search protocols in this paper describe querying a single protein and large numbers of proteins, interacting with the CDD though its web interfaces or programmatically. You may also want to try Batch CD-Search as an interface for scripted data retrieval. A query can be submitted as either an HTTP GET or an HTTP POST request. An HTTP GET request is submitted as a URL. The program performs the search, collects all the data into a master data structure, and extracts the subset of information you have requested for the final output. The Base URL, valid parameters, and examples of URLs for HTTP GET requests, as well as sample PERL scripts for HTTP POST operations, can be found at: https://www.ncbi.nlm.nih.gov/Structure/cdd/cdd_help.shtml#BatchRPSBWebAPI
Time Considerations
Note that unlike running CD-Search and Batch CD-Search, running RPSBLAST is time consuming. It takes 2 s on average to process one protein or nucleotide sequence; thus, for instance, if you have 10,000 sequences in your FASTA file, it may take 5 to 6 hr to finish. However, the rpsbproc processing is fairly quick: it takes only >30 s to process the RPS-BLAST output of 10,000 protein sequences.
Troubleshooting
Help documentation is provided in Table 1.
Acknowledgments
The authors would like to acknowledge the following additional members of the CDD team for their excellent contributions to our resource: CDD curators: Farideh Chitsaz, Noreen R. Gonzales, Marc Gwadz, Gabriele H. Marchler, James S. Song, Narmada Thanki, and Chanjuan Zheng; CDD programmers: David I. Hurwitz, Christopher J. Lanczycki, Shennan Lu, Jiyao Wang, and Dachuan Zhang; and Renata Geer for composing the comprehensive online CDD Help documentation.
This work was supported by the Intramural Research Program of the National Library of Medicine at National Institutes of Health/DHHS. Funding to pay the Open Access publication charges for this article was provided by the Intramural Research Program of the National Library of Medicine at the National Institutes of Health/DHHS.
Supporting Information
| Filename | Description |
|---|---|
| cpbi90-sup-0001-SuppMat.docx21.4 KB |
Please note: The publisher is not responsible for the content or functionality of any supporting information supplied by the authors. Any queries (other than missing content) should be directed to the corresponding author for the article.
Literature Cited
- Apweiler, R., Attwood, T. K., Bairoch, A., Bateman, A., Birney, E., Biswas, M., … Zdobnov, E. M. (2000). InterPro: An integrated documentation resource for protein families, domains and functional sites. Bioinformatics , 16, 1145–1150. doi: 10.1093/bioinformatics/16.12.1145.
- Derbyshire, M. K., Lanczycki, C. J., Bryant, S. H., & Marchler-Bauer, A. (2012). Annotation of functional sites with the Conserved Domain Database. Database , 2012, bar058. doi: 10.1093/database/bar058.
- Finn, R. D., Coggill, P., Eberhardt, R. Y., Eddy, S. R., Mistry, J., Mitchell, A. L., … Bateman, A. (2016). The Pfam protein families database: Towards a more sustainable future. Nucleic Acids Research , 44, D279–D285. doi: 10.1093/nar/gkv1344.
- Geer, L. Y., Domrachev, M., Lipman, D. J., & Bryant, S. H. (2002). CDART: Protein homology by domain architecture. Genome Research , 10, 1619–1623. doi: 10.1101/gr278202.
- Haft, D. H., DiCuccio, M., Badretdin, A., Brover, V., Chetvernin, V., O'Neill, K., … Pruitt, K. D. (2018). RefSeq: An update on Prokaryotic genome annotation and curation. Nucleic Acids Research , 46, D851–D860. doi: 10.1093/nar/gkx1068.
- Haft, D. H., Selengut, J. D., Richter, A. R., Harkins, D., Basu, M. K., & Beck, E. (2013). TIGRFAMs and genome properties in 2013. Nucleic Acids Research , 41, D387–D395. doi: 10.1093/nar/gks1234.
- Klimke, W., Agarwala, R., Badretdin, A., Chetvernin, S., Ciufo, S., Fedorov, B., … Tatusova, T. (2009). The national center for biotechnology information's protein clusters database. Nucleic Acids Research , 37, D216–D223. doi: 10.1093/nar/gkn734.
- Letunic, I., Doerks, T., & Bork, P. (2014). SMART: Recent updates, new developments, and status in 2015. Nucleic Acids Research , 43, D257–D260. doi: 10.1093/nar/gku949.
- Marchler-Bauer, A., Bo, Y., Han, L., He, J., Lanczycki, C. J., Lu, S., … Bryant, S. H. (2017). CDD/SPARCLE: Functional classification of proteins via subfamily domain architectures. Nucleic Acids Research , 45, D200–D203. doi: 10.1093/nar/gkw1129.
- Marchler-Bauer, A., Panchenko, A. R., Shoemaker, B. A., Thiessen, P. A., Geer, L. Y., & Bryant, S. H. (2002). CDD: A database of conserved domain alignments with links to domain three-dimensional structure. Nucleic Acids Research , 30, 281–283. doi: 10.1093/nar/30.1.281.
- Mitchell, A. L., Attwood, T. K., Babbitt, P. C., Blum, M., Bork, P., Bridge, A., … Finn, R. D. (2019). InterPro in 2019: Improving coverage, classification and access to protein sequence annotations. Nucleic Acids Research , 47, D351–D360. doi: 10.1093/nar/gky1100.
- Tatusov, R. L., Natale, D. A., Garkavtsev, I. V., Tatusova, T. A., WorShankavaram, U. T., Rao, B. S., … Koonin, E. V. (2001). The COG Database: New developments in phylogenetic classification of proteins from complete genomes. Nucleic Acids Research , 29, 22–28. doi: 10.1093/nar/29.1.22.
- Tatusov, R. L., Fedorova, N. D., Jackson, J. D., Jacobs, A. R., Kiryutin, B., Koonin, E. V., … Natale, D. A. (2003). The COG Database: An updated version includes eukaryotes. BMC Bioinformatics , 11, 4–41. doi: 10.1186/1471-2105-4-41.
Citing Literature
Number of times cited according to CrossRef: 133
- Steven W. Brugger, Julianne H. Grose, Craig H. Decker, Brett E. Pickett, Mary F. Davis, Genomic Analyses of Major SARS-CoV-2 Variants Predicting Multiple Regions of Pathogenic and Transmissive Importance, Viruses, 10.3390/v16020276, 16 , 2, (276), (2024).
- Lili Wang, Huan He, Jiayin Wang, Zhuang Meng, Lei Wang, Xiang Jin, Jianhang Zhang, Pingping Du, Liyu Zhang, Fei Wang, Hongbin Li, Quanliang Xie, Genome-Wide Identification of the Geranylgeranyl Pyrophosphate Synthase (GGPS) Gene Family Associated with Natural Rubber Synthesis in Taraxacum kok-saghyz L. Rodin, Plants, 10.3390/plants13192788, 13 , 19, (2788), (2024).
- Jinguo Zhang, Dezhuang Meng, Jianfeng Li, Yaling Bao, Peng Yu, Guohui Dou, Jinmeng Guo, Chenghang Tang, Jiaqi Lv, Xinchen Wang, Xingmeng Wang, Fengcai Wu, Yingyao Shi, Analysis of the Rice Raffinose Synthase (OsRS) Gene Family and Haplotype Diversity, International Journal of Molecular Sciences, 10.3390/ijms25189815, 25 , 18, (9815), (2024).
- Fangxue Zhou, Wenmi Feng, Kexin Mou, Zhe Yu, Yicheng Zeng, Wenping Zhang, Yonggang Zhou, Yaxin Li, Hongtao Gao, Keheng Xu, Chen Feng, Yan Jing, Haiyan Li, Genome-Wide Analysis and Expression Profiling of Soybean RbcS Family in Response to Plant Hormones and Functional Identification of GmRbcS8 in Soybean Mosaic Virus, International Journal of Molecular Sciences, 10.3390/ijms25179231, 25 , 17, (9231), (2024).
- Akash Deep, Dhananjay K. Pandey, Genome-Wide Analysis of VILLIN Gene Family Associated with Stress Responses in Cotton (Gossypium spp.), Current Issues in Molecular Biology, 10.3390/cimb46030146, 46 , 3, (2278-2300), (2024).
- Jordan C. Richard, Tim W. Lane, Rose E. Agbalog, Sarah L. Colletti, Tiffany C. Leach, Christopher D. Dunn, Nathan Bollig, Addison R. Plate, Joseph T. Munoz, Eric M. Leis, Susan Knowles, Isaac F. Standish, Diane L. Waller, Tony L. Goldberg, Freshwater Mussel Viromes Increase Rapidly in Diversity and Abundance When Hosts Are Released from Captivity into the Wild, Animals, 10.3390/ani14172531, 14 , 17, (2531), (2024).
- Tian-jiang Liao, Tao Huang, Hui-yan Xiong, Jie-cuo Duo, Jian-zhi Ma, Ming-yang Du, Rui-jun Duan, Genome-wide identification, characterization, and evolutionary analysis of the barley TALE gene family and its expression profiles in response to exogenous hormones, Frontiers in Plant Science, 10.3389/fpls.2024.1421702, 15 , (2024).
- Yuhong Li, Zhengquan He, Jing Xu, Shenyue Jiang, Xiaojiao Han, Longhua Wu, Renying Zhuo, Wenmin Qiu, SpSIZ1 from hyperaccumulator Sedum plumbizincicola orchestrates SpABI5 to fine-tune cadmium tolerance, Frontiers in Plant Science, 10.3389/fpls.2024.1382121, 15 , (2024).
- Xia Zhang, Rong Fan, Zhuo Yu, Xuerun Du, Xinyue Yang, Huiting Wang, Wenfeng Xu, Xiaoxia Yu, Genome-wide identification of GATA transcription factors in tetraploid potato and expression analysis in differently colored potato flesh, Frontiers in Plant Science, 10.3389/fpls.2024.1330559, 15 , (2024).
- Danielle L. Peters, Francis Gaudreault, Wangxue Chen, Functional domains of Acinetobacter bacteriophage tail fibers, Frontiers in Microbiology, 10.3389/fmicb.2024.1230997, 15 , (2024).
- Muhammad Shaban, Riaz Tabassum, Iqrar Ahmad Rana, Rana Muhammad Atif, Muhammad Abubakkar Azmat, Zubair Iqbal, Sajid Majeed, Muhammad Tehseen Azhar, Comparative analysis of SIMILAR to RCD ONE (SRO) family from tetraploid cotton species and their diploid progenitors depict their significance in cotton growth and development, Journal of Cotton Research, 10.1186/s42397-024-00165-2, 7 , 1, (2024).
- Roberto Vera Alvarez, David Landsman, GTax: improving de novo transcriptome assembly by removing foreign RNA contamination, Genome Biology, 10.1186/s13059-023-03141-2, 25 , 1, (2024).
- Hasnaa R. Temsaah, Ahmed F. Azmy, Amr E. Ahmed, Hend Ali Elshebrawy, Nahed Gomaa Kasem, Fatma A. El-Gohary, Cédric Lood, Rob Lavigne, Karim Abdelkader, Characterization and genomic analysis of the lytic bacteriophage vB_EclM_HK6 as a potential approach to biocontrol the spread of Enterobacter cloacae contaminating food, BMC Microbiology, 10.1186/s12866-024-03541-9, 24 , 1, (2024).
- Dengke Han, Suzhen Ma, Chenhong He, Yuxing Yang, Peng Li, Lanfen Lu, Unveiling the genetic architecture and transmission dynamics of a novel multidrug-resistant plasmid harboring blaNDM-5 in E. Coli ST167: implications for antibiotic resistance management, BMC Microbiology, 10.1186/s12866-024-03333-1, 24 , 1, (2024).
- Tao Chen, Long Zhang, Yanyan Zhang, Weidong Gao, Peipei Zhang, Lijian Guo, Delong Yang, Genome-wide identification of the endonuclease family genes implicates potential roles of TaENDO23 in drought-stressed response and grain development in wheat, BMC Genomics, 10.1186/s12864-024-10840-y, 25 , 1, (2024).
- Zhao Ruan, Jiahui Jiao, Junchi Zhao, Jiaxue Liu, Chaoqiong Liang, Xia Yang, Yan Sun, Guanghui Tang, Peiqin Li, Genome sequencing and comparative genomics reveal insights into pathogenicity and evolution of Fusarium zanthoxyli, the causal agent of stem canker in prickly ash, BMC Genomics, 10.1186/s12864-024-10424-w, 25 , 1, (2024).
- Long Zhang, Wanting Sun, Weidong Gao, Yanyan Zhang, Peipei Zhang, Yuan Liu, Tao Chen, Delong Yang, Genome-wide identification and analysis of the GGCT gene family in wheat, BMC Genomics, 10.1186/s12864-023-09934-w, 25 , 1, (2024).
- Qianning Liu, Baiji Wang, Wen Xu, Yuying Yuan, Jinqiu Yu, Guowen Cui, Genome-wide investigation of the PIF gene family in alfalfa (Medicago sativa L.) expression profiles during development and stress, BMC Genomic Data, 10.1186/s12863-024-01264-4, 25 , 1, (2024).
- Jing‐Wen Li, Ping Zhou, Zhi‐Hang Hu, Rui‐Min Teng, Yong‐Xin Wang, Tong Li, Ai‐Sheng Xiong, Xing‐Hui Li, Xuan Chen, Jing Zhuang, CsPAT1, a GRAS transcription factor, promotes lignin accumulation by antagonistic interacting with CsWRKY13 in tea plants, The Plant Journal, 10.1111/tpj.16670, 118 , 5, (1312-1326), (2024).
- Jimmy Jonathan Liunardo, Sebastien Messerli, Ann‐Kathrin Gregotsch, Sonja Lang, Kerstin Schlosser, Christian Rückert‐Reed, Tobias Busche, Jörn Kalinowski, Martin Zischka, Philipp Weller, Imen Nouioui, Meina Neumann‐Schaal, Chandra Risdian, Joachim Wink, Matthias Mack, Isolation, characterisation and description of the roseoflavin producer Streptomyces berlinensis sp. nov., Environmental Microbiology Reports, 10.1111/1758-2229.13266, 16 , 2, (2024).
- João C. Sequeira, Vítor Pereira, M. Madalena Alves, M. Alcina Pereira, Miguel Rocha, Andreia F. Salvador, MOSCA 2.0: A bioinformatics framework for metagenomics, metatranscriptomics and metaproteomics data analysis and visualization, Molecular Ecology Resources, 10.1111/1755-0998.13996, 24 , 7, (2024).
- Md Umar, Titus Susan Merlin, Thavarool Puthiyedathu Sajeevan, Genomic insights into symbiosis and host adaptation of sponge-associated novel bacterium, Rossellomorea orangium sp. nov , FEMS Microbiology Letters, 10.1093/femsle/fnae074, 371 , (2024).
- Rucha M. Wadapurkar, Aruna Sivaram, Renu Vyas, Computational investigations into structure and function impact of novel mutations identified in targeted exons from ovarian cancer cell lines, Journal of Biomolecular Structure and Dynamics, 10.1080/07391102.2024.2310776, (1-15), (2024).
- Cristóbal Uribe, Mariana F. Nery, Kattina Zavala, Gonzalo A. Mardones, Gonzalo Riadi, Juan C. Opazo, Evolution of ion channels in cetaceans: a natural experiment in the tree of life, Scientific Reports, 10.1038/s41598-024-66082-1, 14 , 1, (2024).
- Jason Nomburg, Erin E. Doherty, Nathan Price, Daniel Bellieny-Rabelo, Yong K. Zhu, Jennifer A. Doudna, Birth of protein folds and functions in the virome, Nature, 10.1038/s41586-024-07809-y, 633 , 8030, (710-717), (2024).
- Pok Man Leung, Rhys Grinter, Eve Tudor-Matthew, James P. Lingford, Luis Jimenez, Han-Chung Lee, Michael Milton, Iresha Hanchapola, Erwin Tanuwidjaya, Ashleigh Kropp, Hanna A. Peach, Carlo R. Carere, Matthew B. Stott, Ralf B. Schittenhelm, Chris Greening, Trace gas oxidation sustains energy needs of a thermophilic archaeon at suboptimal temperatures, Nature Communications, 10.1038/s41467-024-47324-2, 15 , 1, (2024).
- Ping Liao, Ting Zeng, Mengyang Huangfu, Cairong Zheng, Jiequn Ren, Changyong Zhou, Yan Zhou, Eureka lemon zinc finger protein ClDOF3.4 interacts with citrus yellow vein clearing virus coat protein to inhibit viral infection, Journal of Integrative Agriculture, 10.1016/j.jia.2024.03.049, 23 , 6, (1979-1993), (2024).
- Thomas Jebastin, M.H. Syed Abuthakir, Ilangovan Santhoshi, Muniraj Gnanaraj, Mansour K. Gatasheh, Anis Ahamed, Velusamy Sharmila, Unveiling the mysteries: Functional insights into hypothetical proteins from Bacteroides fragilis 638R, Heliyon, 10.1016/j.heliyon.2024.e31713, 10 , 11, (e31713), (2024).
- Kauser Parveen, Muhammad Abu Bakar Saddique, Zulfiqar Ali, Shoaib Ur Rehman, undefined Zaib-Un-Nisa, Zulqurnain Khan, Muhammad Waqas, Muhammad Zeeshan Munir, Niaz Hussain, Muhammad Atif Muneer, Genome-wide analysis of Glutathione peroxidase (GPX) gene family in Chickpea (Cicer arietinum L.) under salinity stress, Gene, 10.1016/j.gene.2023.148088, 898 , (148088), (2024).
- Pritish Mitra, Sourav Singha, Payel Roy, Deblina Saha, Sabyasachi Chatterjee, A molecular docking study between heavy metals and hydrophilic Hsp70 protein to explore binding pockets, Journal of Proteins and Proteomics, 10.1007/s42485-024-00150-y, 15 , 3, (413-428), (2024).
- Hong Deng, Zhuang Wen, Qiandong Hou, Runrun Yu, Xiaowei Cai, Ke Liu, Guang Qiao, Genome-wide identification and analysis of the growth-regulating factor (GRF) family in sweet cherry, Genetic Resources and Crop Evolution, 10.1007/s10722-024-01886-8, (2024).
- Urara Miyazaki, Daiki Mizutani, Yurina Hashimoto, Akihiro Tame, Shigeki Sawayama, Junichi Miyazaki, Ken Takai, Satoshi Nakagawa, Helicovermis profundi gen. nov., sp. nov., a novel mesophilic, asporogenous bacterium within the Clostridia isolated from a deep-sea hydrothermal vent chimney, Antonie van Leeuwenhoek, 10.1007/s10482-023-01919-9, 117 , 1, (2024).
- Behnam Davoudnia, Ali Dadkhodaie, Ali Moghadam, Bahram Heidari, Mohsen Yassaie, Transcriptome analysis in Aegilops tauschii unravels further insights into genetic control of stripe rust resistance, Planta, 10.1007/s00425-024-04347-9, 259 , 3, (2024).
- Tao Chen, Yongping Miao, Fanli Jing, Weidong Gao, Yanyan Zhang, Long Zhang, Peipei Zhang, Lijian Guo, Delong Yang, Genomic‐wide analysis reveals seven in absentia genes regulating grain development in wheat (Triticum aestivum L.), The Plant Genome, 10.1002/tpg2.20480, 17 , 3, (2024).
- Anastasia Klemanska, Kelly Dwyer, Gary Walsh, Truncation of a novel C‐terminal domain of a β‐glucanase improves its thermal stability and specific activity, Biotechnology Journal, 10.1002/biot.202400245, 19 , 8, (2024).
- Gengyun Li, Longjie Cheng, Zhilin Li, Yiran Zhao, Yuying Wang, Over-expression of CcMYB24 , encoding a R2R3-MYB transcription factor from a high-leaf-number mutant of Cymbidium , increases the number of leaves in Arabidopsis , PeerJ, 10.7717/peerj.15490, 11 , (e15490), (2023).
- Xingzhi Qian, Wenyin Zheng, Jian Hu, Jinxu Ma, Mengyuan Sun, Yong Li, Nian Liu, Tianhua Chen, Meiqi Wang, Ling Wang, Xinzhe Hou, Qingao Cai, Zhaoshun Ye, Fugui Zhang, Zonghe Zhu, Identification and Expression Analysis of DFR Gene Family in Brassica napus L., Plants, 10.3390/plants12132583, 12 , 13, (2583), (2023).
- Yueqiong Zhou, Liangliang He, Shaoli Zhou, Qing Wu, Xuan Zhou, Yawen Mao, Baolin Zhao, Dongfa Wang, Weiyue Zhao, Ruoruo Wang, Huabin Hu, Jianghua Chen, Genome-Wide Identification and Expression Analysis of the VILLIN Gene Family in Soybean, Plants, 10.3390/plants12112101, 12 , 11, (2101), (2023).
- Qinqin Liu, Zhiyun Guo, Gang Zhu, Ning Li, Guanchen Bai, Meijie Jiang, Genomic Characteristics and Phylogenetic Analyses of a Multiple Drug-Resistant Klebsiella pneumoniae Harboring Plasmid-Mediated MCR-1 Isolated from Tai’an City, China, Pathogens, 10.3390/pathogens12020221, 12 , 2, (221), (2023).
- Jiagui Guo, Yan Yang, Tingting Wang, Yizhen Wang, Xin Zhang, Donghong Min, Xiaohong Zhang, Analysis of Raffinose Synthase Gene Family in Bread Wheat and Identification of Drought Resistance and Salt Tolerance Function of TaRS15-3B, International Journal of Molecular Sciences, 10.3390/ijms241311185, 24 , 13, (11185), (2023).
- Dong-Bin Chen, Run-Xi Xia, Qun Li, Yu-Ping Li, Hui-Ying Cao, Yan-Qun Liu, Genome-Wide Identification of Detoxification Genes in Wild Silkworm Antheraea pernyi and Transcriptional Response to Coumaphos, International Journal of Molecular Sciences, 10.3390/ijms24119775, 24 , 11, (9775), (2023).
- Huaizhi Mu, Xuhong Jin, Songtong Lv, Sheng Long, Yang Liu, Le Chen, Lin Lin, Genome-Wide Identification and Expression Analysis of Auxin Response Factor (ARF) Gene Family in Betula pendula, Horticulturae, 10.3390/horticulturae10010027, 10 , 1, (27), (2023).
- Yifeng Ding, Xiaomeng Wang, Dandan Wang, Liwei Jiang, Jing Xie, Tianle Wang, Lingyu Song, Xiting Zhao, Identification of CmbHLH Transcription Factor Family and Excavation of CmbHLHs Resistant to Necrotrophic Fungus Alternaria in Chrysanthemum, Genes, 10.3390/genes14020275, 14 , 2, (275), (2023).
- Jihong Wang, Lei Zhang, Peiwen Wang, Jinhui Lei, Lingli Zhong, Lei Zhan, Xianfeng Ye, Yan Huang, Xue Luo, Zhongli Cui, Zhoukun Li, Identification and Characterization of Novel Malto-Oligosaccharide-Forming Amylase AmyCf from Cystobacter sp. Strain CF23, Foods, 10.3390/foods12183487, 12 , 18, (3487), (2023).
- Xingze Huang, Ruonan Zhao, Zhiwang Xu, Chuyan Fu, Lei Xie, Shuran Li, Xiaofeng Wang, Yongpu Zhang, gjSOX9 Cloning, Expression, and Comparison with gjSOXs Family Members in Gekko japonicus, Current Issues in Molecular Biology, 10.3390/cimb45110584, 45 , 11, (9328-9341), (2023).
- Li Wang, Jinyu Zhang, Huici Li, Gongzhan Zhang, Dandan Hu, Dan Zhang, Xinjuan Xu, Yuming Yang, Zhongwen Huang, Genome-Wide Identification of the Phytocyanin Gene Family and Its Potential Function in Salt Stress in Soybean (Glycine max (L.) Merr.), Agronomy, 10.3390/agronomy13102484, 13 , 10, (2484), (2023).
- Rui Wu, Wenhui Liu, Kaiqiang Liu, Guoling Liang, Yue Wang, Genome-Wide Identification and Expression of the GRAS Gene Family in Oat (Avena sativa L.), Agronomy, 10.3390/agronomy13071807, 13 , 7, (1807), (2023).
- Xin Yao, Dili Lai, Meiliang Zhou, Jingjun Ruan, Chao Ma, Weijiao Wu, Wenfeng Weng, Yu Fan, Jianping Cheng, Genome-wide identification, evolution and expression pattern analysis of the GATA gene family in Sorghum bicolor, Frontiers in Plant Science, 10.3389/fpls.2023.1163357, 14 , (2023).
- Alsamman M. Alsamman, Khaled H. Mousa, Ahmed E. Nassar, Mostafa M. Faheem, Khaled H. Radwan, Monica H. Adly, Ahmed Hussein, Tawffiq Istanbuli, Morad M. Mokhtar, Tamer Ahmed Elakkad, Zakaria Kehel, Aladdin Hamwieh, Mohamed Abdelsattar, Achraf El Allali, Identification, characterization, and validation of NBS-encoding genes in grass pea, Frontiers in Genetics, 10.3389/fgene.2023.1187597, 14 , (2023).
- Joerg Behnke, Yun Cai, Hong Gu, Julie LaRoche, Short-term response to iron resupply in an iron-limited open ocean diatom reveals rapid decay of iron-responsive transcripts, PLOS ONE, 10.1371/journal.pone.0280827, 18 , 1, (e0280827), (2023).
- Jinqiu Yu, Yuying Yuan, Linling Dong, Guowen Cui, Genome-wide investigation of NLP gene family members in alfalfa (Medicago sativa L.): evolution and expression profiles during development and stress, BMC Genomics, 10.1186/s12864-023-09418-x, 24 , 1, (2023).
- Dake Zhao, Ya Zhang, Huanxing Ren, Yana Shi, Ding Dong, Zonghang Li, Guanghong Cui, Yong Shen, Zongmin Mou, Edward J. Kennelly, Luqi Huang, Jue Ruan, Suiyun Chen, Diqiu Yu, Yupeng Cun, Multi‐omics analysis reveals the evolutionary origin of diterpenoid alkaloid biosynthesis pathways in Aconitum, Journal of Integrative Plant Biology, 10.1111/jipb.13565, 65 , 10, (2320-2335), (2023).
- Jingliang Cheng, Ting Li, Qi Tan, Jiewen Fu, Lianmei Zhang, Luquan Yang, Baixu Zhou, Lisha Yang, Shangyi Fu, Alora Grace Linehan, Junjiang Fu, Novel, pathogenic insertion variant of GSDME associates with autosomal dominant hearing loss in a large Chinese pedigree, Journal of Cellular and Molecular Medicine, 10.1111/jcmm.18004, 28 , 1, (2023).
- Sean Tsz Sum Law, Yifei Yu, Wenyan Nong, Wai Lok So, Yiqian Li, Thomas Swale, David E. K. Ferrier, Jianwen Qiu, Peiyuan Qian, Jerome Ho Lam Hui, The genome of the deep-sea anemone Actinernus sp. contains a mega-array of ANTP-class homeobox genes , Proceedings of the Royal Society B: Biological Sciences, 10.1098/rspb.2023.1563, 290 , 2009, (2023).
- Xichen Lian, Yintao Zhang, Ying Zhou, Xiuna Sun, Shijie Huang, Haibin Dai, Lianyi Han, Feng Zhu, SingPro: a knowledge base providing single-cell proteomic data, Nucleic Acids Research, 10.1093/nar/gkad830, 52 , D1, (D552-D561), (2023).
- Sapna Chandwani, Sahabram Dewala, Sonal Manik Chavan, Dhiraj Paul, Raman Pachaiappan, Muthukaruppan Gopi, Natarajan Amaresan, Complete genome sequencing of Bacillus subtilis (CWTS 5), a siderophore-producing bacterium triggers antagonistic potential against Ralstonia solanacearum , Journal of Applied Microbiology, 10.1093/jambio/lxad066, 134 , 4, (2023).
- Hiroyuki Ichida, Hitoshi Murata, Shin Hatakeyama, Akiyoshi Yamada, Akira Ohta, Near-complete de novo assembly of Tricholoma bakamatsutake chromosomes revealed the structural divergence and differentiation of Tricholoma genomes , G3: Genes, Genomes, Genetics, 10.1093/g3journal/jkad198, 13 , 11, (2023).
- Himisha Dixit, Mahesh Kulharia, Shailender Kumar Verma, Metalloproteome of human-infective RNA viruses: a study towards understanding the role of metal ions in virology, Pathogens and Disease, 10.1093/femspd/ftad020, 81 , (2023).
- Harpreet Kaur, Syed Azmal Ali, Sarah P. Short, Christopher S. Williams, Jeremy A. Goettel, M. Kay Washington, Richard M. Peek, Sari A. Acra, Fang Yan, Identification of a functional peptide of a probiotic bacterium-derived protein for the sustained effect on preventing colitis, Gut Microbes, 10.1080/19490976.2023.2264456, 15 , 2, (2023).
- Li Li, Mengping Nie, Jing Lu, Cailin He, Qi Wu, Genome-wide identification, expression analysis and gene duplication analysis of Dof transcription factors between quinoa and its ancestral diploid sub-genome species reveal key CDF homologs involved in photoperiodic flowering regulation, South African Journal of Botany, 10.1016/j.sajb.2023.09.051, 162 , (545-558), (2023).
- Ravindra Pal Singh, Jayashree Niharika, Raksha Thakur, Ben A. Wagstaff, Gulshan Kumar, Rikuya Kurata, Dhaval Patel, Colin W. Levy, Takatsugu Miyazaki, Robert A. Field, Utilization of dietary mixed-linkage β-glucans by the Firmicute Blautia producta, Journal of Biological Chemistry, 10.1016/j.jbc.2023.104806, 299 , 6, (104806), (2023).
- Ziyi Ye, Fei Hu, Weimeng Zhang, Da Fang, Kui Dong, Jun Cao, Amino acid transporters of Brassica napus: Identification, evolution, expression and response to various stresses, Industrial Crops and Products, 10.1016/j.indcrop.2023.116338, 194 , (116338), (2023).
- Mohd. Kashif, Bhupendra Kumar, Akhilendra Pratap Bharati, Hisham Altayeb, Mohd. Asalam, Mohd Sohail Akhtar, Mohammad Imran Khan, Abrar Ahmad, Hani Chaudhary, Salman Bakr Hosawi, Mazin A. Zamzami, Othman A. Baothman, Association of peptidyl prolyl cis/trans isomerase Rrd1 with C terminal domain of RNA polymerase II, International Journal of Biological Macromolecules, 10.1016/j.ijbiomac.2023.124653, 242 , (124653), (2023).
- Weimeng Zhang, Da Fang, Ziyi Ye, Fei Hu, Xiuzhu Cheng, Jun Cao, Identification and molecular evolution of the La and LARP genes in 16 plant species: A focus on the Gossypium hirsutum, International Journal of Biological Macromolecules, 10.1016/j.ijbiomac.2022.10.195, 224 , (1101-1117), (2023).
- Leonard Whye Kit Lim, K-ras proto-oncogene (KRAS): Evolutionary dissection on the indispensable predictive and prognostic cancer biomarker across 32 primates, Animal Gene, 10.1016/j.angen.2023.200158, 30 , (200158), (2023).
- Gaoyuan Zhang, Bingqiang Wei, Characterization of 14-3-3 gene family and their expression patterns under abiotic and biotic stresses in melon (Cucumis melo L.), Horticulture, Environment, and Biotechnology, 10.1007/s13580-023-00532-z, 64 , 6, (1039-1054), (2023).
- Senthilkumar K. Muthusamy, P. Pushpitha, T. Makeshkumar, M. N. Sheela, Genome-wide identification and expression analysis of Hsp70 family genes in Cassava (Manihot esculenta Crantz), 3 Biotech, 10.1007/s13205-023-03760-3, 13 , 10, (2023).
- Sapna Chandwani, Sahabram Dewala, Sonal Manik Chavan, Dhiraj Paul, Krishna Kumar, Natarajan Amaresan, Genomic, LC–MS, and FTIR Analysis of Plant Probiotic Potential of Bacillus albus for Managing Xanthomonas oryzae via Different Modes of Application in Rice (Oryza sativa L.), Probiotics and Antimicrobial Proteins, 10.1007/s12602-023-10120-3, 16 , 5, (1541-1552), (2023).
- Jing-Wen Li, Ping Zhou, Yuan-Jie Deng, Zhi-Hang Hu, Xing-Hui Li, Xuan Chen, Ai-Sheng Xiong, Jing Zhuang, Overexpressing CsPSY1 Gene of Tea Plant, Encoding a Phytoene Synthase, Improves α-Carotene and β-Carotene Contents in Carrot, Molecular Biotechnology, 10.1007/s12033-023-00942-5, (2023).
- Muhammad Farooq, Rafiq Ahmad, Muhammad Shahzad, Saad Ur Rehman, Yasar Sajjad, Amjad Hassan, Mohammad Maroof Shah, Amber Afroz, Sabaz Ali Khan, Real-time expression and in silico characterization of pea genes involved in salt and water-deficit stress, Molecular Biology Reports, 10.1007/s11033-023-09064-2, 51 , 1, (2023).
- Essam H. Houssein, Mosa E. Hosney, Marwa M. Emam, Eman M. G. Younis, Abdelmgeid A. Ali, Waleed M. Mohamed, Soft computing techniques for biomedical data analysis: open issues and challenges, Artificial Intelligence Review, 10.1007/s10462-023-10585-2, 56 , S2, (2599-2649), (2023).
- Urara Miyazaki, Masaru Sanari, Akihiro Tame, Masaaki Kitajima, Akihiro Okamoto, Shigeki Sawayama, Junichi Miyazaki, Ken Takai, Satoshi Nakagawa, Pyrofollis japonicus gen. nov. sp. nov., a novel member of the family Pyrodictiaceae isolated from the Iheya North hydrothermal field, Extremophiles, 10.1007/s00792-023-01316-0, 27 , 3, (2023).
- Christophe Penno, Julien Tremblay, Mary O’Connell Motherway, Virginie Daburon, Abdelhak El Amrani, Analysis of Small Non-coding RNAs as Signaling Intermediates of Environmentally Integrated Responses to Abiotic Stress, Plant Abiotic Stress Signaling, 10.1007/978-1-0716-3044-0_22, (403-427), (2023).
- Mohammad Mahmoudi Gomari, Seyed Shahriar Arab, Saeed Balalaie, Sorour Ramezanpour, Arshad Hosseini, Nikolay V. Dokholyan, Parastoo Tarighi, Rational peptide design for targeting cancer cell invasion, Proteins: Structure, Function, and Bioinformatics, 10.1002/prot.26580, 92 , 1, (76-95), (2023).
- Chayan Sharma, Sumeeta Khurana, Amit Arora, Alka Bhatia, Amit Gupta, An Insight into the Genome of Pathogenic and Non-Pathogenic Acanthamoeba, Pathogens, 10.3390/pathogens11121558, 11 , 12, (1558), (2022).
- Yi Yin, Gangcheng Chen, Myat Htut Nyunt, Meihua Zhang, Yaobao Liu, Guoding Zhu, Xinlong He, Fang Tian, Jun Cao, Eun-taek Han, Feng Lu, Prevalence of pvmrp1 Polymorphisms and Its Contribution to Antimalarial Response, Microorganisms, 10.3390/microorganisms10081482, 10 , 8, (1482), (2022).
- Yeqing He, Guandi He, Fei Lou, Zheng Zhou, Yao Liu, Yule Zhang, Tengbing He, Identification of the Major Effector StSROs in Potato: A Potential StWRKY-SRO6 Regulatory Pathway Enhances Plant Tolerance to Cadmium Stress, International Journal of Molecular Sciences, 10.3390/ijms232214318, 23 , 22, (14318), (2022).
- Candy Yuriria Ramírez-Zavaleta, Laura Jeannette García-Barrera, Lizette Liliana Rodríguez-Verástegui, Daniela Arrieta-Flores, Josefat Gregorio-Jorge, An Overview of PRR- and NLR-Mediated Immunities: Conserved Signaling Components across the Plant Kingdom That Communicate Both Pathways, International Journal of Molecular Sciences, 10.3390/ijms232112974, 23 , 21, (12974), (2022).
- Xin Yao, Meiliang Zhou, Jingjun Ruan, Ailing He, Chao Ma, Weijiao Wu, Dili Lai, Yu Fan, Anjing Gao, Wenfeng Weng, Jianping Cheng, Genome-Wide Identification, Evolution, and Expression Pattern Analysis of the GATA Gene Family in Tartary Buckwheat (Fagopyrum tataricum), International Journal of Molecular Sciences, 10.3390/ijms232012434, 23 , 20, (12434), (2022).
- Alexander V. Shabaev, Konstantin V. Moiseenko, Olga A. Glazunova, Olga S. Savinova, Tatyana V. Fedorova, Comparative Analysis of Peniophora lycii and Trametes hirsuta Exoproteomes Demonstrates “Shades of Gray” in the Concept of White-Rotting Fungi, International Journal of Molecular Sciences, 10.3390/ijms231810322, 23 , 18, (10322), (2022).
- Xiaoshan Xue, Runqing Li, Caijuan Zhang, Wenna Li, Lin Li, Suying Hu, Junfeng Niu, Xiaoyan Cao, Donghao Wang, Zhezhi Wang, Identification and Characterization of Jasmonic Acid Biosynthetic Genes in Salvia miltiorrhiza Bunge, International Journal of Molecular Sciences, 10.3390/ijms23169384, 23 , 16, (9384), (2022).
- Lu-Yu Yan, Jia-Gui Guo, Xin Zhang, Yang Liu, Xin-Xin Xiong, Yu-Xuan Han, Li-Li Zhang, Xiao-Hong Zhang, Dong-Hong Min, Genome-Wide Analysis of the Peptidase M24 Superfamily in Triticum aestivum Demonstrates That TaM24-9 Is Involved in Abiotic Stress Response, International Journal of Molecular Sciences, 10.3390/ijms23136904, 23 , 13, (6904), (2022).
- Mingyang Wang, Lanxin Wu, Shouhong Zhu, Wei Chen, Jinbo Yao, Yan Li, Tengyu Li, Haihong Shang, Yongshan Zhang, Evolutionary Relationships and Divergence of Filamin Gene Family Involved in Development and Stress in Cotton (Gossypium hirsutum L.), Genes, 10.3390/genes13122313, 13 , 12, (2313), (2022).
- Jinwan Fan, Gang Nie, Jieyu Ma, Ruchang Hu, Jie He, Feifei Wu, Zhongfu Yang, Sainan Ma, Xin Zhang, Xinquan Zhang, The Identification and Characterization of the KNOX Gene Family as an Active Regulator of Leaf Development in Trifolium repens, Genes, 10.3390/genes13101778, 13 , 10, (1778), (2022).
- Hajra Maqsood, Faiza Munir, Rabia Amir, Alvina Gul, Genome-wide identification, comprehensive characterization of transcription factors, cis-regulatory elements, protein homology, and protein interaction network of DREB gene family in Solanum lycopersicum, Frontiers in Plant Science, 10.3389/fpls.2022.1031679, 13 , (2022).
- Xiaofei Liang, Shiqiang Mei, Haodong Yu, Song Zhang, Jiaxing Wu, Mengji Cao, Mixed infection of an emaravirus, a crinivirus, and a begomovirus in Pueraria lobata (Willd) Ohwi, Frontiers in Microbiology, 10.3389/fmicb.2022.926724, 13 , (2022).
- Zhaoqing Yu, Yang Fu, Wei Zhang, Li Zhu, Wen Yin, Shan-Ho Chou, Jin He, The RNA Chaperone Protein Hfq Regulates the Characteristic Sporulation and Insecticidal Activity of Bacillus thuringiensis, Frontiers in Microbiology, 10.3389/fmicb.2022.884528, 13 , (2022).
- Moritz Koch, Avery J. C. Noonan, Yilin Qiu, Kalen Dofher, Brandon Kieft, Soheyl Mottahedeh, Manisha Shastri, Steven J. Hallam, The survivor strain: isolation and characterization of Phormidium yuhuli AB48, a filamentous phototactic cyanobacterium with biotechnological potential, Frontiers in Bioengineering and Biotechnology, 10.3389/fbioe.2022.932695, 10 , (2022).
- Yuxuan Han, Lili Zhang, Luyu Yan, Xinxin Xiong, Wenjing Wang, Xiao-Hong Zhang, Dong-Hong Min, Genome-wide analysis of TALE superfamily in Triticum aestivum reveals TaKNOX11-A is involved in abiotic stress response, BMC Genomics, 10.1186/s12864-022-08324-y, 23 , 1, (2022).
- Kshitij Tandon, Yu-Jing Chiou, Sheng-Ping Yu, Hernyi Justin Hsieh, Chih-Ying Lu, Ming-Tsung Hsu, Pei-Wen Chiang, Hsing-Ju Chen, Naohisa Wada, Sen-Lin Tang, Microbiome Restructuring: Dominant Coral Bacterium Endozoicomonas Species Respond Differentially to Environmental Changes , mSystems, 10.1128/msystems.00359-22, 7 , 4, (2022).
- Megan Kirchhoff, Carlos Ortega, James Clark, Tram Le, Ben Burrowes, Mei Liu, Complete Genome Sequence of Stenotrophomonas maltophilia Podophage Piffle, Microbiology Resource Announcements, 10.1128/mra.00159-22, 11 , 4, (2022).
- Zdeněk Knejzlík, Michal Doležal, Klára Herkommerová, Kamila Clarova, Martin Klíma, Matteo Dedola, Eva Zborníková, Dominik Rejman, Iva Pichová, The mycobacterial guaB1 gene encodes a guanosine 5′‐monophosphate reductase with a cystathionine‐β‐synthase domain, The FEBS Journal, 10.1111/febs.16448, 289 , 18, (5571-5598), (2022).
- Ruilin Huang, Ruirui Ding, Yu Liu, Fuli Li, Zhaohui Zhang, Shi’an Wang, GATA transcription factor WC2 regulates the biosynthesis of astaxanthin in yeast Xanthophyllomyces dendrorhous, Microbial Biotechnology, 10.1111/1751-7915.14115, 15 , 10, (2578-2593), (2022).
- Yun Feng, Qin-yu Gou, Wei-hong Yang, Wei-chen Wu, Juan Wang, Edward C Holmes, Guodong Liang, Mang Shi, A time-series meta-transcriptomic analysis reveals the seasonal, host, and gender structure of mosquito viromes, Virus Evolution, 10.1093/ve/veac006, 8 , 1, (2022).
- Mario Rodríguez Mestre, Linyi Alex Gao, Shiraz A Shah, Adrián López-Beltrán, Alejandro González-Delgado, Francisco Martínez-Abarca, Jaime Iranzo, Modesto Redrejo-Rodríguez, Feng Zhang, Nicolás Toro, UG/Abi: a highly diverse family of prokaryotic reverse transcriptases associated with defense functions, Nucleic Acids Research, 10.1093/nar/gkac467, 50 , 11, (6084-6101), (2022).
- Karim Benzerara, Elodie Duprat, Tristan Bitard-Feildel, Géraldine Caumes, Corinne Cassier-Chauvat, Franck Chauvat, Manuela Dezi, Seydina Issa Diop, Geoffroy Gaschignard, Sigrid Görgen, Muriel Gugger, Purificación López-García, Maxime Millet, Fériel Skouri-Panet, David Moreira, Isabelle Callebaut, A New Gene Family Diagnostic for Intracellular Biomineralization of Amorphous Ca Carbonates by Cyanobacteria, Genome Biology and Evolution, 10.1093/gbe/evac026, 14 , 3, (2022).
- Mossaab Maaloum, Cheikh Ibrahima Lo, Sokhna Ndongo, Marine Makoa Meng, Rachid Saile, Stéphane Alibar, Didier Raoult, Pierre-Edouard Fournier, Ottowia massiliensis sp. nov., a new bacterium isolated from a fresh, healthy human fecal sample , FEMS Microbiology Letters, 10.1093/femsle/fnac086, 369 , 1, (2022).
- Mossaab Maaloum, Pamela Afouda, Cheikh Ibrahima Lo, Gregory Dubourg, Thi Tien Nguyen, Anthony Levasseur, Rachid Saile, Didier Raoult, Pierre-Edouard Fournier, Prevotella merdae sp. nov., a new bacterial species isolated from human faeces , FEMS Microbiology Letters, 10.1093/femsle/fnac066, 369 , 1, (2022).
- Meng Wu, Fulei Nie, Haibin Liu, Tianyang Zhang, Miaomiao Li, Xiaoming Song, Wei Chen, The evolution of N6-methyladenosine regulators in plants, Methods, 10.1016/j.ymeth.2021.11.013, 203 , (268-275), (2022).
- Kyra Dougherty, Katalin A. Hudak, Phylogeny and domain architecture of plant ribosome inactivating proteins, Phytochemistry, 10.1016/j.phytochem.2022.113337, 202 , (113337), (2022).
- See more

