Multiparametric Analyses of Hepatocellular Carcinoma Somatic Mouse Models and Their Associated Tumor Microenvironment
Leila Akkari, Leila Akkari, Daniel Taranto, Daniel Taranto, Christel F.A. Ramirez, Christel F.A. Ramirez, Serena Vegna, Serena Vegna, Marnix H.P. de Groot, Marnix H.P. de Groot, Niels de Wit, Niels de Wit, Martijn Van Baalen, Martijn Van Baalen, Sjoerd Klarenbeek, Sjoerd Klarenbeek
hepatocellular carcinoma
somatic murine models
multi-parametric flow cytometry
tumor microenvironment
Abstract
The rising incidence and increasing mortality of hepatocellular carcinoma (HCC), combined with its high tumor heterogeneity, lack of druggable targets, and tendency to develop resistance to chemotherapeutics, make the development of better models for this cancer an urgent challenge. To better mimic the high diversity within the HCC genetic landscape, versatile somatic murine models have recently been developed using the hydrodynamic tail vein injection (HDTVi) system. These represent novel in vivo tools to interrogate HCC phenotype and response to therapy, and importantly, allow further analyses of the associated tumor microenvironment (TME) shaped by distinct genetic backgrounds. Here, we describe several optimized protocols to generate, collect, and experimentally utilize various samples obtained from HCC somatic mouse models generated by HDTVi. More specifically, we focus on techniques relevant to ex vivo analyses of the complex liver TME using multiparameter flow cytometric analyses of over 21 markers, immunohistochemistry, immunofluorescence, and histochemistry. We describe the transcriptional assessment of whole tissue, or of isolated immune subsets by flow-cytometry-based cell sorting, and other protein-oriented analyses. Together, these streamlined protocols allow the optimal use of each HCC murine model of interest and will assist researchers in deciphering the relations between cancer cell genetics and systemic and local changes in immune cell landscapes in the context of HCC progression. © 2021 The Authors. Current Protocols published by Wiley Periodicals LLC.
Basic Protocol 1 : Generation of HCC mouse models by hydrodynamic tail vein injection
Basic Protocol 2 : Assessment of HCC tumor progression by magnetic resonance imaging
Basic Protocol 3 : Mouse sacrifice and sample collection in HCC mouse models
Support Protocol 1 : Preparation of serum or plasma from blood
Basic Protocol 4 : Single-cell preparation and HCC immune landscape phenotyping by flow cytometry
Alternate Protocol 1 : Flow cytometric analysis of circulating immune cells
Support Protocol 2 : Generation, maintenance, and characterization of HCC cell lines
Support Protocol 3 : Fluorescence-activated cell sorting of liver single-cell preparation
Basic Protocol 5 : Preparation and immunohistochemical analysis of tumor tissues from HCC-bearing liver
Alternate Protocol 2 : Preparation and analyses for immunofluorescence staining of HCC-bearing liver
Support Protocol 4 : Liver-specific phenotypic analyses of liver sections
Support Protocol 5 : Immunohistochemical quantification in liver sections
Basic Protocol 6 : Preparation of snap-frozen tumor tissue from extracted liver and transcriptional analyses of bulk tumor or sorted cells
Alternate Protocol 3 : Protein analyses from HCC samples and serum or plasma
INTRODUCTION
Despite a general decline in cancer-related death, the incidence of hepatocellular carcinoma (HCC) has risen in developed countries, and its associated mortality is growing at an alarming rate (Gadiparthi et al., 2019). Challenges in HCC treatment lie in the high intra- and intertumor heterogeneity, lack of druggable targets, and rapid emergence of resistance in patients ineligible for curative approaches (Llovet, Montal, Sia, & Fingn, 2018; Zucman-Rossi, Villanueva, Nault, & Llovet, 2015). Patients with HCC exhibit a highly heterogeneous mutational landscape (Schulze, Nault, & Villanueva, 2016). Therefore, studies aiming at dissecting the effects of the genetic background of HCC cancer cells on the composition and phenotype of the tumor microenvironment (TME) must consider both intra- and interpatient heterogeneity as well as the precise biological processes that may be altered in these patients. Although studies have reported immune cell analyses in different contexts, complete methods for examining genetically distinct HCC phenotypic features and for the optimal use of each tumor tissue, including in-depth TME characterization, remained to be compiled.
In this article, we describe precise procedures to generate, collect, and analyze HCC mouse model samples designed to bear distinct genetic backgrounds. HCC mouse models are generated by overexpressing oncogenes relevant to HCC using the Sleeping Beauty transposon system in combination with knockout of tumor-suppressor genes using CRISPR-Cas9, both delivered by hydrodynamic tail injection (HDTVi; Bell et al., 2007; Xue et al., 2014; Zhang, Budker, & Wolff, 1999). Genetic combinations can be chosen from among the several mutations reported in patients with HCC.
We detail optimized steps to measure tumor volume over the course of HCC development by magnetic resonance imaging (MRI), as well procedures to collect murine material for follow-up experiments (Basic Protocols 1, 2, and 3). Applications for these samples include complex flow-cytometry-based immune cell content analyses in the HCC microenvironment, with antibodies optimized for applying gating strategies underlying cell sorting (Basic Protocol 4, Alternate Protocol 1, and Support Protocol 3); generation of HCC cell lines (Basic Protocol 4 and Support Protocol 2); preparation of tissue samples for immunohistochemistry, immunofluorescence, and histochemistry (Basic Protocol 5, Alternate Protocol 2, and Support Protocol 4); quantification of immune cells in tissue using high-throughput software (Support Protocol 5); and transcriptomic and proteomic analyses (Support Protocol 1, Basic Protocol 6, and Alternate Protocol 3). These broad and versatile techniques can then be applied to genetically distinct HCC through careful experimental design and planning and represent mainstay approaches in cancer biology (Fig. 1).
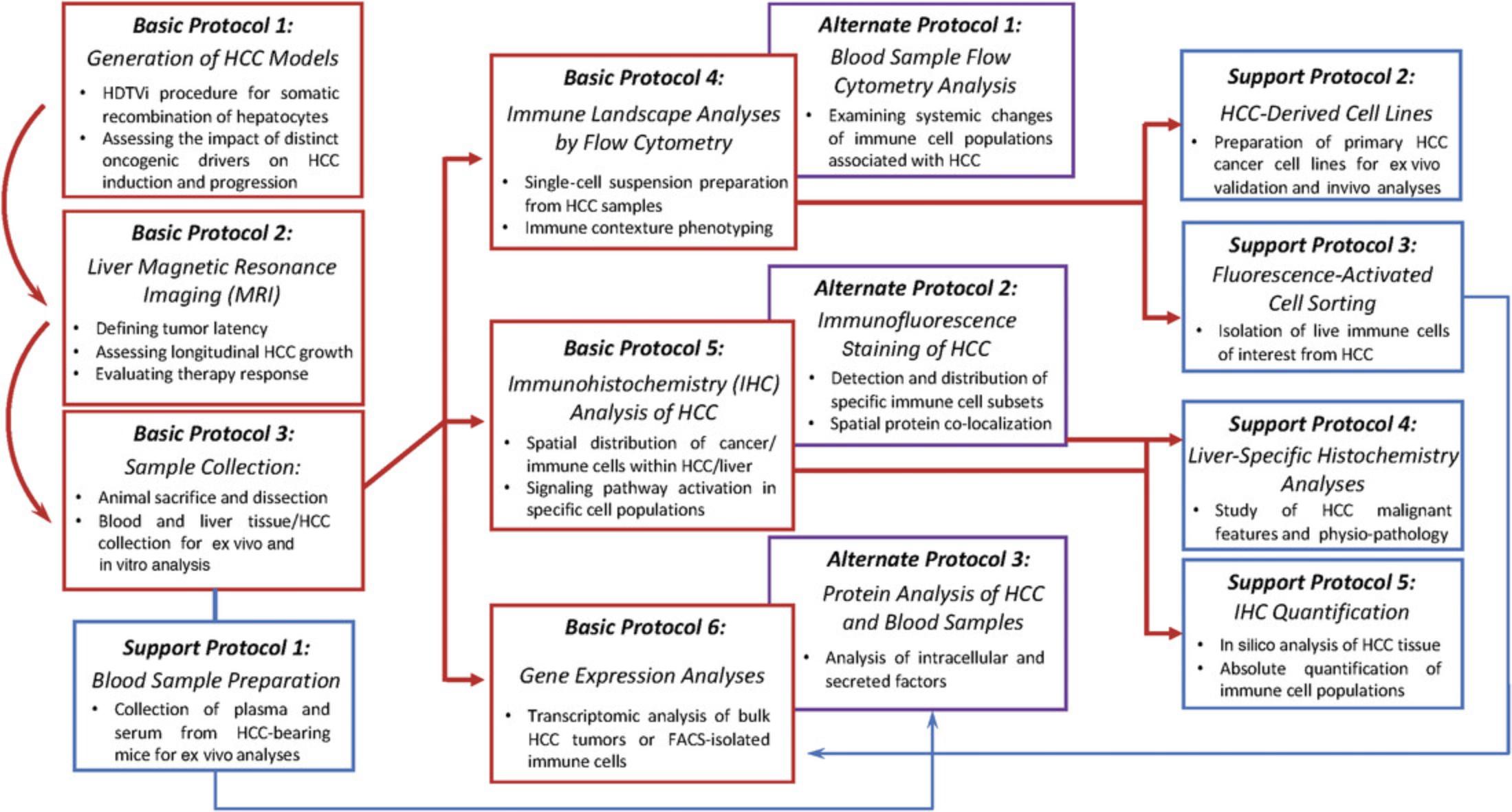
Basic Protocol 1: GENERATION OF HCC MOUSE MODELS BY HYDRODYNAMIC TAIL VEIN INJECTION
This protocol contains a brief description of the HDTVi procedure for generating HCC mouse models (Zhang et al., 1999). Examples of HCC mouse models obtained by this procedure and further applications of this technique have been reported in the literature (Molina-Sánchez et al., 2020; Wang et al., 2019). This approach allows the stable transduction of mouse hepatocytes to induce HCC by tail vein injection of a large volume of saline solution containing a combination of a Sleeping Beauty transposon for oncogene somatic integration, a vector encoding the Sleeping Beauty transposase, and a third vector expressing a single guide RNA (sgRNA) targeting the tumor-suppressor gene of interest, depending on the HCC oncogenic drivers of interest. A procedure for following HCC growth kinetics by measuring tumor volume using MRI are also described, along with steps to extract blood and HCC-bearing liver samples from mice upon reaching the predefined experimental endpoint.
NOTE : The instructions given in this protocol pertain to mice housed in individually ventilated cage (IVC) units; therefore, it is advisable to work under sterile conditions.
Materials
-
Animals: age (6-8 weeks)-, gender-, and strain-matched mice
-
Mixture of the plasmids of interest (selected according to the guidelines in step 1), in sterile saline solution (0.9% [w/v] NaCl) in a 50-ml tube
-
Virkon S (Biosecurity B.V.)
-
70% (v/v) ethanol
-
Warm (37°C) water in a 50-ml tube
-
Laminar flow hood
-
Food, cage, water, and housing material for mice
-
Highly absorbent diaper for animals
-
Scale (Sartorius Entris 4202-1S)
-
Heating lamp (IR100R PAR Philips)
-
Mouse restrainer (Bioseb Lab Instruments)
-
Three 26-G, 5/8-inch needles (BD Microlance)
-
3-ml syringes (BD Emerald)
-
Paper tissues
-
Heating pad (Digital thermostat TS-5000 comfort)
1.Before initiating the experiment, weigh the mice on a scale to determine the final volume of saline solution required for injections. The final volume should provide for an injection of 10% of each mouse body weight, plus enough excess for three mice to avoid not having enough material. Then, in a 50-ml tube, prepare the sterile saline solution containing the plasmids of interest:
1.10 µg per mouse of Sleeping Beauty transposon pT3 vector containing the oncogene of interest; 2.10 µg per mouse of pX330 vector expressing the sgRNA targeting the tumor-suppressor gene of interest for CRISPR-Cas9-mediated gene knockout; 3.5 µg per mouse of Sleeping Beauty transposase; 4.Sterile 0.9% (w/v) NaCl solution (amount equivalent to 10% of each mouse's weight).
The average mouse weight can be considered 20 g for this preparation.
The concentration of vectors injected can be changed according to the researcher's need. Injecting smaller amounts of plasmids may be useful when slower-developing/growing liver tumors are sought, to better assess treatment response, for instance.
The Sleeping Beauty transposon vectors can be purchased from Addgene and complemented, if desired, with a fluorescent reporter gene (e.g., gfp) through cloning for the detection of cancer cells.
2.Before beginning to work with the mice, clean a laminar flow hood using Virkon S and 70% ethanol. Place all required equipment in the flow hood and add a diaper to absorb any spilled bodily fluids.
3.Transfer the mice to the clean hood prepared in step 2.
4.Place the heating lamp above the open cages. Place the mice under the heater for 15 min before injection to allow vein dilation.
5.Immediately before injection, insert the mouse tail in warm water (37°C) for 15 s to further dilate the tail vein.
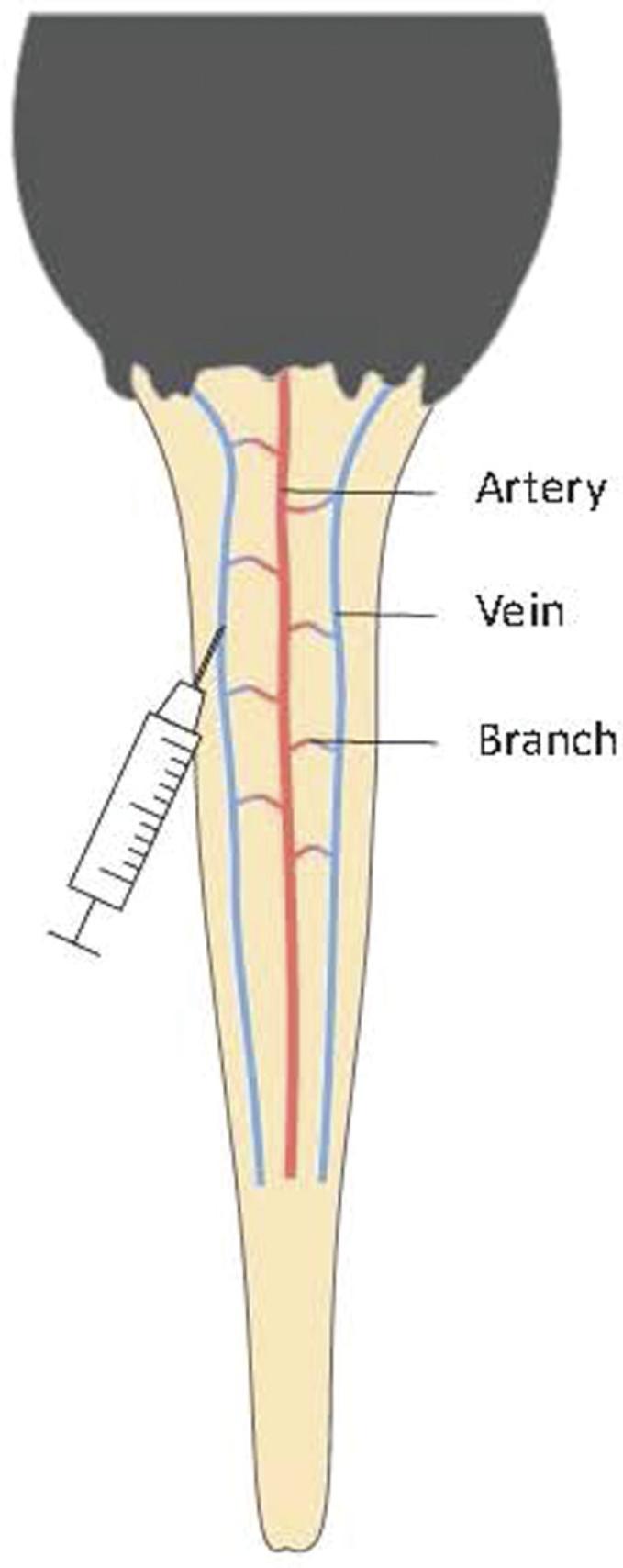
6.Accommodate the mouse in the rotating mouse restrainer, identify the caudal vein, and inject the saline solution containing the plasmids (10% of the mouse body weight) with a 3-ml syringe connected to a 26-G, 5/8-inch needle (Fig. 2). The injection point should be near the bottom of the tail, and the duration of the injection should be 8-10 s. Clean off any blood or bodily fluids with a paper tissue wetted with ethanol.
7.After the procedure, the mouse will be immobile in a resting state. Carefully transfer the mouse to a heating pad recovery station.
8.After a recovery time of 30 min, mice can be transferred to fresh cages with food, water, and housing material. Wet food should be placed at the bottom of the cage for 48 hr after the HDTVi procedure.
9.Monitor mice daily for a week after the procedure for any signs of distress or poor health. Warning signs include reduced mobility, piloerection, hunched gait, and weight loss.
10.Weigh mice twice a week to monitor possible weight loss following the HDTVi procedure.
Basic Protocol 2: ASSESSMENT OF HCC TUMOR PROGRESSION BY MAGNETIC RESONANCE IMAGING
After the injection of the saline solution containing the plasmids of interest via HDTVi, the mice should be monitored regularly for any symptoms of tumor growth, with tumor penetrance and aggressivity dependent on the combination of oncogene and tumor-suppressor knockout chosen. A key readout is the enlargement of the mouse abdomen, which increases as HCC progress and can affect the mouse's mobility and ability to ingest food and water.
Orthogonal to these clinical signs, tumor progression should be quantified using MRI. This procedure should start 2 weeks after injection and be repeated weekly or biweekly, adjusted according to the tumor growth kinetics of the combination of oncogenic drivers chosen.
Longitudinal tumor volume measurement can be used to determine longitudinal HCC tumor growth and to assess the efficacy of drug treatment in the context of preclinical intervention trials.
Materials
-
HCC model mice (Basic Protocol 1)
-
Opthalmic ointment (Alcon Duratears)
-
Laminar flow hood
-
Filled isoflurane station to provide 1.8% isoflurane
-
Respiratory sensor (Bruker)
-
Highly absorbent diaper for animals
-
Lubricated rectal thermometer probe (SA Instruments Inc.) and probe protectors
-
Anesthesia induction chamber
-
7-Tesla MRI instrument (Bruker Biospec 70/20)
-
Bruker Paravision 6.0.1
-
1H transmit-receive volume coil
-
Silicone warm water heating mat connected to a heating bath set at 35°C, to cover the mouse during scanning (SA Instruments Inc.)
-
Surgical tape
-
MIPAV (Medical Image Processing, Analysis and Visualization) open-source software (US National Institutes of Health, nih.gov) for use on Java-enabled platform
1.Check and refill the isoflurane stations if necessary. Open the valves for isoflurane, providing O2 at ∼35% and air at ∼65%.
2.Place the respiratory sensor on the animal bed and secure it with a piece of tape.
3.Lay a highly absorbent diaper on the animal bed inside the machine. Place a fresh lubricated probe protector around the rectal probe and ensure that the eye ointment is ready to be used.
4.In a laminar flow hood, open the mouse cage and place the mouse in an anesthesia induction chamber.
5.From the isoflurane station, open only the valve to the anesthesia induction chamber, ensuring that it is standing under the ventilation arm. Turn the isoflurane to 1.8%.
6.Confirm with a toe pinch that the mouse is properly anesthetized (after ∼3 min with isoflurane). Open the valve to the MRI and close the one to the anesthesia induction chamber.
7.Move the mouse to the animal bed, apply ophthalmic ointment to eyes, and secure the mouse with the tooth in the toothbar in the prone position (Fig. 3A). Ensure that the front paws are pointing backwards. Tape the silicone heating blanket over the mouse.
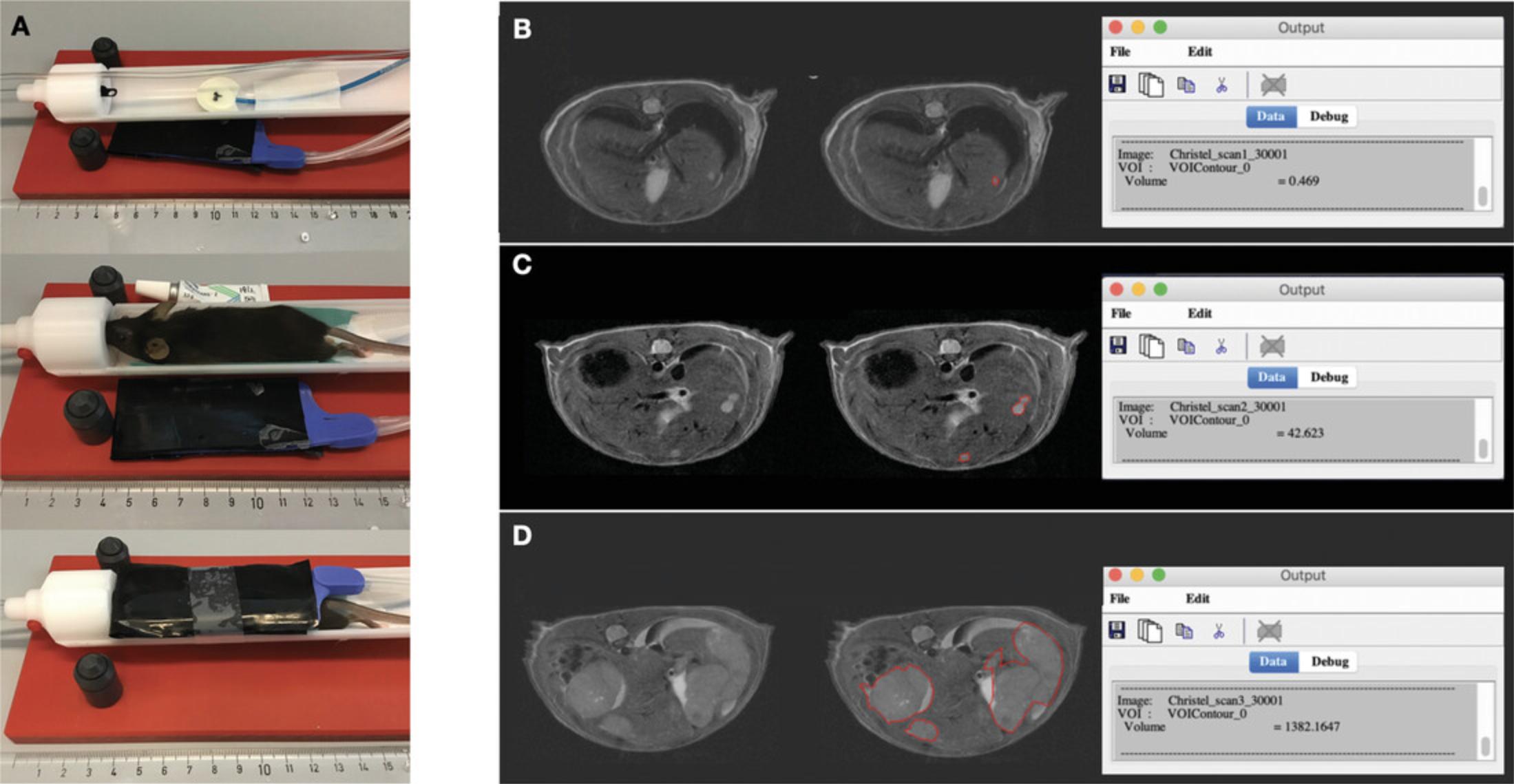
8.Carefully insert the bed until the abdomen of the mouse is in the center of the coil and magnetic field. Change the height of the animal bed if needed.
9.To start the scanning using the program Bruker ParaVision 6.0.1, select File (upper left) > New Study and fill in the (bold) fields. Click Create followed by Exam.
10.In the Palette > Explorer window, you can select Object, Region, and Application. Select the protocol optimized for the liver. For this, the following parameters are used: respiratory gated sequence with TR/TE = 2500/25 ms, 32 × 24-mm field of view (320 × 240 matrix, resolution of 0.1 mm), 30 × 0.7-mm axial slices and 4 averages (scan duration = 10 min).
11.Drag the localizer to the scan queue. Press Continue on the bottom of the queue to start the scan.
12.When the scan is finished, drag the user protocol into the examination window. Right-click the protocol to duplicate it. Adjust the geometry in the examination window to obtain a symmetric image. Click Continue to start the scan.
13.Repeat step 10 until the image is symmetric. Physically adjust the bed until your region of interest is in the middle of the scanned images.
14.Before proceeding to the first high-quality scan, ensure that the signal is fine-tuned by shimming. Some receivers do this automatically; otherwise, follow the instructions for the MRI machine in use.
15.Drag the desired T2 liver imaging protocol from the palette window into the queue. Select the finished scan protocol with the best geometry and drag it onto your new protocol. Confirm that the geometry should be copied, and press Continue.
16.When finished with the scan, ensure that the Export to DICOM macro is enabled, and close the examination window.
17.When the MRI scanning is finished:
-
Quit Paravision.
-
Unplug the respiratory battery.
-
Turn off the gas and isoflurane, and close the valves.
-
Turn off the warm bath.
-
Clean the animal bed thoroughly to avoid cross-contamination.
18.Repeat steps 1-17 for each mouse needed.
19.To measure the HCC tumor volume, use the software MIPAV (Bazin et al., 2007). The volume of interest (VOI) area is calculated after manual delineation of the tumor areas in each scan section. A step-by-step approach for tumor quantification can be found below and is shown in Figure 3B-D.
-
Draw the tumor area with the software tool that best fits the tumor contour. The option Draw level set VOI identifies contrast differences automatically and is commonly the optimal choice but can be adjusted manually if required.
-
If the contrast difference between tumor and healthy tissue is not clear, circle the tumor manually using the Ellipse VOI or Bspline VOI function.
-
Calculate the tumor volume by clicking on the drawn area, selecting View VOI Properties, and then selecting Volume in the next pop-up. Press Calculate. The tumor volume in mm3can be found in the Output pop-up.
Longitudinal tumor volume measurement for each mouse should be recorded in a charted format to allow the experimenter to generate growth curves illustrating the progressive increase in tumor volume through time, to compare the outgrowth of HCC based on the genetic makeup of cancer cells of interest, or to assess efficacy of treatment when applicable.
Basic Protocol 3: MOUSE SACRIFICE AND SAMPLE COLLECTION IN HCC MOUSE MODELS
When they reach a predefined endpoint, such as clinical symptoms, treatment cessation, or endpoint tumor size (2000 mm³, defined by MRI), mice are euthanized. The required steps to collect HCC tumors and blood for all subsequent sections are presented here.
Materials
-
HCC model mice (Basic Protocol 1)
-
Virkon S (Biosecurity B.V.)
-
70% (v/v) ethanol
-
1× PBS (see recipe)
-
Complete medium 10% (see recipe)
-
Laminar flow hood
-
Food, cage, water, and housing material for mice
-
Highly absorbent diaper for animals
-
CO2 hose to supply 10% of mouse cage volume per minute
-
Dissection plank with pin needles
-
Forceps, straight/serrated (Fine Science Tools)
-
Titanium Bulldog Spring Clamps (Fine Science Tools)
-
Scissors: straight, 8.5 cm (Fine Science Tools)
-
10-ml syringes (BD Emerald)
-
Butterfly needles (Terumo Surflo winged infusion set, 25-G)
-
EDTA blood collection tubes (Sarstedt Inc.)
-
50-ml tubes
-
Ice
1.Prepare a sterile laminar flow hood and clean with Virkon S and 70% ethanol. Place a clean diaper to absorb any spilled bodily fluids.
2.Clean surgical tools thoroughly, spray all required equipment with 70% ethanol, and place them inside the flow hood.
3.Transfer the mouse cage containing mice to be sacrificed to the hood. Transfer any littermates to a fresh cage with food, housing material, and water.
4.Connect a CO2 hose to the IVC cage. Adjust the system so the CO2 flow rate displaces ∼10% of the cage volume per minute.
5.After the mouse is immobile, check for any toe reflex to ensure that the mouse has been euthanatized successfully, and dislocate the neck to ensure sacrifice.
6.Place the mouse on a dissection plate with its ventral side up. Stretch out its legs, and use pin needles to fix the mouse's limbs.
7.Spray the abdomen with 70% ethanol, lift the skin with forceps, and carefully cut the skin open with scissors. Proceed with the incision from the lower abdomen up to the diaphragm and sternum, being careful not to puncture any organs. The liver should be readily visible after the initial incision is made.
8.Hold the sternum with forceps and carefully cut the diaphragm. Hold the sternum up with a clamp to expose the heart, which will be used for blood collection and PBS perfusion.
9.Before starting with the organ perfusion, prepare a 10-ml syringe filled with PBS and the EDTA tube for blood collection.
10.Connect a butterfly needle to an empty 1-ml syringe. Carefully puncture the right ventricle of the heart with the needle and pull the desired volume of blood from the heart. Immediately transfer this to the EDTA tube and place it on ice.
11.Remove the blood collection butterfly needle. Place a fresh butterfly needle on a 10-ml syringe previously loaded with PBS. Carefully puncture the left heart ventricle with the needle, and gently push the plunger to start perfusing the mouse with 10 ml PBS.
12.After ∼2 ml of PBS has been injected, perform a small cut with scissors of the left atrium and continue perfusing the remaining PBS. This procedure will ensure better circulation of the PBS to the peripheral organs. If the perfusion is successful, the liver should slowly decolorize after the injection of the 10 ml PBS.
13.To macrodissect the liver, gently push the gut and stomach aside and cut the supporting connective tissues. Be careful not to damage the neighboring organs. This step is crucial, as the anatomy can be significantly disrupted with the presence of tumors, and this could compromise the sterile environment, thus contaminating further steps of the protocol.
14.When the liver is successfully extracted, place the organ in a 50-ml tube containing 10 ml complete medium 10% on ice. This material will be used in Basic Protocols 4-6 and Alternate Protocol 2.
15.Dispose of the mouse body and clean the flow hood, surgical tools, and dissection plank with Virkon S and 70% ethanol.
Support Protocol 1: PREPARATION OF SERUM OR PLASMA FROM BLOOD
Serum and plasma can be prepared using the blood samples obtained in Basic Protocol 3 (step 10). The choice of serum versus plasma preparation is determined ahead of time depending on the research of interest. These samples have to be prepared immediately after blood withdrawal. Serum or plasma can be divided into aliquots and stored at −80°C, and subsequently used for protein analyses as described in Alternate Protocol 3. Other possible applications for these samples include measurement of the activity of hepatic enzymes (such as alanine transaminase) and the expression of HCC-related biomarkers (such as α-fetoprotein).
Materials
- Blood samples from HCC-bearing mice, in EDTA collection tubes (see Basic Protocol 3, step 10)
- 1.5-ml microcentrifuge tubes
- Microcentrifuge, 4°C
Preparation of plasma
1a. Centrifuge the blood sample in the EDTA collection tube for 20 min at 2000 × g , 4°C.
2a. A transparent solution of plasma will form on top of the red blood cells, along with a smooth buffy coat of white blood cells. Transfer the clear supernatant solution to a fresh 1.5-ml microcentrifuge tube.
Preparation of serum
1b. Transfer 100 µl of the blood sample from the EDTA collection tube to a 1.5-ml microcentrifuge tube and let it rest for 15-30 min at room temperature to allow it to clot.
2b. Centrifuge the 1.5-ml tube for 20 min at 2000 × g , 4°C.
3b. Transfer the clear supernatant (seric phase) to a fresh 1.5-ml tube.
Basic Protocol 4: SINGLE-CELL PREPARATION AND HCC IMMUNE LANDSCAPE PHENOTYPING BY FLOW CYTOMETRY
In this section, the steps required to generate a single-cell suspension from mouse HCC samples are described, along with the necessary steps for HCC immune landscape analysis by flow cytometry. The immune context analyses described below are tailored to characterize the changes in liver immune cell subsets associated with HCC development and corresponding to the chosen genetic heterogeneity of HCC. These changes can encompass differences in cell population content, as well as distinct markers of activation, proliferation, and polarization. The single-cell preparation described in this protocol can also be used for preparing HCC cell lines (Support Protocol 2).
NOTE : If planning to use the preparation for the isolation of HCC cell lines (Support Protocol 2), perform this procedure under sterile conditions in a laminar flow hood and with sterile solutions.
Materials
-
Liver Dissociation Kit (Miltenyi)
-
HCC-bearing mouse liver (see Basic Protocol 3, step 14)
-
Complete medium 10% (see recipe)
-
Liver medium (see recipe)
-
FACS buffer (see recipe), sterile, 4°C
-
Erylysis buffer (see recipe)
-
Primary antibodies (see Tables 2 and 3)
-
BD Brilliant Buffer stain (BD Biosciences)
-
1× PBS (see recipe)
-
UltraComp eBeads (Thermo Fisher), diluted 1/10 in FACS buffer
-
Live/dead staining solution (see recipe)
-
Fixation/permeabilization solution: 1 part eBioscience™ Fixation/Permeabilization Concentrate and 3 parts eBioscience™ Fixation/Permeabilization Dilute (Invitrogen)
-
Permeabilization solution: Permeabilization Buffer (10×; Invitrogen), diluted 1:10 in deionized water
-
gentleMACS C-Tubes (Miltenyi)
-
Laminar flow hood
-
10-cm Petri dish
-
Forceps, straight/serrated (Fine Science Tools)
-
Disposable scalpel blades (Swann Worton)
-
gentleMACS Octo Dissociator with heaters (Miltenyi)
-
50-ml tubes
-
100-µm filter (Falcon)
-
Centrifuge for 15- and 50-ml tubes
-
1.5-ml microcentrifuge tubes
-
96-well U-bottom plates
-
Flow cytometer (preferably BD Biosciences Fortessa) capable of detecting 16 fluorescent labels and with 4 lasers (violet, 405 nm; blue, 488 nm; green, 561 nm; red, 640 nm) or superior; see Table 1 for recommended configuration and fluorochromes
-
Centrifuge for cell plates
-
Aluminum foil
-
Ice
-
Paper towel
-
FACS tubes: 1.40-ml U-bottom tubes (Micronic)
-
Appropriate software: e.g., FACSDiva, FlowJo, and optionally software for automation or high-dimensional reduction analysis (see step 37)
| Laser | Position | Mirror | Filter | Fluorescent markers that can be used in each channel |
|---|---|---|---|---|
| 405 nm (violet) | A | 750 LP | 780/60 | BV786 |
| B | 690 LP | 710/50 | BV711, SB702 | |
| C | 650 LP | 670/30 | BV650, QD655 | |
| D | 595 LP | 610/20 | BV605 | |
| E | 505 LP | 525/50 | BV510, QD525 | |
| F | Empty | 450/50 | BV421, eF450, AF405, Pacific Blue, BFP, Hoechst, DAPI | |
| 488 nm (blue) | A | 735 LP | 780/40 | - |
| B | 650 LP | 695/40 | BB700, PerCP, PerCP-Cy5.5, PerCP-eFluor710 | |
| C | 585 LP | 610/20 | PI | |
| D | 505 LP | 530/30 | BB515, AF488, FITC, CFSE, GFP, YFP | |
| E | Empty | 488/10 | - | |
| 561 nm (green) | A | 750 LP | 780/60 | PE-Cy7 |
| B | 690 LP | 710/50 | PE-Cy5.5 | |
| C | 650 LP | 670/30 | PE-Cy5, 7-AAD | |
| D | 595 LP | 610/20 | PE-CF594, AF594, PE-Dazzle, mCherry, PI | |
| E | Empty | 586/15 | PE, tdTomato | |
| 638 nm (red) | A | 755 LP | 780/60 | APC-Cy7, IR-Dye |
| B | 690 LP | 730/45 | APC-R700, AF700 | |
| C | Empty | 670/30 | APC, AF647 |
Single-cell preparation
1.Before starting the processing of HCC-bearing or control liver, prepare liver dissociation solution according to the manufacturer's instructions for the Liver Dissociation Kit in a gentleMACS C-Tube, using 200 µl enzyme D, 100 µl enzyme R, 20 µl enzyme A, and 4.7 ml DMEM per reaction. Prepare the solution immediately before use or store in 4°C for use on the same day.
2.In a laminar flow hood, transfer the HCC-bearing liver isolated in Basic Protocol 3 (step 14) to a sterile 10-cm Petri dish containing enough complete medium 10% to submerge the liver.
3.Remove the gallbladder with forceps and a scalpel. Be careful to not burst the organ open.
4.Carefully dissect the tumor with a scalpel, gently clamping the organ parts with forceps to expose the tumor nodules. The tumor may be fibrotic, mushy, or necrotic, which can interfere with the dissection procedure.
5.Cut off several small tumor pieces with a scalpel. If more than one nodule is present, take a small piece of each nodule to account for any cancer cell or microenvironmental heterogeneity.
6.Transfer ∼25 mg of tumor pieces to a C-Tube containing the 5 ml of liver dissociation solution from step 1.Further pipet up and down to ensure that samples are collected from the Petri dish.
7.Place C-Tubes on the gentleMACS dissociator. Run the program for liver digestion, 37C_m_LIDK_1 , which runs for 37 min.
8.While the digestion is taking place, prepare for the next steps by placing a 100-µm filter on a 50-ml tube. Rinse the filter with 1 ml liver medium.
9.When the liver digestion 37C_m_LIDK_1 program has ended, remove the C-Tubes from the gentleMACS dissociator. Working inside the laminar flow hood, transfer the digested samples to the 50-ml tube through the 100-µm filter. Gently dissociate any leftover tissue/tumor clumps caught in the filter and rinse the filter with 10 ml liver medium.
10.Discard the filter. Add 10 ml liver medium to the digested solution and centrifuge 10 min at 300 × g , 4°C.
11.Remove the supernatant and wash the pellet in 10 ml 4°C sterile FACS buffer. Centrifuge 10 min at 300 × g , 4°C.
12.Remove the supernatant. The single-cell preparation can be further processed to generate HCC cell lines (Support Protocol 2) or subjected to flow cytometry analysis of the HCC immune landscape as described in the steps that follow.
Primary antibody staining and single stain preparation
Ensure that all the following steps are performed on ice and protected from light (using aluminum foil, for instance). Antibody panels for myeloid and lymphoid immune populations, along with the fluorochrome and recommended dilution of each antibody, are listed in Tables 2 and 3.
| Marker | Clone | Fluorochrome | Product reference | Recommended dilution |
|---|---|---|---|---|
| Cell surface antigens | ||||
| CD16/CD32 (Fc block) | 2.4G2 | - | BD Biosciences, cat. no. 553142 | 1:400 |
| CD11b | M1/70 | BV421 | BD Biosciences, cat. no. 562605 | 1:1200 |
| CD45 | 30-F11 | AF700 | Biolegend, cat. no. 103128 | 1:200 |
| Ly6G | 1A8 | APC | Biolegend, cat. no. 127614 | 1:150 |
| Ly6C | HK1.4 | BV605 | Biolegend, cat. no. 128036 | 1:600 |
| F4/80 | BM8 | PE-Cy7 | Biolegend, cat. no. 123114 | 1:400 |
| MHCII | M5/114.15.2 | BV650 | BD Biosciences, cat. no. 563415 | 1:200 |
| CD11c | N418 | PE-Cy5.5 | Life Technologies, cat. no. 35-0114-82 | 1:400 |
| CD115 | AFS98 | PE/Dazzle | Biolegend, cat. no. 135528 | 1:200 |
| PD-L1 | 10F.9G2 | PE | Biolegend, cat. no. 124308 | 1:400 |
| Intracellular antigens | ||||
| Ki67 | B56 | BV786 | BD Biosciences, cat. no. 563756 | 1:200 |
| Live/dead staining | ||||
| Zombie NIR Fixable Viability Kit | - | - | Biolegend, cat. no. 423105 | 1:1000 |
| Marker | Clone | Fluorochrome | Product reference | Recommended dilution |
|---|---|---|---|---|
| Cell surface antigens | ||||
| CD16/CD32 (Fc Block) | 2.4G2 | - | BD Biosciences, cat. no. 553142 | 1:400 |
| CD11b | M1/70 | BV650 | BD Biosciences, cat. no. 563402 | 1:400 |
| CD45 | 30-F11 | AF700 | Biolegend, cat. no. 103128 | 1:200 |
| CD19 | 6D5 | BV510 | Biolegend, cat. no. 115546 | 1:200 |
| CD3 | 500A2 | PerCP-eF710 | eBioScience, 46-0033-82 | 1:300 |
| NK1.1 | PK136 | PE-Cy7 | Biolegend, cat. no. 108714 | 1:200 |
| CD8 | 53-6.7 | FITC | Life Technologies, cat. no. 11-0081-82 | 1:200 |
| CD4 | GK1.5 | APC | Biolegend, cat. no. 100412 | 1:200 |
| PD-1 | J43 | BV421 | BD Biosciences, cat. no. 562584 | 1:500 |
| CD44 | IM7 | BV605 | Biolegend, cat. no. 103047 | 1:400 |
| CD69 | H1.2F3 | PE/Dazzle | Biolegend, cat. no. 104536 | 1:200 |
| Intracellular antigens | ||||
| FoxP3 | FJK-16s | PE-Cy5.5 | Life Technologies, cat. no. 35-5773-82 | 1:200 |
| Granzyme B | QA16A02 | PE | Biolegend, cat. no. 372208 | 1:200 |
| Ki67 | B56 | BV786 | BD Biosciences, cat. no. 563756 | 1:200 |
| Live/dead staining | ||||
| Zombie NIR Fixable Viability Kit | - | - | Biolegend, cat. no. 423105 | 1:1000 |
NOTE : If the liver perfusion was successful and only few red blood cells are present in the sample (pellet is not red), erythrocyte lysis (erylysis; steps 13 and 14) is optional.
13.Optional : Resuspend single-cell suspension from step 12 in 1 ml per sample of erylysis buffer.
14.Optional : Incubate 5 min at room temperature. Centrifuge samples 10 min at 300 × g , 4°C, and remove the supernatant.
15.Resuspend the single-cell suspension from step 12 (or step 14 if erylysis is performed) in 1 ml FACS buffer.
16.Transfer 200 µl of each single-cell suspension to individual wells of a 96-well plate for each staining panel to be applied. Keep 100 µl of one sample to use in the compensation setup (step 23).
17.Wash cells by adding 100 µl 4°C FACS buffer to each well and centrifuging the 96-well plate 2 min at 350 × g , 4°C. Discard supernatants and dry off any excess liquid by inverting the plate on a paper towel.
18.Repeat step 17.
19.Add 75 µl of Fc block prepared in FACS buffer (see Tables 2 and 3 for dilution) to each single-cell HCC suspension sample (∼5 × 106 cells each).
20.Resuspend cells and incubate 15-30 min on ice. Repeat step 17.
21.Add 75 µl of primary antibody solution prepared in BD Brilliant Buffer (using myeloid and lymphoid panels if needed; see Tables 2 and 3 for antibodies and dilutions) to each HCC single-cell suspension sample.
22.Resuspend cells up and down and incubate 15-30 min on ice, protected from light in aluminum foil.
23.While cells are incubating, prepare the compensation samples:
-
Add 100 µl diluted UltraComp eBeads per fluorophore used per well of a 96-well plate.
-
Add 0.5 µl of each primary antibody used in each well. Mix up and down with a pipet.
-
Incubate plate, protected from light, for 5 min at room temperature.
-
Wash compensation beads according to step 17 and resuspend in 100 µl FACS buffer. They can now be used as compensation samples (see step 35).
-
For live/dead compensation, divide 100 µl of the cell suspension prepared in step 16 between two separate wells containing 50 µl of each sample. Proceed to the steps below, in which the live/dead compensation samples will be prepared along with the main samples to be stained.
24.After the primary antibody incubation (step 22) is complete, wash cells with 4°C 100 µl PBS and centrifuge 2 min at 350 × g , 4°C. Discard supernatant and dry off any excess liquid by inverting the plate on a paper towel. Repeat this step twice.
Live/dead staining
25.Add 100 µl of live/dead staining solution to each HCC single-cell suspension sample. For live/dead compensation, add 100 µl live/dead staining solution to one compensation well and 100 µl PBS to the other. Resuspend all samples up and down to homogenize the solutions.
26.Incubate samples 20-30 min on ice, protected from light.
27.Wash cells twice according to step 17.
28.Depending on the application, proceed as indicated:
- a.If live cells are required for flow cytometry analyses or sorting:
- Resuspend cells in 100 µl FACS buffer; 2.Transfer the cell suspension to FACS tubes and proceed to flow cytometry (step 36) or cell sorting (Support Protocol 3).
- b.If cells need to be fixed for short-term storage or if intracellular staining is desired: Continue to steps 29-35 (these also include the preparation of live/dead compensation samples).
Fixation and intracellular staining
29.Resuspend cells and live/dead compensation wells in 100 µl fixation/permeabilization solution and incubate 30 min at room temperature, protected from light.
30.Wash cells and live/dead compensation wells with 100 µl permeabilization solution and centrifuge 2 min at 350 × g , 4°C. Discard supernatant and dry off any excess liquid by inverting the plate on a paper towel.
31.Repeat washing step 17.
32.For live/dead compensation, resuspend both unstained and stained compensation samples in 50 µl FACS buffer each and mix them into one well. This live/dead sample is now ready to be used for compensation.
33.Add 75 µl of the intracellular primary antibody solution diluted in permeabilization solution to each single-cell suspension sample (including myeloid and lymphoid panels if needed; see Tables 2 and 3 for antibodies and dilutions).
34.Repeat washing step 17 twice.
Sample acquisition
35.Resuspend cells in 100 µl FACS buffer and proceed to flow cytometry for analysis of the samples, along with the stained compensation beads (step 23) and live/dead compensation sample (step 32). Gating strategies designed for the analysis of lymphoid and myeloid cell populations for the hepatic microenvironment can be found in Figures 4 and 5.
![Details are in the caption following the image Representative flow cytometry plots of the liver immune landscape analyses, depicting the gating strategy applied for sequential identification of myeloid populations in a mouse liver that underwent HDTVi with empty vectors. Total cell populations were gated on forward and side scatter to identify single-cell and live-cell events (Zombie-NIR-negative cells). Immune cells were identified by their high expression of CD45. The myeloid cells were further gated as double-positive CD45<sup>+</sup>CD11b<sup>+</sup> cells and the lymphoid cells as CD45<sup>+</sup>CD11b<sup>+</sup> cells. The subpopulations of myeloid cells (dendritic cells [DCs], classical monocytes, nonclassical monocytes, neutrophils, total macrophages, Kupffer cells [KCs], and bone-marrow-derived macrophages [BMDMs]) were identified based on marker combinations, as represented. The myeloid panel was optimized with titrated antibodies, and fluorescence minus one (FMO) controls were used to set up positive and negative populations.](https://static.yanyin.tech/literature_test/cpz1147-fig-0004-m.jpg)
![Details are in the caption following the image Representative flow cytometry plots of the liver immune landscape analyses, depicting the gating strategy applied for sequential identification of lymphoid cell populations in a mouse liver that underwent HDTVi with empty vectors. Total cell populations were gated on forward and side scatter to identify single-cell and live-cell events (Zombie-NIR-negative cells). Immune cells were identified by their high expression of CD45. The myeloid cells were further gated as double positive CD45<sup>+</sup>CD11b<sup>+</sup> cells and the lymphoid cells as CD45<sup>+</sup>CD11b<sup>–</sup> cells. The subpopulations of lymphoid cells (NK cells, NKT cells, B cells, CD4<sup>+</sup> T cells, CD8<sup>+</sup> T cells, conventional CD4<sup>+</sup> T [Conv CD4], and regulatory T cells [Tregs]) were identified based on marker combinations, as represented. The lymphoid panel was optimized with titrated antibodies, and fluorescence minus one (FMO) controls were used to set up positive and negative populations.](https://static.yanyin.tech/literature_test/cpz1147-fig-0005-m.jpg)
36.The samples can be acquired using software such as FACSDiva. During sample acquisition and compensation setup:
-
Ensure that the voltage settings for forward versus side scatter (SSC-A vs. FSC-A) are optimal to include all cells.
-
Ensure that the voltage used for the fluorescence channels is within the instrument's detection range and that it satisfactorily separates the positive and negative signals for each fluorescence channel.
-
Acquire a predefined minimum number of cells per gate of interest. As a general recommendation, acquire at least 100,000 singlet live cells for liver tumor samples.
37.The acquired data can be analyzed using several methods.
-
When the numbers of samples, experimental conditions, and markers are relatively low, analysis can be done manually using commercially available software packages (such as FlowJo and FACSDiva).
-
When the data require high-dimensional reduction analysis or automated approaches (Cheung et al.,2021), software packages such as viSNE, UMAP, FlowSOM, SPADE, or CITRUS (Saeys, Van Gassen, & Lambrecht,2016) may be employed.
Alternate Protocol 1: FLOW CYTOMETRIC ANALYSIS OF CIRCULATING IMMUNE CELLS
The blood acquired in Basic Protocol 3 (step 10) can be used in this flow cytometry protocol to assess systemic changes of immune cell populations in HCC tumor-bearing mice by using the panels reported in Tables 2 and 3.
NOTE : Ensure that all the following steps are performed on ice with the samples protected from light (using aluminum foil, for instance).
Materials
-
Blood samples from HCC-bearing mice, in EDTA collection tubes (Basic Protocol 3, step 10)
-
Erylysis buffer (see recipe)
-
FACS buffer (see recipe)
-
Primary antibodies (see Tables 2 and 3)
-
BD Brilliant Buffer stain (BD Biosciences)
-
UltraComp eBeads (Thermo Fisher), diluted 1/10 in FACS buffer
-
Live/dead staining solution (see recipe)
-
1× PBS (see recipe)
-
Fixation/permeabilization solution: 1 part eBioscience™ Fixation/Permeabilization Concentrate and 3 parts eBioscience™ Fixation/Permeabilization Dilute (both from Invitrogen)
-
Permeabilization solution: Permeabilization Buffer (10×; Invitrogen), diluted 1:10 in deionized water
-
96-well U-bottom plates
-
Centrifuge for cell plates
-
Paper towel
-
Ice
-
Aluminum foil
-
FACS tubes: 1.40-ml U-bottom tubes (Micronic)
-
Flow cytometer (preferably BD Biosciences Fortessa) capable of detecting 16 fluorescent labels and with 4 lasers (violet, 405 nm; blue, 488 nm; green, 561 nm; red, 640 nm) or superior; see Table 1 for recommended configuration and fluorochromes
-
Appropriate software: e.g., FACSDiva, FlowJo
Erythrocyte lysis (erylysis)
1.Transfer 50 µl of each blood sample into separate wells of a 96-well plate for each staining panel to be applied. Keep 100 µl of one sample for the compensation setup (discussed in step 10e).
2.Resuspend the blood samples in 100 µl erylysis buffer per sample.
3.Incubate 10 min at room temperature.
4.Wash cells with 100 µl 4°C FACS buffer and centrifuge the 96-well plate 2 min at 350 × g , 4°C. Discard supernatant and dry off any excess liquid by inverting the plate on a paper towel.
5.Repeat step 4.
Primary antibody staining and single stain preparation
6.Add 25 µl of Fc block prepared in FACS buffer (see Tables 2 and 3 for dilution) to each single-cell suspension sample (∼5 × 106 cells each).
7.Resuspend cells and incubate 15-30 min on ice. Repeat washing step 4.
8.Add 25 µl of primary antibody solution prepared in BD Brilliant Buffer (include myeloid and lymphoid panels if needed; see Tables 2 and 3 for antibodies and dilutions) to each single-cell suspension sample.
9.Resuspend cells up and down and incubate for 15-30 min on ice, protected from light with aluminum foil.
10.While cells are incubating, prepare the compensation samples:
-
Add 100 µl of diluted UltraComp eBeads per fluorophore used to each well of a 96-well plate.
-
Add 0.5 µl of each primary antibody used to the appropriate well. Mix up and down with a pipet.
-
Incubate plate, protected from light with aluminum foil, for 5 min at room temperature.
-
Wash compensation beads according to step 4 and resuspend in 100 µl FACS buffer. They can now be used as compensation samples.
-
For live/dead compensation, divide 100 µl of the cell suspension prepared in step 1 into two separate wells containing 50 µl of each sample. Proceed to the next steps, in which the live/dead compensation samples will be prepared along with the main samples to be stained.
11.After the primary antibody incubation (step 9) is complete, wash cells with 4°C 100 µl PBS and centrifuge 2 min at 350 × g , 4°C. Discard supernatant and dry off any excess liquid by inverting the plate on a paper towel. Repeat this step twice.
Live/dead staining
12.Add 50 µl live/dead staining solution to each single-cell suspension sample. For live/dead compensation, add 100 µl live/dead staining solution to one compensation well and 100 µl PBS to the other. Resuspend all samples up and down to homogenize the solutions.
13.Incubate samples for 20-30 min on ice and protected from light.
14.Wash cells twice as in step 4.
15.Depending on the application, proceed as indicated:
1.Resuspend the cells in 100 µl FACS buffer; 2.Transfer the cell suspension to FACS tubes and proceed to flow cytometry (step 23) or cell sorting (Support Protocol 3).
If the cells need to be fixed for short-term storage or if intracellular staining is desired: Continue to the following steps.
Fixation and intracellular stain
16.Resuspend cells and live/dead compensation samples in 100 µl fixation/permeabilization solution and incubate 30 min at room temperature, protected from light.
17.Wash cells and live/dead compensation wells with 100 µl permeabilization solution and centrifuge 2 min at 350 × g , 4°C. Discard supernatant and dry off any excess liquid by inverting the plate on a paper towel.
18.Repeat step 4.
19.For live/dead compensation, resuspend both unstained and stained compensation samples in 50 µl each of FACS buffer and mix them into one well. This live/dead sample is ready to be used for compensation.
20.Add 50 µl of the intracellular primary antibody solution diluted in permeabilization solution to each single-cell suspension sample (include myeloid and lymphoid panels if needed; see Tables 2 and 3 for antibodies and dilutions).
21.Repeat washing step 4 twice.
Sample acquisition
22.Resuspend cells in 100 µl FACS buffer and proceed to flow cytometry for analysis of the samples, along with the stained compensation beads (step 10) and live/dead compensation sample (step 19). Gating strategies designed for the analysis of lymphoid and myeloid cell populations can be found in Figures 4 and 5.
23.The samples can be acquired using software such as FACSDiva. During sample acquisition and compensation setup:
-
Ensure that the voltage settings for forward versus side scatter (SSC-A vs. FSC-A) are optimal to include all cells.
-
Ensure that the voltage used for the fluorescence channels is within the instrument's detection range and that it satisfactorily separates the positive and negative signals for each fluorescence channel.
-
Acquire a predefined minimum number of cells per gate of interest. As a general recommendation, acquire at least 10,000 singlet live cells for blood samples.
Support Protocol 2: GENERATION, MAINTENANCE, AND CHARACTERIZATION OF HCC CELL LINES
In this protocol, the steps required to ensure the establishment and maintenance of ex vivo cell lines obtained from the single-cell suspension from Basic Protocol 4 are described. These cell lines are useful tools for interrogating how the genetic background of HCC can influence intrinsic factors in cancer cells, such as proliferative capacity and up- or downregulation of signaling pathways. These cell lines can also be used to assess the heterotypic interactions of HCC cell lines with immune cells in co-culture settings, thus providing an additional way to study the influence of the genetic makeup of HCC cancer cells on the liver immune microenvironment.
NOTE : Perform this procedure under sterile conditions in a laminar flow hood and using sterile solutions.
Materials
-
Collagen solution (see recipe)
-
1× PBS (see recipe)
-
HCC single-cell preparation (see Basic Protocol 4, step 12)
-
Complete medium 10% (see recipe)
-
Complete medium 20% (see recipe)
-
Trypsin/EDTA: Trypsin/EDTA 0.5% (Gibco), diluted 1:10 in 1× PBS
-
Freezing medium (see recipe), 4°C
-
Laminar flow hood
-
10-cm Petri dishes
-
Culture flasks (e.g., T75)
-
6-well plates
-
37°C, 5% CO2 incubator cell incubator
-
Water bath, 37°C
-
15- and 50-ml tubes
-
Centrifuge for 15- and 50-ml tubes
-
1.8-ml cryotubes
-
Cell freezing container (Biocision)
-
−80°C and (optionally) −150°C freezers
1.Before proceeding to cell line plating, coat culture flasks and Petri dishes with collagen solution to provide optimal cell culture conditions. Flasks coated with collagen are also used for the maintenance of cell lines.
-
Add 3 ml collagen solution to each 10-cm Petri dish needed, and 5 ml collagen solution to each T75 flask (for cell line maintenance).
-
Incubate flasks and Petri dish 1 hr at 37°C.
-
Recover the collagen solution for reuse, or discard if it has been used more than three times.
-
Add 10 ml PBS to each flask or Petri dish. Keep for later use in the incubator or proceed to the next step.
If storing coated dishes or flasks, remove the PBS before use.
2.Resuspend the single-cell suspension (Basic Protocol 4, step 12) in 1 ml complete medium 10% onto a precoated Petri dish containing an additional 9 ml complete medium 10%.
3.Incubate cells overnight in 10 ml complete medium 10% in a 37°C, 5% CO2 incubator.
4.On day 1, remove 5 ml medium and add 5 ml fresh complete medium 10%.
5.On day 2, if cells are attached, remove all medium from the Petri dish and add 10 ml fresh complete medium 10%. If cells are not yet attached, repeat step 4 instead.
6.From this point onwards, closely monitor the growth of cell clumps. Replenish medium every 3 days.
-
Wash cells twice with 10 ml prewarmed PBS.
-
Add 1 ml prewarmed trypsin/EDTA and incubate cells 5 min at 37°C, 5% CO2.
-
Add 9 ml complete medium 10% to inactivate trypsin. Transfer 2 ml of the cells into precoated 6-well plates and monitor cell behavior and growth. Replenish medium every 3 days.
7.When the cells are confluent, transfer cells to a T75 culture flask for expansion. The dilution needs to be optimized, but should permit cells to maintain close contacts. For cell transfer:
-
Wash cells twice with 10 ml prewarmed PBS.
-
Add 1 ml prewarmed trypsin/EDTA and incubate cells 5 min at 37°C, 5% CO2.
-
Add 9 ml complete medium 10% to inactivate trypsin. Transfer the cells into a precoated T75 flask and monitor cell behavior and growth.
8.After cells are expanded satisfactorily, the usual passage rate is twice weekly. This can be adjusted according to the cell growth rate. To split cells from T75 flasks for maintenance, repeat step 7.The split ratio must be optimized for each cell line.
9.Once the cell line is established (passage 4-5), assess cell line purity by flow cytometry (see Table 5 for a list of the cell populations that should be investigated in the established HCC cell lines and the proposed markers to identify these cells).
10.To freeze cells for long-term storage:
-
Wash cells twice with 10 ml prewarmed PBS.
-
Add 1 ml prewarmed trypsin/EDTA and incubate cells 5 min at 37°C, 5% CO2.
-
Add 9 ml complete medium 10% to inactivate trypsin.
-
Transfer cell suspension to a 15-ml tube and centrifuge 5 min at 300 ×g, room temperature.
-
Remove the supernatant and resuspend the cell pellet in 5 ml cold (4°C) freezing medium to obtain a concentration of 5-10 × 106cells/ml.
-
Distribute 1 ml each into 1.8-ml cryotubes, transfer to a cell freezing container, and place in a −80°C freezer for 2 hr.
-
Transfer cryotubes to a −150°C freezer for long-term storage or keep in −80°C freezer for medium-term use.
Support Protocol 3: FLUORESCENCE-ACTIVATED CELL SORTING OF LIVER SINGLE-CELL PREPARATION
Cells obtained from single-cell suspensions generated from the extracted HCC samples and prepared for flow cytometry in Basic Protocol 4 can be sorted based on surface expression markers. This step is essential for the subsequent preparation of mRNA and transcriptional analyses of cell populations shown in Figures 4 and 5.
Materials
-
Unfixed (live) stained HCC samples (see Basic Protocol 4, step 28)
-
FACS buffer (see recipe)
-
UltraComp eBeads (Thermo Fisher), diluted 1/10 in FACS buffer, to compensate the panels designed using the antibodies listed in Tables 2 and 3
-
TRIzol LS reagent (Thermo Fisher)
-
5-ml round-bottom polystyrene tubes with strainer cap (Stemcell Technologies)
-
1.5-ml microcentrifuge tubes
-
Flow cytometry cell sorter (e.g., BD FACSAria Fusion Cell Sorter)
-
Microcentrifuge
-
Fume hood
1.Filter the unfixed HCC cell suspension samples in the strainer cap of a 5-ml round-bottom tube to remove cell clumps.
2.Prepare the compensation beads and live/dead compensation samples for each fluorochrome using UltraComp eBeads (see Basic Protocol 4, step 23).
3.Bring the cell suspension, stained compensation beads, and live/dead compensation samples to the FACS machine. Define the gates to sort the cell populations of interest based on Figures 4 and 5.The number of sorted cells should be optimal for the desired application.
4.Sort the population of interest in 350 µl FACS buffer in 1.5-ml tubes.
5.Centrifuge cells 10 min at 300 × g , 4°C.
6.Remove the supernatant and resuspend the cell pellet in 1 ml of TRlzol in a fume hood.
7.Transfer sample to −80°C freezer for long-term storage or proceed to Basic Protocol 6 for mRNA extraction and subsequent reverse transcription quantitative polymerase chain reaction (RT-qPCR) analyses.
Basic Protocol 5: PREPARATION AND IMMUNOHISTOCHEMICAL ANALYSIS OF TUMOR TISSUES FROM HCC-BEARING LIVER
Immunohistochemistry (IHC) can be used to assess the absolute number and spatial distribution of immune cell populations of interest within the tumor tissue. The following protocol describes the general steps to create FFPE blocks from HCC-bearing mouse liver (see Basic Protocol 3, step 14) and optimized steps for IHC staining of antigens commonly used to identify different immune cell populations, investigate the activation of signaling pathways frequently involved in hepatocarcinogenesis, and confirm the expression or knockout of chosen target proteins in HCC for markers described in Table 4.
| Antigen retrieval | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Marker | Fixative | Primary purpose | Buffer | Method | Temp. (°C) | Time (min) | Cooldown time (min) | Blocking buffer | Incubation buffer | Primary antibody and dilution (by fixative) | Secondary antibody | Marker uses streptavidin/H RP? |
| B220 | Formalin or EAF | Identify B cells | Citrate buffer | Water bath | 96 | 30 | 30 | II | I | B220 (BD Pharmingen; 557390), 1:4000 in formalin- and EAF-fixed | Goat-α-Rat-Bio (Biotech; 3052-08), 1:150 | Yes |
| c-Myc | Formalin or EAF | Assess expression of c-Myc oncogene | Tris-EDTA | Pressure cooker | 98 | 20 | - | I | I | C-MYC (Abcam; ab32072), 1:4000 in formalin- or 1:100 in EAF-fixed | EnVision+ System–HRP Labelled Polymer Anti-Rabbit (Dako) | No |
| CD4 | Formalin or EAF | Identify CD4+ T cells | Tris-EDTA | Heat-induced epitope retrieval machine | 98 | 30 | - | I | I | CD4 (eBiosience; 14-976682), 1:1000 in formalin- or 1:2000 in EAF-fixed | Goat-α-Rat-Bio (Southern Biotech; 305208), 1:100 | Yes |
| CD8 | Formalin or EAF | Identify CD8+ T cells | Tris-EDTA | Heat-induced epitope retrieval machine | 98 | 20 | - | I | I | CD8 (eBiosience; 14-080882), 1:2000 in formalin- or 1:6000 in EAF-fixed | Goat-α-Rat-Bio (Biotech; 3052-08), 1:150 | Yes |
| F4/80 | Formalin or EAF | Identify macrophages | Tris-EDTA | Heat-induced epitope retrieval machine | 98 | 30 | - | II | I | F4/80 (Abcam; ab240946), 1:1000 for formalin- or 1:500 in EAF-fixed | EnVision+ System–HRP Labelled Polymer AntiRabbit (Dako) | No |
| Ly-6G | Formalin or EAF | Identify neutrophils | Proteinase K | Stove | 37 | 20 | - | II | I | LY6-G (BD Pharmingen; 551459), 1:150 in formalin- or 1:400 in EAF-fixed | Goat-α-Rat-Bio (Biotech; 3052-08), 1:150 | Yes |
| NKp46 | Formalin or EAF | Identify NK cells | Tris-EDTA | Pressure cooker | 110 | 30 | 30 | II | II | NKp46 (R&D systems; AF2225), 1:400 in formalin- or EAF-fixed | Rabbit-α-Goat-Bio (Dako; E0466), 1:100 | Yes |
| P53 | Formalin or EAF | Assess expression of p53 tumor suppressor | Tris-EDTA | Heat-induced epitope retrieval machine | 98 | 20 | - | II | I | P53 (GeneTex; GTX102965), 1:500 in formalin- or EAF-fixed | EnVision+ System–HRP Labelled Polymer Anti-Rabbit (Dako) | No |
| p-AKT | EAF | Assess activation of PI3K pathway | Citrate buffer | Water bath | 96 | 30 | 30 | II | I | P-AKT (Cell Signaling; 4060), 1:150 in EAF-fixed | Goat-α-Rabbit-Bio (Dako; E0432), 1:1000 | Yes |
| p-ERK | Formalin or EAF | Assess activation of MAPK/ERK pathway | Tris-EDTA | Heat-induced epitope retrieval machine | 98 | 20 | - | I | I | P-P44/42 (Cell Signaling; 4370), 1:200 in formalin- or EAF-fixed | EnVision+ System–HRP Labelled Polymer Anti-Rabbit (Dako) | No |
| PTEN | Formalin or EAF | Assess expression of pTEN tumor suppressor | Citrate buffer | Water bath | 99 | 30 | 30 | I | I | PTEN (Cell Signaling; 9559), 1:100 in formalin- or in EAF-fixed | Goat-α-Rabbit-Bio (Dako; E0432), 1:1000 | Yes |
| Target cell population | Marker |
|---|---|
| Immune cells | CD45 |
| Endothelial cells | CD31 |
| Fibroblasts | CD140a |
| Cancer cells | Fluorescent reporter protein (e.g., GFP)a |
- a
This suggestion applies if the fluorescent reporter gene was included in the pT3 Sleeping Beauty vector used in the HDTVi procedure.
NOTE : Except for p-AKT, which only stains satisfactorily in EAF-fixed tissue, all antigens described in Table 4 can be stained successfully using both formalin and EAF fixation methods.
Materials
-
HCC-bearing liver pieces (see Basic Protocol 3, step 14)
-
10% neutral buffered formalin or EAF (see recipe)
-
Xylene
-
Paraffin
-
100%, 95%, and 70% (v/v) ethanol
-
Optional : Materials for antigen retrieval (varying depending on the marker, as described in Table 4); in this protocol:
- 1× TE (10 mM Tris·Cl/1.25 mM EDTA), pH 9
- Citrate buffer (Scytek)
- 20 μg/ml proteinase K (Sigma-Aldrich)
-
Blocking buffers I and II (see recipes; use buffer appropriate for antigen, as detailed in Table 4)
-
Washing buffer I (see recipe)
-
3% (v/v) H2O2 in methanol
-
Streptavidin/HRP (Dako), for markers not using EnVision+ System–HRP Labelled Polymer Anti-Rabbit (Dako; see Table 4)
-
Biotin-blocking system (Agilent), for markers using streptavidin/HRP
-
Incubation buffers I and II (see recipes; use buffers appropriate for antigen depending on host species of primary antibody, as detailed in Table 4)
-
Primary antibodies (see Table 4)
-
Secondary antibodies (or EnVision+ System–HRP Labelled Polymer Anti-Rabbit [Dako], as certain markers were optimized to use this as a secondary antibody construct; see Table 4)
-
Liquid DAB+ (Dako): substrate buffer + chromogen
-
Hematoxylin (Klinipath), diluted 1:1 in deionized water
-
Entellan mounting medium
-
Disposable scalpel blades (Swann Worton)
-
Optional : Tissue processor machine
-
Microtome
-
Glass slides
-
Cassettes for formalin-fixed, paraffin-embedded (FFPE) tissue
-
Plastic slide container with rack for staining glass slides
-
Optional : Equipment for antigen retrieval (depending on marker; see Table 4):
- Pressure cooker, stove, and/or water bath
- Heat-induced epitope retrieval machine (e.g., Dako PT Link)
-
Glass cover slips
-
Microscope for tissue slides (e.g., Aperio ScanScope)
Preparation of FFPE-fixed tumor tissue slides
1.From HCC-bearing liver, separate one liver lobe containing both tumor and neighboring healthy tissue.
2.Transfer the tissue/tumor pieces to a vial containing formalin or EAF (see Table 4). The choice of EAF or formalin as a fixative depends on the antibodies to be used subsequently (see Table 4), as both fixatives preserve liver morphological details.
3.Incubate sample in formalin for 48 hr at room temperature or in EAF for 24 hr at room temperature.
4.Cut the fixed tumor tissue in half with a scalpel. Place the tissue with the flat surface on a cassette for FFPE.
5.Proceed to dehydrate, clear, and paraffinize the tissue with serial ethanol dilutions, xylene, and paraffin according to previously well-established protocols (Slaoui & Fiette, 2011). This step can be done with a tissue processor machine.
6.Store the FFPE block at room temperature for long-term storage or proceed to the next steps for IHC staining.
7.Cut FFPE block into 4-µm slices using a microtome and mount on glass slides.
8.Place slides on a rack and proceed to rehydrate and deparaffinize them following the steps below:
-
Incubate slides with xylene for 10 min three times.
-
Incubate slides with 100% ethanol for 5 min.
-
Incubate slides with 95% ethanol for 5 min.
-
Incubate slides with 70% ethanol for 5 min.
9.Rinse off the ethanol using deionized water.
Antigen retrieval (optional)
This step is advised to improve the quality of the later staining. This can be especially useful for the liver. The antigen retrieval buffer and technique vary by marker. Table 4 describes the solutions, equipment, and antigen retrieval conditions for each marker based on optimized steps performed in our lab.
10.If desired, perform antigen retrieval to unmask epitopes.
-
Add the appropriate antigen retrieval buffer solutions for each antibody staining as indicated in Table4.
-
Place the slides at the indicated temperatures in the indicated containers as listed in Table4.
-
Incubate slides at the appropriate temperatures for the indicated amount of time as presented in Table4.
-
Allow slides to cool down for the amount of time indicated in Table4.
Washing and blocking of slides
11.Wash slides three times, 5 min each, with washing buffer I.
12.Incubate slides 20 min with 3% H2O2 in methanol to inactivate endogenous peroxidase.
13.Repeat washing step 11.
14.Block slides with blocking buffer I or II (depending on antigens; see Table 4) for 30 min at room temperature.
15.For antigens using streptavidin/HRP (see Table 4), an avidin/biotin blocking step is advised. For this, incubate slides in avidin and biotin for 10 min each at room temperature, following the instructions for the biotin-blocking system kit. Repeat washing step 11 after each incubation.
Antibody incubation and staining of slides
16.Incubate slides overnight at 4°C with the primary antibody of choice in 200 µl of incubation buffer I or II (see Table 4 for the recommended incubation buffers and dilutions for each suggested antibody depending on the fixative used).
17.On day 2, repeat washing step 11.
18.Incubate slides at 4°C with the secondary antibody of choice in 200 µl of incubation buffer I or II (see Table 4) for 30 min. For markers using EnVision+ System–HRP Labelled Polymer, instead incubate in 200 µl of EnVision+ System–HRP Labelled Polymer for 30 min.
19.Repeat washing step 11.
20.For markers using streptavidin/HRP (see Table 4 ) : Incubate slides in 200 µl streptavidin/HRP diluted 1:200 in incubation buffer I or II for 30 min. Repeat washing step 11.
21.Incubate slides with Liquid DAB+ for 3 min.
22.Transfer slides to washing buffer I to assess whether the staining worked properly. If the slides are still light brown, repeat steps 11 and 21.If slides are dark brown, proceed to the next step.
23.Rinse slides in deionized water for 2 min.
24.Counterstain slides with hematoxylin for 1 min.
25.Wash slides with tap water for 2 min.
26.Place slides on a rack and proceed to dehydrate them following the steps below:
-
Incubate slides 5 min with 70% ethanol.
-
Incubate slides 5 min with 100% ethanol, twice.
-
Incubate slides 5 min with xylene.
27.Mount slides with Entellan and cover slides with glass cover slips.
28.Scan slides using a microscope of choice, such as the Aperio ScanScope. To proceed with quantifying the images of immune cell content staining, follow Support Protocol 5.
Alternate Protocol 2: PREPARATION AND ANALYSES FOR IMMUNOFLUORESCENCE STAINING OF HCC-BEARING LIVER
Immunofluorescence (IF) is a useful technique with two or more antigens detecting proteins of interest staining in the same sample and/or in the same cell, providing spatial information on the cell types expressing these markers in the HCC microenvironment. This is particularly important when analyzing immune cell populations that require more than one antigen to be fully characterized.
Materials
-
HCC-bearing liver pieces (see Basic Protocol 3, step 14)
-
O.C.T. Compound (Tissue-Tek)
-
4% paraformaldehyde (PFA)
-
Washing buffer II (see recipe)
-
1× PBS (see recipe)
-
Blocking buffer III (see recipe)
-
Primary and secondary antibodies of interest
-
5 mg/ml DAPI solution
-
Dako mounting medium (Agilent)
-
Nail polish
-
Standard vinyl cryomold cassettes (Tissue-Tek)
-
Cryotome
-
Dry ice
-
Glass slides
-
Humidity chamber for glass slides, covered with Whatman paper soaked in distilled water or 1× PBS
-
Plastic slide container with rack for staining glass slides
-
Paper tissue
-
Platform shaker
-
Super PAP pen (Thermo Fisher)
-
Glass coverslips
-
Fluorescence microscope for tissue slides (e.g., Axioscan SlideScanner, Zeiss)
Generation of cryoblocks and cryosections from HCC liver samples
1.Separate a liver lobe containing both tumor and neighboring healthy tissue from Basic Protocol 3 (step 14); cut in half if the tissue piece is too large to fit into the vinyl cryomold cassette.
2.Add O.C.T. to the vinyl cassette until the cassette bottom is fully covered. Lay down the cassette on dry ice for 10 s and then carefully place the liver lobe sample in the cassette.
3.Top up the cassette with additional O.C.T. until the tissue piece is fully embedded. Wait until the tissue is fully frozen on dry ice.
4.Store cryoblock at −80°C for later use or proceed to the next steps for sectioning and staining.
5.Cut cryoblock into 10-µm slices using a cryotome and mount on glass slides.
6.Fix the samples by placing the glass slides on the grid of the humidity chamber, adding enough 4% PFA to cover the tissue (200-300 µl), and letting stand 10 min at room temperature.
7.Remove PFA by gently tapping the slides on a piece of paper tissue.
Immunostaining
8.Transfer the glass slides to the slide container and fill with washing buffer II. Place the staining container on a shaker and incubate slides for 10 min.
9.Remove washing buffer II by discarding the solution. Repeat step 8.
10.Remove washing buffer II and add PBS to fully cover slides in the plastic container. Place staining container on the shaker and incubate slides for 10 min.
11.Remove the slides from the staining container and gently dry by tipping slides on a piece of paper tissue. Do not let the slide dry out completely.
12.Transfer slides to wet chamber and circle the tissue of interest using a hydrophobic super PAP pen.
13.Add 200 µl blocking buffer III to each circled section and incubate 1 hr at room temperature, protected from light.
14.In the meantime, prepare the primary antibody mix solution of choice, making 200 µl per circled tissue section.
15.Remove blocking buffer III by gently tipping slides on a paper towel. Add 200 µl of primary antibody solution in blocking buffer III to each circled section and incubate in the wet chamber for 4 hr at room temperature or overnight at 4°C.
16.Remove primary antibody solution by gently tipping slides on a paper towel. Transfer slides to the slide container.
17.Fill up staining container with PBS and incubate slides. Place it on a shaker for 5 min.
18.Wash by removing and replenishing the staining container with PBS, and place on the shaker for 3 min.
19.Repeat washing step 18 twice more.
20.Prepare the secondary antibody mix solution in blocking buffer III (200 µl per circled section).
21.Dry slides with paper towel and transfer them to the wet chamber. Add 200 µl of the secondary antibody solution per circled slide and incubate 1 hr at room temperature, protected from light.
22.Remove the secondary antibody solution by gently tipping slides on a paper towel. Add 200 µl of DAPI diluted 1:1000 in blocking buffer III per circled slide and incubate 5 min at room temperature, protected from light.
Mounting of stained slides
23.Remove DAPI solution by gently tipping slides on a paper towel. Transfer slides to the staining container.
24.Repeat washing step 18 four times.
25.Wash slides by placing inside the staining container and covering with distilled water for 30 s, and repeat twice more (three times total).
26.Dry slides one by one with a paper towel and transfer to the wet chamber.
27.Add 1 drop of Dako mounting medium per circled section. Carefully remove bubbles by gentle contact with a cover slip.
28.Carefully cover the section with a glass coverslip.
29.Carefully dry slides with paper tissue, removing any excess mounting medium.
30.Use nail polish or other similar reagent to seal the borders of the cover slip. Let it dry protected from light.
31.Repeat steps 26-30 for each slide of interest.
32.Store slides at 4°C or proceed to scan slides in a microscope of choice, such as the Slide Scanner AxioScan (Zeiss).
Support Protocol 4: LIVER-SPECIFIC PHENOTYPIC ANALYSES OF LIVER SECTIONS
Analyses of certain aspects of liver histology in the context of hepatocellular carcinoma can be performed by histochemistry (HC). This technique is useful to identify the development of fibrosis and steatosis, which are linked to the deposition of collagen and fat, respectively. This protocol details two separate sets of steps: the staining of collagen in FFPE-fixed slides using Masson's trichrome staining and the staining of lipids and neutral triglycerides using Oil red O in frozen sections.
Materials
-
Tissue slides cut from FFPE blocks (see Basic Protocol 5, step 7) for trichrome staining or tissue slides containing frozen sections from cryoblocks (see Alternate Protocol 2, step 5) for Oil red O staining
-
For trichrome staining:
- Bouin's solution (see recipe)
- Hematoxylin Weigert (Elastica van Gieson kit)
- Solutions A, B, and C (see recipes)
- 1% (v/v) acetic acid in deionized water
- 96% (v/v) ethanol
- Xylene
-
For Oil red O staining:
- Oil red O solution (see recipe)
- Hematoxylin (Klinipath), diluted 1:1 in deionized water
-
Entellan mounting medium
-
Plastic slide container with rack for staining glass slides
-
Glass coverslips
-
Paper tissue
-
Microscope for tissue slides (e.g., Aperio ScanScope)
Trichrome staining
1a. Prepare deparaffinized tissue slides cut from HCC FFPE blocks of choice (see Basic Protocol 5, step 9).
2a. Fix the tissue by submerging the slides in Bouin's solution at 65°C for 1 hr.
3a. Wash slides with tap water for 5 min.
4a. Counterstain slides with hematoxylin for 8 min.
5a. Wash slides with tap water for 10 min.
6a. Wash slides with deionized water for 5 min.
7a. Submerge slides in solution A for 5 min.
8a. Repeat washing step 6a.
9a. Submerge slides in solution B for 5 min.
10a. Let the liquid run off the slides with a paper tissue and stain with solution C for 5 min.
11a. Repeat washing step 6a.
12a. Wash slides with 1% acetic acid in deionized water for 2 min.
13a. Place slides on a rack and proceed to dehydrate them as follows:
- Incubate slides 5 min in 96% ethanol;
- Incubate slides 5 min in xylene.
14a. Mount slides with Entellan and cover with glass coverslips.
15a. Scan slides using a microscope of choice, such as the Aperio ScanScope. Upon analysis, the nuclei will be stained black; the cytoplasm, erythrocytes, and fibrins red; and the collagen deposits blue (Fig. 6).
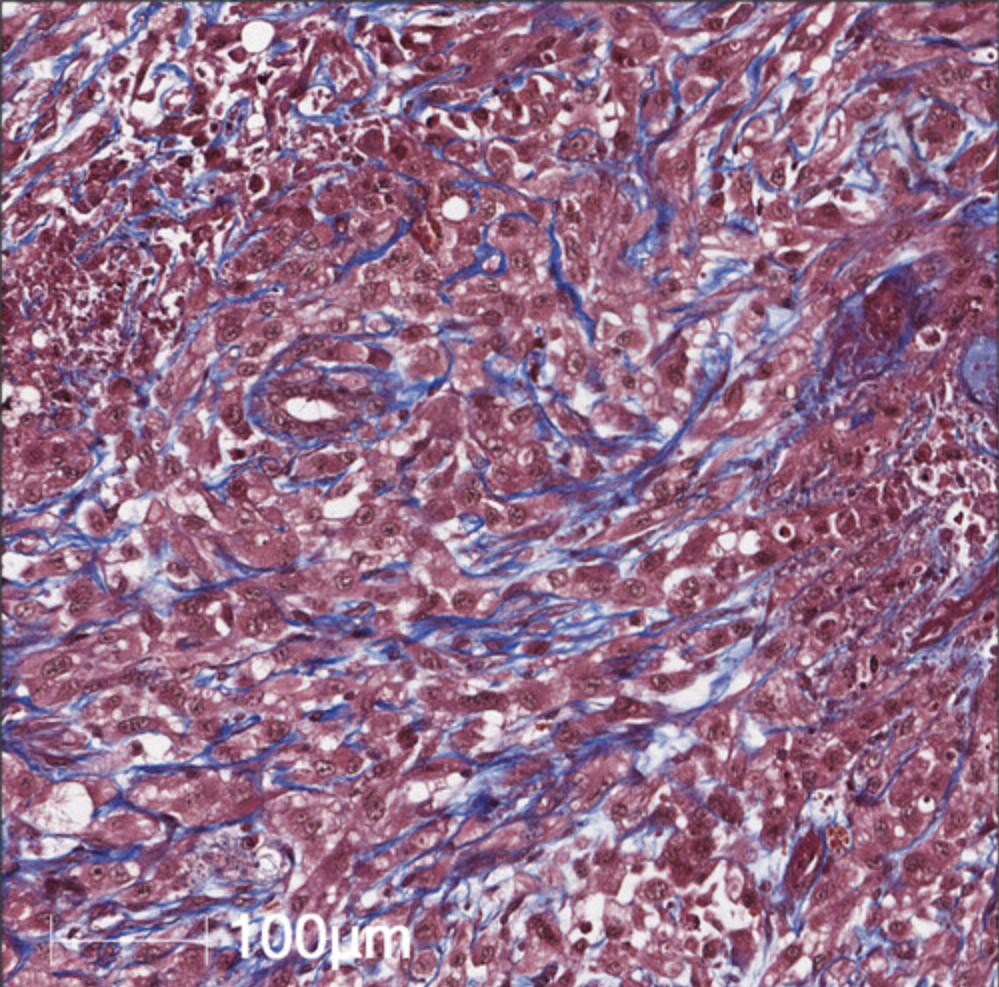
Oil red O staining
The following procedure (steps 1b-8b) should be performed at room temperature.
1b. Prepare frozen, fixed tissue slides cut from HCC cryoblocks of choice (see Alternate Protocol 2, step 5).
2b. Submerge the tissue slides in Oil red O solution for 10 min.
3b. Wash slides with deionized water for 2 min.
4b. Counterstain slides with hematoxylin for 2 min.
5b. Wash slides with tap water for 3 min.
6b. Repeat washing step 3b.
7b. Mount and cover slides with glass coverslips.
8b. Scan slides using a microscope of choice, such as the Aperio ScanScope. Upon analysis, the nuclei are stained blue and the neutral triglycerides and lipid deposits are stained red (Fig. 7).
Support Protocol 5: IMMUNOHISTOCHEMICAL QUANTIFICATION IN LIVER SECTIONS
Stained IHC slides can be used to quantify the number of cells that are positive for a specific marker per tumor nodule or per other area of interest. This is a useful strategy to calculate the absolute number of immune cell populations in scanned IHC slides from Basic Protocol 5 and complement the flow cytometry analyses described in Basic Protocol 4.The steps below consider the IHC quantification using the software HALO from Indica Labs, a technique discussed in other scientific publications (Thommen et al., 2018; Wang et al., 2019).
Materials
- Stained and scanned IHC slides (see Basic Protocol 5)
- HALO software (Indica Labs)
1.Design the appropriate algorithm based on the Multiplex IHC v1.2 module available in the HALO platform.
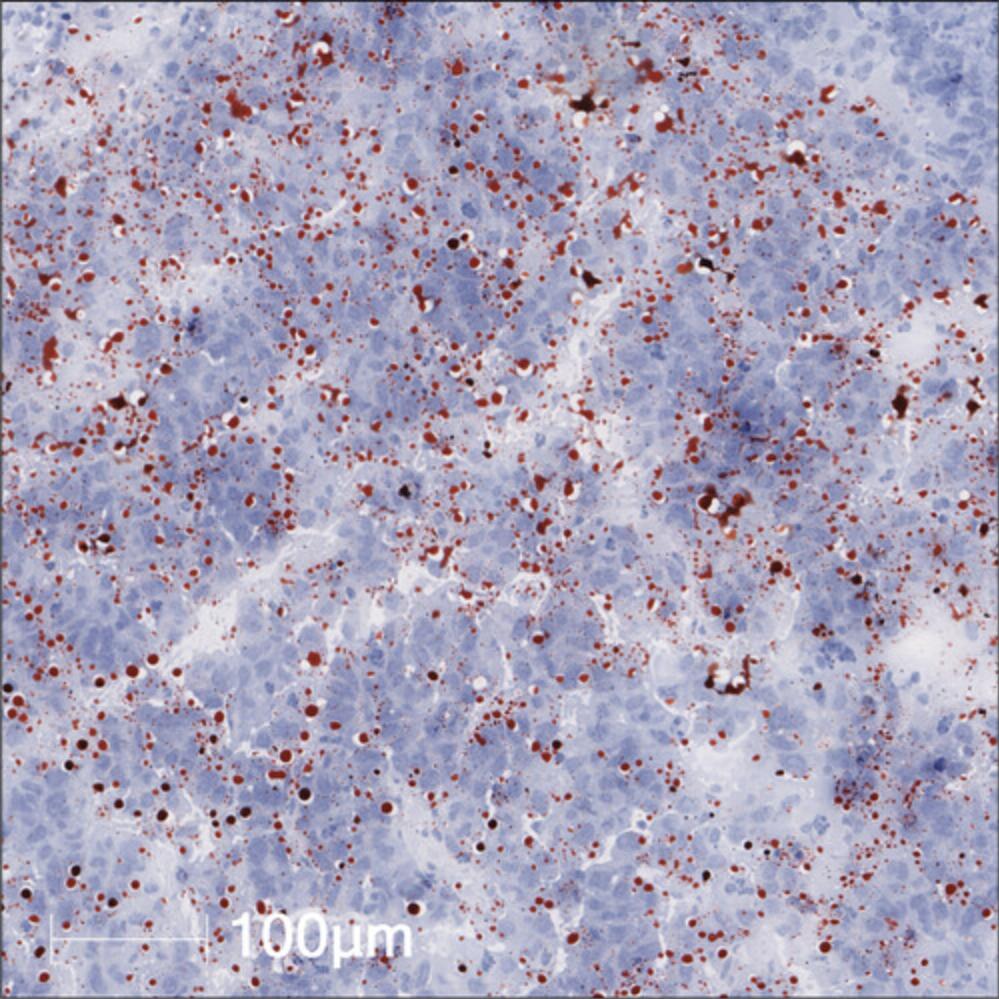
2.Using the “Real-time” tuning feature, select the threshold for cell recognition, positive cell detection, and nuclear segmentation for the liver tumor nodules. An example of this algorithm can be seen in a CD4-stained IHC sample for a MycOE/Trp53KO-driven HCC tumor region in Figure 8.
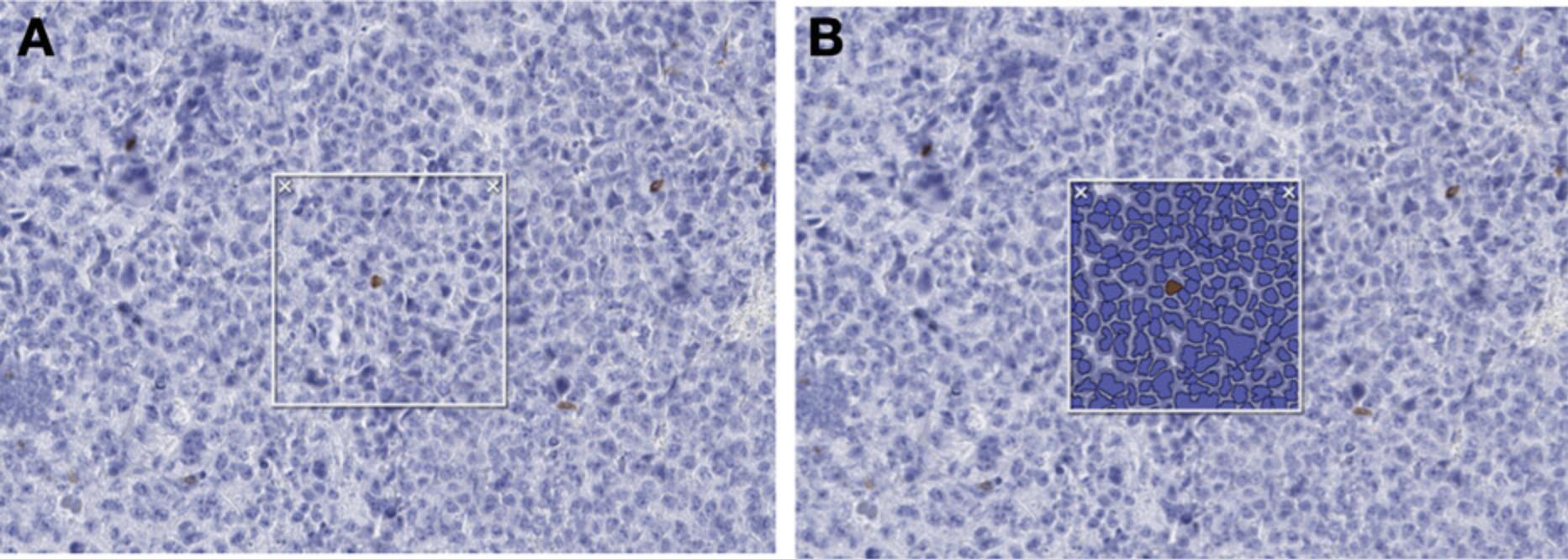
3.Circle the tumor nodules as shown in Figure 9 using the drawing tool (symbolized as a pen), where A represents the circled nodule region and B includes the nuclear segmentation algorithm applied.
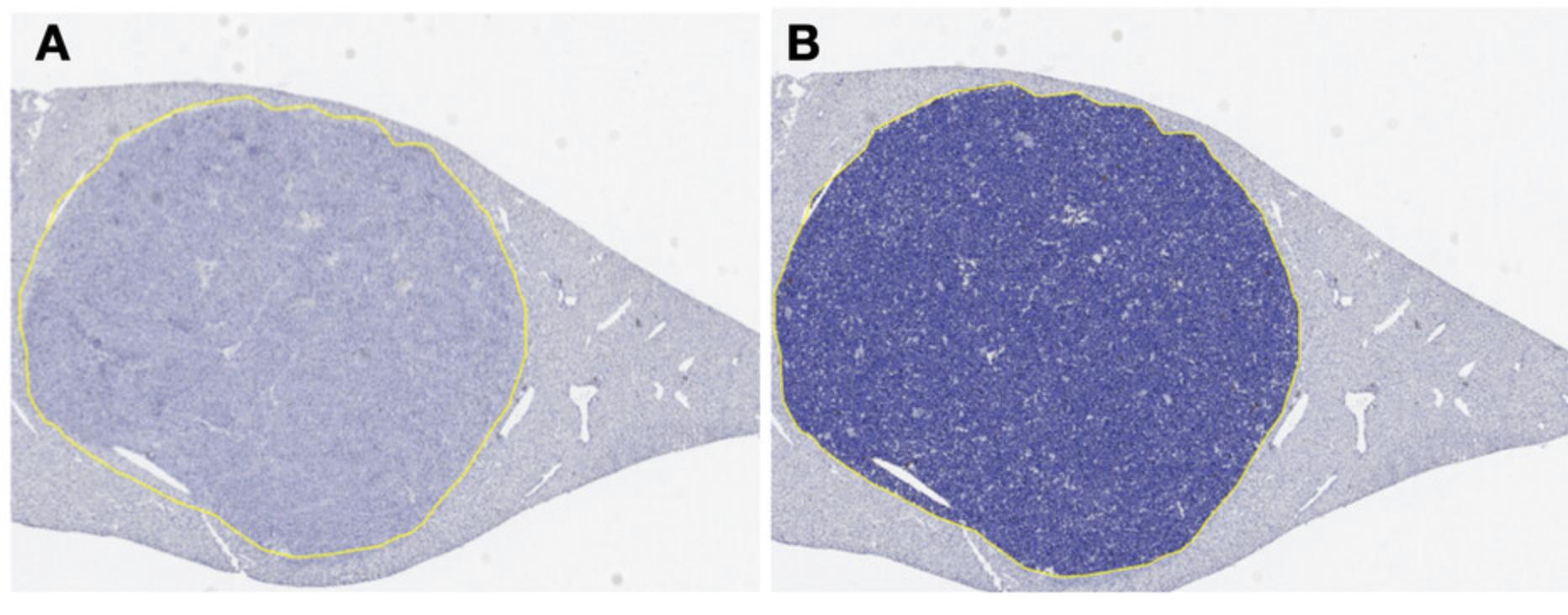
4.Ensure that tissue areas subject to technical problems, such as tissue folds or tissue edges, are not considered.
Basic Protocol 6: PREPARATION OF SNAP-FROZEN TUMOR TISSUE FROM EXTRACTED LIVER AND TRANSCRIPTIONAL ANALYSES OF BULK TUMOR OR SORTED CELLS
Transcriptional analyses of HCC tumor nodules makes it possible to assess gene expression regulation in liver cancer cells or immune cells participating to tumor malignancy, for example that of genes related to metabolism, inflammation, angiogenesis, and others. In this protocol, we describe the process of preparing snap-frozen samples from tumors (Basic Protocol 3) and subsequent RNA extraction and transcriptional analyses from bulk tumor samples or sorted cells (see Support Protocol 3).
Materials
-
Pieces of HCC-bearing liver (see Basic Protocol 3, step 14) and/or sorted immune cells in TRIzol (see Support Protocol 3, step 7)
-
RNaseZap RNase Decontamination solution (Thermo Fisher)
-
TRIzol LS Reagent (Thermo Fisher)
-
DEPC-treated water (Thermo Fisher)
-
75% (v/v) ethanol in DEPC-treated water
-
Chloroform
-
Isopropanol
-
Optional : Glycoblue Coprecipitant (Thermo Fisher)
-
High-Capacity cDNA Reverse Transcriptase kit (Thermo Fisher)
-
DEPC-treated water (Thermo Fisher)
-
Taqman probes of interest (Thermo Fisher)
-
TaqMan Fast Advanced Master mix (Thermo Fisher)
-
1.5-ml microcentrifuge tubes
-
Microcentrifuge for 1.5-ml tubes
-
Dry ice
-
−80°C freezer
-
10-cm Petri dish
-
Disposable scalpel blades (Swann Worton)
-
Forceps, straight/serrated (Fine Science Tools)
-
10-ml round-bottom tubes
-
Ice
-
Fume hood
-
Standing tissue homogenizer
-
Nanodrop quantification machine
-
0.2-ml polypropylene tube strips
-
PCR machine (e.g., C1000 Series Touch Thermal Cycler, Bio-Rad)
-
384-well PCR plates
-
Adhesive PCR plate sealers
-
Centrifuge for plates
-
Real-time PCR System (e.g., QuantStudio 6)
Snap freezing
1.When working with pieces of HCC-bearing liver, separate tumor pieces isolated in Basic Protocol 3 (step 14) from the surrounding liver tissue into small (∼10 mg) sections in one or more 1.5-ml tubes.
2.Transfer the tube immediately to dry ice and leave until fully frozen.
3.Store the snap-frozen tumor pieces in −80°C for long-term storage or further process them for mRNA extraction and transcriptional analysis as described below. They can also be used to generate protein lysate for protein analysis (see Alternate Protocol 3).
Extraction of RNA from snap-frozen tumor and sorted cells
4.Clean all tools with RNaseZAP to avoid RNA degradation.
5.Working on dry ice, cut ∼5 mg of tumor/tissue with a scalpel from the snap-frozen tumor of interest on a 10-cm Petri dish.
6.Transfer the tissue piece into a 10-ml round-bottom tube. Working under a fume hood, add 1 ml TRIzol.
7.Use a standing tissue homogenizer to grind the tumor piece. Clean the homogenizer thoroughly with 75% ethanol in DEPC-treated water and proceed to the next sample.
8.Transfer the homogenate to a 1.5-ml tube.
9.Working on ice and in a fume hood, add 200 µl chloroform to bulk tumor homogenates in TRIzol and/or to sorted cells in TRIzol (see Support Protocol 3, step 7). Invert tubes three times (do not vortex) and incubate 10 min on ice.
10.Centrifuge tubes 15 min at 12,000 × g , 4°C. A clear upper layer will form after centrifugation.
11.Carefully transfer 300-600 µl of the clear upper layer to a fresh 1.5-ml tube without touching the red bottom layer or the white middle ring.
12.Add 500 µl isopropanol to the extracted upper layer. Invert tubes three times (do not vortex) and incubate for 10 min on ice.
13.Optional : Add 1 µl Glycoblue Coprecipitant to each tube to better visualize the RNA.
14.Centrifuge tubes 10 min at 12,000 × g , 4°C.
15.Carefully remove and discard the supernatant from each tube without disturbing the pellet.
16.Wash pellet with 75% ethanol diluted in DEPC-treated water and vortex tubes.
17.Centrifuge tubes 5 min at 7500 × g , 4°C.
18.Carefully remove and discard the supernatant from each tube without disturbing the pellet.
19.Air dry the pellet for 20 min on ice. To facilitate later RNA dilution, do not dry the pellet completely.
20.Dilute pellet in 20 µl DEPC -treated water. Quantify RNA using a Nanodrop instrument. Dilute sample if necessary (especially bulk tumor samples).
21.Store RNA at −80°C for long-term storage or proceed to the following section for cDNA preparation using reverse transcriptase and transcriptional analysis using quantitative polymerase chain reaction (RT-qPCR).
RT-qPCR
22.Calculate a volume from bulk tumor or sorted cells containing 100-1000 ng of RNA in a maximum of 10 µl, maintaining the same amount of RNA between the samples that are to be compared. Transfer the calculated volume of each sample to 200-µl tube strips. Add DEPC-treated water to top up to a total of 10 µl if needed.
23.Prepare reverse transcriptase solution according to the manufacturer's instructions for the High-Capacity cDNA Reverse Transcriptase kit. Prepare enough solution to add 10 µl per sample to a total of 20 µl.
24.Vortex and spin down samples, and then load onto PCR machine.
25.Run PCR cycle according to the manufacturer's instructions for the High-Capacity cDNA Reverse Transcriptase kit.
26.After the PCR cycle is complete, dilute samples with DEPC-treated water to 5 ng/µl cDNA final.
27.Choose the genes of interest and the housekeeping genes and prepare the probe solutions. The probe solutions should be prepared with 20 µl of Taqman Master Mix, 2 µl of the Taqman probe of interest, and 14 µl of DEPC-treated water to a total of 36 µl of solution per sample.
28.Add 9 µl per well of probe solutions per sample in three wells of a 384-well plate to form triplicates.
29.Add 1 μl of 5 ng/µl cDNA solution to the probe solutions to be assessed in each triplicate well.
30.Cover the plate with an adhesive PCR plate seal, vortex, and centrifuge plate 1 min at 300 × g , 4°C.
31.Assess gene expression using comparative CT in a QuantStudio 6 PCR system or similar.
32.For the relative gene expression quantification analysis, follow the instructions in Table 6.
| cDNA sample (from sorted cells or tissue lysates) | Gene | Sample identification | CT | ΔCT | ΔΔCT | Fold change |
|---|---|---|---|---|---|---|
| Control sample | Housekeeping gene | A | CT(A) | - | - | - |
| Control sample | X | B | CT(B) | ΔCT(B) = CT(B) – CT(A) | ΔΔCT(B) = ΔCT(B) – ΔCT(B) | 1 |
| Sample from HCC-bearing mouse | Housekeeping gene | C | CT(C) | - | - | - |
| Sample from HCC-bearing mouse | X | D | CT(D) | ΔCT(D) = CT(D) – CT(C) | ΔΔCT(D) = ΔCT(D) – ΔCT(B) | 2-ΔΔCT(D) |
-
The calculation considers the comparison between one control sample and one sample from ab HCC-bearing mouse. CT, cycle threshold.
Alternate Protocol 3: PROTEIN ANALYSES FROM HCC SAMPLES AND SERUM OR PLASMA
Protein analyses from HCC tumor samples allow the correlation of findings obtained at the transcriptional level with protein expression. Several methods can be used to assess the activation of signaling pathways and the presence of cytokines and chemokines of interest. The assays described below include western blotting and the enzyme-linked immunosorbent assay (ELISA). Other protein assays, such as cytokine arrays, can also be performed using the techniques described here, but are beyond the scope of this protocol.
Materials
-
Snap-frozen HCC pieces (see Basic Protocol 6, step 3) or serum or plasma of interest (see Support Protocol 1)
-
Lysis buffer (cOmplete™ Lysis-M) with protease inhibitor cocktail (Roche), ice cold
-
Phosphatase inhibitor cocktail (Roche)
-
Pierce BCA kit (Thermo Fisher)
-
ELISA kit for the protein of interest (Thermo Fisher)
-
Running buffer: Bolt™ MES SDS Running Buffer (Invitrogen), diluted 1:20 in deionized water
-
1× TBS-T (see recipe)
-
Appropriate SDS-polyacrylamide gel: Bolt™ 4%-12%, Bis-Tris, 12-well, 1.0 mm Mini Protein Gel (Thermo Fisher), or other gel of appropriate concentration
-
10× Bolt™ Sample Reducing Agent (Invitrogen)
-
4× Bolt™ LDS Sample Buffer (Invitrogen)
-
Optional : 0.1% Ponceau S, (Sigma Aldrich P7170-1L)
-
Blocking buffer IV (see recipe)
-
Unconjugated primary antibody of interest
-
Fluorescent secondary antibody of interest
-
Optional : Restore™ Fluorescent Western Blot Stripping Buffer
-
10-cm Petri dish
-
Disposable scalpel blades (Swann Worton)
-
Dry ice
-
1.5-ml microcentrifuge tubes
-
Ice
-
Pestle for tissue homogenization
-
Sonicator (Diagenode Bioruptor)
-
Infinite 200 Pro microplate reader (Tecan) or similar
-
1.5-ml microcentrifuge tube
-
Heating block, 95°C
-
Microcentrifuge for 1.5-ml tubes
-
Mini Gel Tank (Invitrogen) or similar western blotting system
-
PowerPac 300 Electrophoresis power supply (Bio-Rad) or similar
-
Trans-Blot Turbo transfer pack (Bio-Rad)
-
Trans-Blot Turbo transfer system (Bio-Rad)
-
Dark 50-ml tubes
-
50-ml tube rotator
-
Odyssey fluorescence reader (LI-COR) or similar
Preparation of protein lysates (if starting with snap-frozen HCC samples)
1.Working on dry ice, dissect ∼5 mg of tumor tissue from snap-frozen samples (see Basic Protocol 6, step 3) with a scalpel on a Petri dish.
2.Transfer tumor piece to a 1.5-ml tube and add 300 µl ice-cold lysis buffer with protease inhibitor according to the manufacturer's instructions.
3.Homogenize tumor piece with a pestle. Repeat for each sample using clean pestles.
4.Incubate samples for 30 min on ice, vortexing every 5 min.
5.Sonicate samples on ice for three cycles of 1 min sonication and 45 s rest, to avoid overheating (for a total of 3 min of sonication).
6.Centrifuge samples 20 min at 15,000 × g , 4°C.
7.Collect supernatant in a fresh 1.5-ml tube on ice and discard the pellet.
8.Proceed to measure the protein concentration with the Pierce BCA assay according to the manual instructions or store protein lysates at −80°C for later use.
9.Dilute concentrated samples in lysis buffer if necessary.
Enzyme-linked immunosorbent assay (ELISA)
This experiment can be performed using either the protein lysates obtained in step 9 or serum or plasma obtained from Support Protocol 1.For the protein lysates, a concentration of 100-1000 µg/ml can be used as an estimate for samples to fall within the standard curve for most protein targets. For the serum/plasma, dilutions of 1/5 to 1/10 can serve as a starting points that will be within the standard curve for most protein tests. For optimal results, test multiple dilutions for both sample types.
10.Perform ELISA using the Thermo Fisher ELISA kit according to the manufacturer's instructions. Some personalized tips and steps follow:
-
Incubating samples overnight at 4°C usually gives better results and is recommended.
-
If working with tissue/tumor sample protein lysates, use corresponding lysis buffer as a negative control for the samples.
Western blotting, staining, and imaging of proteins
Western blotting can be performed to quantify proteins of interest and to investigate active signaling pathways in bulk tumor protein lysate samples. The percentage of polyacrylamide in the gel should be chosen based on the weight of the proteins of interest. As a general rule, higher-percentage gels are used when a better separation of lighter proteins is desired; lower-percentage gels are more suitable for separation of heavier proteins. If proteins of mixed molecular weight are probed, the 4%-12% gel listed in Materials is recommended.
The number of wells in the gel must be chosen according to the number of samples to be loaded, the amount of protein per sample, and the total volume of sample loaded per well. This protocol describes a sample volume of 20 μl, which is optimal for a 12-well gel. If samples are too dilute, consider using a gel with fewer wells.
11.Determine the sample volume needed to contain 20-40 µg protein from samples generated in step 9 and transfer to a 1.5-ml microcentrifuge tube.
12.Prepare the loading samples according to Table 7.The calculation considers a final volume of 20 µl per well, with a final protein concentration of 1-2 µg/µl.
| Component | Volume (µl) per loading sample |
| Protein lysate | Variable; should contain 20-40 µg protein |
| 10× Bolt™ Sample Reducing Agent | 2 |
| 4× Bolt™ LDS Sample Buffer | 5 |
| Lysis buffer | Used as a diluent to complete a total volume of 20 µl after addition of the above components |
-
The volume of reagents stated above should be multiplied by the number of samples to be generated
13.Heat the loading samples for 5 min in 95°C heating block.
14.Centrifuge the loading samples for 10 s at 10,000 × g , room temperature.
15.Open gel, remove protective label, and mount it on the western blot Mini Gel Talk system.
16.Add running buffer to the chambers of the western blot system. Ensure that there is no leakage in the system.
17.Remove protective comb from the gel, and rinse wells with running buffer.
18.Add 20 µl of loading samples per well, making sure the samples do not spill into neighboring wells.
19.Fill front chamber with running buffer up to the marked limit.
20.Run loading samples at 200 V on ice.
21.Unmount the cast gel. Open cast gel carefully with a blunt knife, rinse gel with distilled water, and carefully detach the gel.
22.Carefully transfer gel to a Trans-Blot Turbo transfer pack membrane. Set the transfer program according to the protein weight, following the instructions in the manual for the Trans-Blot Turbo transfer system.
23.Optional : Check protein transfer quality by incubating the membrane in 0.1% Ponceau S for 5 min and visually assessing the stained proteins.
24.Carefully transfer membrane to a light-protected, dark 50-ml tube.
25.Block membrane with blocking buffer IV for 1-2 hr at room temperature and continuous rotation.
26.Remove blocking buffer IV and add 5 ml of the primary antibody solution of interest in blocking solution IV. The antibody concentration must be optimized.
27.Incubate overnight at 4°C with continuous rotation.
28.The next day, remove primary antibody solution and wash membranes three times with TBS-T for 5 min with constant shaking.
29.Remove washing solution and add the solution of secondary antibody in blocking buffer IV. The antibody concentration must be optimized.
30.Incubate 30 min at room temperature with continuous shaking.
31.Repeat washing step 28.
32.Wash twice with PBS for 5 min and constant shaking.
33.Rinse blot with distilled water. Scan membranes using an Odyssey fluorescence reader or similar.
34.After scanning, the images can be analyzed and quantified using software such as ImageStudio. Blots can be stored in TBS-T at 4°C for short-term if rescanning is necessary.
35.Optional : If desired, the membranes can be stripped and then reprobed with an additional protein. For this, we recommend using Restore™ Fluorescent Western Blot Stripping Buffer (Thermo Fisher). The stripping time and concentration must be optimized according to the manufacturer's instructions.
REAGENTS AND SOLUTIONS
Acetic acid, 0.2 N, sterile
In a glass bottle, add 572 µl of 100% acetic acid to 500 ml distilled water. Filter the solution using a 500-ml Vacuum Filter/Storage Bottle System (Corning) to sterilize. Store at room temperature until needed.
Blocking buffer I
In a 50-ml tube, dissolve 2 g bovine serum albumin (BSA) and 2.5 ml normal goat serum (NGS) in 47.5 ml 1× PBS (see recipe) to make a 4% BSA/5% NGS/PBS solution. Mix solution until the BSA is fully dissolved. Store at 4°C until needed.
Blocking buffer II
In a 50-ml tube, dissolve 5 g skimmed milk powder in 50 ml of 1× PBS (see recipe) to make a 10% skimmed milk powder/PBS solution. Mix solution until the milk powder is fully dissolved. Store at 4°C until needed.
Blocking buffer III
In a 50-ml tube, dissolve 1 g BSA in 50 ml 1× PBS (see recipe) and add 50 µl of Tween 20 to make a 2% BSA/0.1% Tween 20/PBS solution. Mix the solution in a shaker until fully homogeneous. Store at 4°C until needed.
Blocking buffer IV
In a 50-ml tube, dissolve 2.5 g bovine serum albumin (BSA) in 50 ml of 1× TBS-T (see recipe) to make a TBS-T/5% BSA solution. Mix the solution in a shaker until fully homogeneous. Store at 4°C until needed.
Bouin's solution
Working in a fume hood, in a glass bottle, mix 300 ml saturated picric acid, 100 ml of 37% formaldehyde, and 20 ml of 100% acetic acid.
Collagen solution
In a sterile 50-ml tube, add 676 µl of Collagen Type I Rat tail (Corning) to 50 ml of 0.2 N sterile acetic acid (see recipe) to obtain a final solution of 50 µg/ml. Store at 4°C until needed. This solution can be reused up to three times.
Complete medium 10%
Add 50 ml heat-inactivated fetal bovine serum (FBS) and 1 ml of 500× penicillin and streptomycin (Roche) to 450 ml of DMEM (Gibco). Store at 4°C until needed.
Complete medium 20%
Add 100 ml heat-inactivated FBS and 1 ml of 500× penicillin and streptomycin (Roche) to 400 ml DMEM (Gibco). Store at 4°C until needed.
EAF (ethanol/acetic acid/formaldehyde)
To prepare 9.5 L of the fixative, in a fume hood, add 40.5 g NaCl to 4.5 L deionized water. Add 4 L of 100% ethanol. Add 1 L of 37% formaldehyde. This solution is stable for long term. To finish making EAF, add 100% acetic acid to make a 5% (v/v) solution. After addition of acetic acid, the solution should be used within 14 days.
Erylysis (erythrocyte lysis) buffer
In a glass bottle, dissolve 8.4 g NH4Cl, 1.2 g NaHCO3, and 0.2 ml of 0.5 M EDTA in 1 L distilled water. Store at 4°C until needed.
FACS buffer
In a glass bottle, add 10 ml FBS to 490 ml of 1× PBS (see recipe). Degas buffer before use. Store at 4°C until needed.
Freezing medium
In a sterile 50-ml tube, combine 45 ml FBS and 5 ml DMSO. Store at 4°C until needed.
Incubation buffer I
In a 50-ml tube, dissolve 0.5 g BSA in 49.37 ml of 1× PBS (see recipe) and add 625 µl NGS to make a 1% BSA/1.25% NGS/PBS solution. Mix the solution until fully homogeneous. Store at 4°C until needed.
Incubation buffer II
In a 50-ml tube, dissolve 0.5 g BSA in 49.37 ml of 1× PBS (see recipe) and add 625 µl normal rat serum (NRS) to make a 1% BSA/1.25% NRS/PBS solution. Mix the solution until fully homogeneous. Store at 4°C until needed.
Live/dead staining solution
Prepare the live/staining solution by diluting the Zombie NIR™ Fixable Viability Kit (Biolegend) vial according to the manufacturer's instructions. Prepare 1-µl aliquots and store at −20°C in the dark until needed. Upon use, dilute aliquots 1:1000 in 1× PBS (see recipe).
Liver medium
Add 1 ml of 500× penicillin and streptomycin (Roche) to 500 ml DMEM (Gibco). Store at 4°C until needed.
Oil red O solution
In a glass bottle, dissolve 0.5 g of crystal Oil red O in 100 ml isopropanol. Stir and boil the solution for 5 min inside a fume hood. Dilute 60 ml of the solution in 40 ml distilled water. Let the diluted solution settle overnight. On day 2, filter with a filter paper to remove undissolved particles. Use the same day.
Phosphate-buffered saline (PBS), 1×, sterile
In a glass bottle, add 1 PBS tablet (Gibco) to 500 ml deionized water. Leave to dissolve. Autoclave to sterilize and store at room temperature until needed.
Solution A
In a glass bottle, dissolve 0.5 g acid fuchsin in a mixture of 100 ml distilled water and 0.5 ml of 100% acetic acid.
Solution B
In a glass bottle, dissolve 1 g phosphomolybdic acid in 100 ml distilled water. Prepare this solution for immediate use.
Solution C
In a glass bottle, dissolve 2 g methyl blue in a mixture of 100 ml distilled water and 2.5 ml of 100% acetic acid.
TBS-T, 1×
In a glass bottle, add 100 ml of 10× TBS (see recipe) to 900 ml deionized water and 100 µl Tween 20. Store at room temperature.
Tris-buffered saline (TBS), 10×
In a glass bottle, add 24.2 g Tris base and 80 g NaCl to 1 L deionized water. Adjust the pH to 7.6 using 0.1 M acetic acid. Store at room temperature.
Washing buffer I
In a glass bottle, combine 500 ml of 1× PBS (see recipe) and 250 µl Tween 20 to make a 0.05% Tween 20/PBS solution. Store at room temperature.
Washing buffer II
In a glass bottle, add 500 ml of 1× PBS (see recipe ) and 1 ml Triton X-100 to make a 0.2% Triton X-100/PBS solution. Store at room temperature.
COMMENTARY
Background Information
In the protocols detailed in this article, we sought to delineate the effective and streamlined use of hepatocellular carcinoma tumors generated using the HDTVi somatic system of gene delivery (Wang et al., 2019; Zhang et al., 1999). Technological advancements in the generation of somatic mouse models have permitted the development of genetically distinct tumor models within the same organ (Xue et al., 2014), a strategy recently applied to the generation of hepatocellular carcinoma (HCC; Brown, Heinrich, & Greten, 2018; Molina-Sánchez et al., 2020; Wang et al., 2019). HCC is genetically highly heterogeneous, but none of the major identified classes of HCC is predictive of therapeutic outcome, nor have easily targetable mutant drivers been found (EASL-EORTC, 2012). So far, T-cell-centric immunotherapy trials have shown limited but encouraging results, priming the design of combination therapies for advanced HCC (El-Khoueiry, 2017) and stimulating the growing interest in employing immunomodulation in HCC treatment (Ringelhan, Pfister, O'Connor, Pikarsky, & Heikenwalder, 2018). We recently documented the intertwined relationship between cancer cell genetics and therapy response together with modulation of the local and systemic TME (Wang et al., 2019). This emerging evidence suggests that cancer genetic aberrations strongly influence the content, nature, and activation phenotype of immune cells at the systemic and local level. Thus, in-depth and high-throughput methods to generate, follow, and analyze the progression of HCC in somatic mouse models, as well as to examine their heterogeneous immune landscape in an integrated fashion, are essential to interrogate immune cell diversity connection to tumor cell mutational status (Wellenstein & de Visser, 2018).
Investigations have been conducted to analyze the TME of different solid tumor types (Giraldo et al., 2019; Kielbassa, Vegna, Ramirez, & Akkari, 2019) by either flow cytometry or immunohistochemistry. Our detailed, descriptive, and methodical protocols provide several key features compared to previously reported analyses. First, our protocols allow for the use of each tumor sample in multiple assays with minimal material required. In addition to further detailing the procedure of hydrodynamic tail vein injection (HDTVi; Niola, Dagnæs-Hansen, & Frödin, 2019) for the development of liver tumors, we provide here an optimized protocol for monitoring their longitudinal tumor growth by MRI and subsequently quantifying tumor volume. This is an essential step to determine the consequences of different genetic backgrounds introduced in hepatocyte cancer cells. From the liver tumors generated, our protocols describe the completion of systemic peripheral blood analyses, the assessment of local TME composition by flow cytometry using optimized complex panels of markers validated to efficiently label liver immune cells, and the spatial quantification of immune cells. This latter feature includes immunohistochemistry markers of myeloid and lymphoid cells in liver HCC with their associated methods of fixation and quantification using high-throughput software, which can then be cross-compared with flow cytometry results for the same HCC-bearing mice. In order to assess not only the content of immune cells but also their phenotypic activation, we detail transcriptomic and biochemical analyses of sorted immune cell populations, each applicable at different stages of tumor progression. In addition to the immune-related changes in the liver microenvironment, we present protocols to assess histological alterations associated with liver disease, such as fibrosis and steatosis, both key features of HCC progression. Results obtained from applying these diverse and complementary analyses thus allow a complete evaluation of dynamic changes occurring in HCC and makes it possible to correlate alterations of the immune landscape with cancer cell genetic background and liver tumor malignancy in each mouse model generated.
Critical Parameters
The steps described in these protocols have carefully been optimized for application to HCC tumors generated using the HDTVi-mediated somatic integration of gene overexpression or gene knockouts of interest. In generating HCC models (Basic Protocol 1), careful attention should be paid on the age, strain, and gender of the animals used, as these may influence the phenotype of liver tumors and associated immune landscape. We recommend using 6- to 8-week-old C57/BL6J mice to enable the comparison of different HCC genetic backgrounds to what has been previously reported in the literature (Molina-Sánchez et al., 2020; Wang et al., 2019), and working in sterile conditions during the preparation of the plasmid mix in endotoxin-free solutions, and the subsequent HDTVi, using an IVC flow.
The choice of oncogenic drivers and concentrations is central to the experimental planning and should reflect the specific goal of the experiments. The ratio of plasmids injected during the HDTVi procedure (Basic Protocol 1) should consistently be 2:2:1 (SB vectors, sgRNA, and SB transposase, respectively) for successful integration of the SB vectors and sgRNA gene targets into hepatocytes. Although this ratio should be consistent, increasing the concentration of each vector will lead to accelerated tumor development, if desired. It is thus advisable to maintain the same concentrations of plasmids when injecting them into different mice.
To successfully monitor the outgrowth of liver tumors (Basic Protocol 2), the condition of the mice should be optimal for MRI scanning, and breathing and body temperature should be monitored during the procedure, especially in mice bearing advanced tumors. The mouse should be positioned appropriately on its back and fully anesthetized to control breathing movement, which may blur scan acquisition.
During the mouse sacrifice procedure (Basic Protocol 3), efficient heart perfusion will limit blood erythrocyte content and lead to better IHC and IF image quality after staining. This latter point can easily be confirmed by the discoloration of the liver to a pale beige when it is performed successfully. To successfully employ the diverse methods of processing HCC liver samples described in this protocol, it is essential to rapidly process the liver tumor samples after mouse sacrifice (<2 hr post isolation) and to continuously work on ice to maintain cell viability.
In circumstances where the HCC model studied gives rise to multiple tumor nodules, interlesion heterogeneity should be accounted for in follow-up analyses, and pieces of each nodule should be pooled together for all applications presented in this protocol.
Basic Protocol 4 presents the optimal methods to prepare, digest, and stain HCC samples for flow cytometry/cell sorting applications, which were optimized to maintain the integrity of specific epitopes (presented in Tables 2 and 3). If alternative or additional markers are to be applied to the suggested flow cytometry staining panel, it is essential to confirm that the proposed liver tumor digestion method does not compromise the integrity of these epitopes. Additionally, all antibodies used in the protocols listed in these methods should be properly titrated before use to ensure staining quality and limit the overuse of antibodies.
In order to identify and isolate specific cell populations after flow cytometry, samples must be processed without fixation, and sufficient cell numbers must be sorted out of the tumor-bearing livers. For either RT-qPCR or RNA sequencing analyses of sorted immune cells (Basic Protocol 6), a minimum of 5000 cells is recommended. To ensure proper RNA quality and reproducibility of results, samples must be kept on ice at all times and RNase-free tools and tubes should be used; moreover, once sorted, the samples should be immediately frozen on dry ice and stored at −80.
Regarding the processing of samples for protein extraction and western-blot/ELISA analyses (Alternate Protocol 3), the use of protease or phosphatase inhibitors is strongly recommended when examining the activation of kinases. Additionally, using sonication will greatly enhance the quality of the samples and limit the appearance of smear bands. Cycle of freezing and thawing should be avoided to ensure reproducible results and publication-quality images.
Proper equipment and protective measures must be used when performing this protocol, as some of the chemicals listed here are carcinogenic. Additionally, the safety and protocol data sheets for all reagents should be carefully read before starting the protocols listed here.
Troubleshooting
Troubleshooting experiences and solutions for potential problems pertaining to each individual protocol are presented in Table 8.
| Issues | Potential causes/solutions |
|---|---|
| Basic Protocol Basic Protocol 1 | |
| HDTVi injections do not result in consistent tumor outgrowth |
The injected plasmids are defective: Ensure that the plasmids concentration is consistent between experiments and prepared in endotoxin free solution. Verify the purity of these plasmids by enzymatic digestions. The HDTVi were not performed successfully: Check the effectiveness of HDTVi by making sure the injection takes 6-8 s in heated mice, and by observing the behavior of the mice after HDTVi. Mice should slow sluggishness and lack of movement for ∼20 min, which is a sign of successful entry of the injected solution into the liver parenchyma and thus into hepatocytes. |
| Basic Protocol Basic Protocol 2 | |
| MRI images are uninterpretable (high background, blurry, contrast issues) |
Ensure the correct imaging configuration is applied. Use the localizer to see whether the region of interest is in the center of the coil, and that the coil is in the center of the B0/field of view. If not, physically correct this by adjusting the animal/bed position. When everything is centered, repeat the setup procedure (wobble adjustment, all shims, basic frequency, reference power, receiver gain, B0 map). Redoing this setup can resolve image quality issues that may arise during acquisition, especially when other parameters remain constant. Check if the respiratory trigger is on and calibrated correctly. To avoid motion artifacts, the mouse must be correctly anesthetized during the procedure, with respiration between 40 and 100 breaths per minute. A respiratory level that is too low can result in hiccups and one that is too high in the mouse waking up. To ensure the proper degree of anesthesia, induce anesthesia 5 min before placing the mouse in the MR instrument. Ensure that the correct imaging configuration is applied, with the right TR/TE/flip angle and amount of averages; if needed, increase the amount of averages, which should improves image quality. Finally, make sure no one enters the MRI room during image acquisition to prevent loss of quality due to disturbance of the magnetic fields. |
| Basic Protocol Basic Protocol 3 | |
| Unsuccessful heart perfusion | Intracardiac heart perfusion is a necessary step for clearing the liver and preparing it for analyses. This procedure should cause in livers to develop a pale beige color after perfusion. If this is not achieved, the perfusion butterfly needle needs to be cleared of air and repositioned into the heart left ventricle toward the heart apex. Be sure to increase the volume of PBS to 20 ml when repeating the procedure. |
| Support Protocol Support Protocol 1 | |
| Blood clogging | EDTA collection tubes should be placed on ice before and after collection and inverted regularly until the preparation of the sample to avoid clogging. |
| Basic Protocol Basic Protocol 4 | |
| Tissue digestion incomplete | Cut the tumor/tissue in multiple small pieces to be digested in independent tubes, each with its own digestion mix. This is particularly relevant to HCC tumors that are very fibrotic. |
| Basic Protocol Basic Protocol 4 and Alternate Protocol Alternate Protocol 1 | |
| Low signal/absence of signal upon flow cytometry |
Assuming that it has been confirmed by other means (IHC/IF) that the target protein is expressed in the samples, insufficient amount/not optimized antibody concentration, dim fluorochrome conjugation, or inaccurate compensation matrix are the most likely causes for these problems. Antibody optimization: Optimize the concentration of each antibody concentration on liver tissue independently; concentration range should be 1/50-1/1000 and a final optimization with the concentration of choice should be performed in the complete panel presented in Tables 2 and 3. Dim/poorly detectable fluorophore: If an antibody is known to have worked well previously (e.g., those optimized and listed in Tables 2 and 3), it is likely that the conjugated antibody has been left exposed to light and the fluorescence conjugation has become unstable; in that case, re-order fresh antibody. When working with a new antibody added to the panel listed in Tables 2 and 3, it is likely that the conjugated fluorophore is not properly detected; in that case, confirm that the problem cannot be solved by adjusting the compensation matrix, and then switch to a more visible fluorophore. Compensation matrix gives a high percentage of errors to compensate for: Correct the matrix manually with the support of a flow cytometry technician or experienced user. |
| Basic Protocol Basic Protocol 5 and Alternate Protocol Alternate Protocol 2 | |
| Weak staining |
The antibody concentration may be inadequate: perform dilution series with positive control using tonsils or spleen. Ensure that the target and host species are compatible. If problems continue, section thinner slides (5 µm) instead of the 10 µm suggested for the cryosections |
| High background |
Try troubleshooting above (antibody concentration and host-species compatibility). Repeat washing steps and increase blocking time. Apply the optional antigen retrieval step. |
| Basic Protocol Basic Protocol 6 | |
| RNA degradation | Ensure all used reagents and tool used are RNase free. Selected tumor pieces with limited necrosis which will affect the RNA stability. Omit the TRIzol extraction and use a column-base RNA isolation kit. Proceed to RNA extraction with this commercial kit. |
| Low RNA yield | For sorted cell RNA preparation, increase the number of sorted cells per sample. Omit the TRIzol extraction and proceed to sort the cells in lysis buffer from a column-based RNA isolation kit. Proceed to RNA extraction with this commercial kit. |
| Unexpected gene expression | Redesign the Taqman probe used in the qPCR assay. Use multiple housekeeping genes to control for RNA/cDNA quantity between samples. Reduce the number of detection cycles. |
Understanding Results
HDTV injections of SB and CRISPR-Cas9 plasmids will give rise to steady liver tumors outgrowth, with different longitudinal kinetics depending on the concentration of plasmid chosen and the combination of oncogenic drivers. Thus, the weekly or biweekly MRI measurements presented in Figure 3B-D will be variable based on these considerations and should be initiated 2 weeks after HDTVi. To properly determine tumor outgrowth, the same experimenter should measure the volume of each mouse tumor longitudinally to identify emerging tumors based on contrasting white areas in the liver. A possible consequence of this protocol is that liver tumors will present distinct features, including different outgrowth kinetics and single/multiple nodules of changing contrast within the same mice. It is advisable to seek confirmation of the measured tumor areas from an MRI technician or experienced user in case of doubts. Additionally, certain combinations of plasmids may not result in tumor development; in that case, the mouse should be kept and imaged less frequently until a determined time point (30 weeks is suggested) to confirm that no late outgrowth emerges. Humane endpoints should be considered in these experiments, in which a tumor volume exceeding 2000 mm3 may result in animal discomfort, and requires sacrifice. Of note, metastases to distant organs are rarely seen in HDTVi somatic models of HCC, but could be an explanation for early symptoms not correlated with tumor volume measured by MRI. In that case, biopsies of lungs and lymph nodes will shed light on the cause of a potential early death of themouse.
The isolation of liver tumors and subsequent single-cell preparation should give rise to clear populations identified by flow cytometry after staining, and thus follow-up quantification analyses are expected to be specific for the HDTVi-induced HCC models, based on the oncogenic drivers chosen. As an example, Figures 4 and 5 depict the proportions of seven different populations of myeloid cells and six populations of lymphoid cells based on marker expression in a control liver of a mouse injected with empty vectors via HDTVi. These proportions will be differentially enriched in the context of HCC outgrowth, depending on the combination of oncogenic drivers chosen, and through time. These analyses will make it possible to connect the genetic landscape of HCC tumor cells and their stage of development to the associated immune cell context. Furthermore, the transcriptional analyses of immune cell subsets described in Basic Protocol 6 will shed light into the education features of these cells in a cancer-cell-genetics- and time-dependent manner. It is thus expected that heterogeneous responses will occur in immune cells, which should translate into cell- and stage-specific differences between HCC models generated by HDTVi.
From the single-cell preparation, we present methods to isolate liver cancer cell lines that can be maintained in vitro for further analyses of their proteomic, transcriptional, and secretomic landscapes. This protocol should yield pure cancer cell lines without contamination from fibroblasts or other immune cells, which is clearly identifiable based on cell morphology. The established cancer cell lines should present epithelial and not mesenchymal-like features, as expected from stromal fibroblasts. However, we recommend confirming the purity of these isolated cell lines, as discussed in Support Protocol 2.
The protocols for IHC and IF staining and analyses (Basic Protocol 5 and Alternate Protocol 2) will complement the flow cytometric quantification of specific immune cells in liver tumors. Coexpression of markers within the same cell type can readily be detected by flow cytometric gating, as discussed above, and will be visible from IF co-staining of markers present in distinct or within the same cell type. Clear IF signals may be obstructed by background fluorescence of the liver; it is thus standard to optimize the fixation procedure and concentration of antibodies and to include secondary antibody staining to control for background fluorescence, which threshold can then be removed from the stained sample to obtain publication-quality images. Tissues that do not express the target protein or contain the target cell type will present only background staining. Importantly, orthogonal analyses of IHC/IF and flow cytometry results may yield different considerations. Immune cell quantitation by flow cytometry will determine the proportion of each cell population relative to the parental cell gate chosen (total immune or total liver cells for example), whereas IF/IHC will give rise to absolute number counts. It is thus recommended that both analyses be performed to obtain a clear overview of immune cell content in the liver tumors generated.
Time Considerations
The complete time considerations for the experimental procedures pertaining to Basic Protocol 1 will strongly depend on the penetrance of the HCC model chosen, based on the oncogenic drivers transduced by HDTVi. As an example, the use of the MycOE/P53KO genetic combination (leading to overexpression of c-Myc and knockout of p53 in hepatocytes; Wang et al., 2019) will give rise to HCC detectable by MRI 2 weeks after HDTVi, which will progress into aggressive tumors in an additional 14 days. The HDTVi procedure, including preparing the flow hood for sterile injections conditions, heating the mice to dilate the caudal vein, and performing the HDTVi, requires 1.5 hr total for a ten-mouse cohort. Once the mice have been injected, they should be allowed to rest on a heated bed for an additional 30 min. The MRI procedure described in Basic Protocol 2 has been optimized to give proper, publishable images in 20 min per mouse scanned, to which 15 min should be added for the tumor volume quantification depicted in Figure 3.
The timeframe for the liver dissection and tumor isolation described in Basic Protocol 3 includes the preparation of the sterile environment, mouse euthanasia, cardiac perfusion, and blood collection, which altogether take 40 min per mouse. Whole liver isolated through this procedure should be processed within 1 hr after sacrifice for follow-up analyses delineated in Basic Protocols 4, 5, and 6, or can be kept at −80°C for up to 2 years (as snap-frozen samples). The tumor processing for flow cytometry in Basic Protocol 4 entails tumor digestion, single-cell preparation, and staining, for a total of 3.5 hr per sample, although several samples can be processed simultaneously. For follow-up live cell sorting (Support Protocol 3), samples are to be run on a FACS sorter to isolate the cell subsets of choice, which will require 30 min per sample, not including the time needed to set up the compensation matrix (1 hr). However, if a rare cell population is sought, the sorting time could increase up to 1 hr. After fixation, flow cytometry samples can be kept at 4°C in the dark, to maintain the fluorescent signal, for a maximum of 48 hr before flow cytometry analyses, which will take 20 min per sample.
Basic Protocols 5 and Alternate Protocol 2 are less hands-on than Basic Protocol 4, but together will take more time—a minimum of 3-4 days if all procedures are performed sequentially without breaks. After isolation of tumor-bearing livers, lobes will be fixed over 1-2 days in the appropriate solution according to Basic Protocol 5 and embedded in cassettes, and they can then be stored at room temperature for up to 5 years as paraffin blocks (for IHC/histochemistry) or at −80°C (frozen blocks for IF) before samples will display high background after staining. After sectioning, IHC or IF staining will take 48 hr, including the overnight primary antibody mix incubation and the secondary staining performed on day 2. Importantly, high-throughput scanning of IHC/histochemistry sections using the AperioScan or IF sections using the Axioscan (Zeiss) allows up to 100 sections to be processed simultaneously, and an average 20 sections per scan will take 30 min using the AperioScan and 3 hr in the Axioscan.
Basic Protocol 6, which includes RNA extraction and RT-qPCR transcriptional analyses of sorted cells after FACS or of snap-frozen tumor samples, will require a full day of work, following the manufacturer's instructions and ensuring that the experimental plan and genes to assess are carefully planned. Similarly, the procedures described in Alternate Protocol 3 pertaining to protein analyses will require 2 days of work for western blotting or ELISA procedures, as it is recommended that the staining be performed overnight.
Acknowledgments
We thank members of the Akkari lab for insightful discussion. This research was supported by the Oncode Institute, the Cancer Genomics Center, and the Dutch Cancer Society (KWF 12049 liters.A.). The authors declare no conflict of interest.
Author Contributions
Daniel Taranto : Conceptualization, investigation, validation, visualization, writing–original draft, writing–review and editing; Christel F.A. Ramirez : Investigation, resources, software, validation, visualization, writing–review and editing; Serena Vegna : Investigation, methodology, resources, validation, visualization, writing–review and editing; Marnix H.P. de Groot : validation, visualization, writing–review and editing; Niels de Wit : Data curation, methodology, software, writing–review and editing; Martijn Van Baalen : Data curation, writing–review and editing; Sjoerd Klarenbeek : Methodology, resources, writing–review and editing; Leila Akkari : Conceptualization, funding acquisition, methodology, project administration, supervision, validation, writing–original draft, writing–review and editing.
Conflict of Interest
JRT is a co-founder of and shareholder in Thelium Therapeutics and has consulted for several biopharmaceutical companies interested in developing approaches to analyze or modulate intestinal barrier function. None of these relationships have direct bearing on or make use of this protocol.
Open Research
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Literature Cited
- Bazin, P.-L., Cuzzocreo, J. L., Yassa, M. A., Gandler, W., McAuliffe, M. J., Bassett, S. S., & Pham, D. L. (2007). Volumetric neuroimage analysis extensions for the MIPAV software package. Journal of Neuroscience Methods , 165(1), 111–121. doi: 10.1016/j.jneumeth.2007.05.024.
- Bell, J. B., Podetz-Pedersen, K. M., Aronovich, E. L., Belur, L. R., McIvor, R. S., & Hackett, P. B. (2007). Preferential delivery of the Sleeping Beauty transposon system to livers of mice by hydrodynamic injection. Nature Protocols , 2(12), 3153–3165. doi: 10.1038/nprot.2007.471.
- Brown, Z. J., Heinrich, B., & Greten, T. F. (2018). Mouse models of hepatocellular carcinoma: An overview and highlights for immunotherapy research. Nature Reviews. Gastroenterology & Hepatology, 15(9), 536–554. doi: 10.1038/s41575-018-0033-6.
- Cheung, M., Campbell, J. J., Whitby, L., Thomas, R. J., Braybrook, J., & Petzing, J. (2021). Current trends in flow cytometry automated data analysis software. Cytometry Part A , n/a(n/a). doi: 10.1002/cyto.a.24320.
- European Association for the Study of the Liver, & European Organisation for Research and Treatment of Cancer (EASL-EORTC). (2012). EASL-EORTC clinical practice guidelines: Management of hepatocellular carcinoma. Journal of Hepatology , 56(4), 908–943. doi: 10.1016/j.jhep.2011.12.001.
- El-Khoueiry, A. (2017). The promise of immunotherapy in the treatment of hepatocellular carcinoma. American Society of Clinical Oncology Educational Book , 37, 311–317. doi: 10.1200/edbk_175230.
- Gadiparthi, C., Yoo, E. R., Are, V. S., Charilaou, P., Kim, D., Cholankeril, G., … Ahmed, A. (2019). Hepatocellular carcinoma is leading in cancer-related disease burden among hospitalized baby boomers. Annals of Hepatology , 18(5), 679–684. doi: 10.1016/j.aohep.2019.04.014.
- Giraldo, N. A., Sanchez-Salas, R., Peske, J. D., Vano, Y., Becht, E., Petitprez, F., … Sautès-Fridman, C. (2019). The clinical role of the TME in solid cancer. British Journal of Cancer , 120(1), 45–53. doi: 10.1038/s41416-018-0327-z.
- Kielbassa, K., Vegna, S., Ramirez, C., & Akkari, L. (2019). Understanding the origin and diversity of macrophages to tailor their targeting in solid cancers. Frontiers in Immunology , 10, 2215. doi: 10.3389/fimmu.2019.02215.
- Llovet, J. M., Montal, R., Sia, D., & Finn, R. S. (2018). Molecular therapies and precision medicine for hepatocellular carcinoma. Nature Reviews Clinical oncology , 15(10), 599–616. doi: 10.1038/s41571-018-0073-4.
- Molina-Sánchez, P., Ruiz de Galarreta, M., Yao, M. A., Lindblad, K. E., Bresnahan, E., Bitterman, E., … Lujambio, A. (2020). Cooperation between distinct cancer driver genes underlies intertumor heterogeneity in hepatocellular carcinoma. Gastroenterology , 159(6), 2203–2220.e2214. doi: 10.1053/j.gastro.2020.08.015.
- Niola, F., Dagnæs-Hansen, F., & Frödin, M. (2019). In vivo editing of the adult mouse liver using CRISPR/Cas9 and hydrodynamic tail vein injection. Methods in Molecular Biology , 1961, 329–341. doi: 10.1007/978-1-4939-9170-9_20.
- Ringelhan, M., Pfister, D., O'Connor, T., Pikarsky, E., & Heikenwalder, M. (2018). The immunology of hepatocellular carcinoma. Nature Immunology , 19(3), 222–232. doi: 10.1038/s41590-018-0044-z.
- Saeys, Y., Van Gassen, S., & Lambrecht, B. N. (2016). Computational flow cytometry: Helping to make sense of high-dimensional immunology data. Nature Reviews Immunology , 16(7), 449–462. doi: 10.1038/nri.2016.56.
- Schulze, K., Nault, J. C., & Villanueva, A. (2016). Genetic profiling of hepatocellular carcinoma using next-generation sequencing. Journal of Hepatology , 65(5), 1031–1042. doi: 10.1016/j.jhep.2016.05.035.
- Slaoui, M., & Fiette, L. (2011). Histopathology procedures: From tissue sampling to histopathological evaluation. Methods in Molecular Biology , 691, 69–82. doi: 10.1007/978-1-60761-849-2_4.
- Thommen, D. S., Koelzer, V. H., Herzig, P., Roller, A., Trefny, M., Dimeloe, S., … Zippelius, A. (2018). A transcriptionally and functionally distinct PD-1+ CD8+ T cell pool with predictive potential in non-small-cell lung cancer treated with PD-1 blockade. Nature Medicine , 24(7), 994–1004. doi: 10.1038/s41591-018-0057-z.
- Wang, C., Vegna, S., Jin, H., Benedict, B., Lieftink, C., Ramirez, C., … Bernards, R. (2019). Inducing and exploiting vulnerabilities for the treatment of liver cancer. Nature , 574(7777), 268–272. doi: 10.1038/s41586-019-1607-3.
- Wellenstein, M. D., & de Visser, K. E. (2018). Cancer-cell-intrinsic mechanisms shaping the tumor immune landscape. Immunity , 48(3), 399–416. doi: 10.1016/j.immuni.2018.03.004.
- Xue, W., Chen, S., Yin, H., Tammela, T., Papagiannakopoulos, T., Joshi, N. S., … Jacks, T. (2014). CRISPR-mediated direct mutation of cancer genes in the mouse liver. Nature , 514(7522), 380–384. doi: 10.1038/nature13589.
- Zhang, G., Budker, V., & Wolff, J. A. (1999). High levels of foreign gene expression in hepatocytes after tail vein injections of naked plasmid DNA. Human Gene Therapy , 10(10), 1735–1737. doi: 10.1089/10430349950017734.
- Zucman-Rossi, J., Villanueva, A., Nault, J. C., & Llovet, J. M. (2015). Genetic landscape and biomarkers of hepatocellular carcinoma. Gastroenterology , 149(5), 1226–1239.e1224. doi: 10.1053/j.gastro.2015.05.061.
Citing Literature
Number of times cited according to CrossRef: 2
- Anthony Lozano, Francois-Régis Souche, Carine Chavey, Valérie Dardalhon, Christel Ramirez, Serena Vegna, Guillaume Desandre, Anaïs Riviere, Amal Zine El Aabidine, Philippe Fort, Leila Akkari, Urszula Hibner, Damien Grégoire, Ras/MAPK signalling intensity defines subclonal fitness in a mouse model of hepatocellular carcinoma, eLife, 10.7554/eLife.76294, 12 , (2023).
- Antonio Mulero‐Sánchez, Christel F. A. Ramirez, Aimée Chatinier, Hui Wang, Sofie J. I. Koomen, Ji‐Ying Song, Marnix H. P. Groot, Cor Lieftink, Astrid Bosma, Artur Burylo, Olaf Tellingen, Roderick L. Beijersbergen, Cun Wang, Leila Akkari, René Bernards, Sara Mainardi, Rational combination of SHP2 and mTOR inhibition for the treatment of hepatocellular carcinoma, Molecular Oncology, 10.1002/1878-0261.13377, 17 , 6, (964-980), (2023).

