Methods to Evaluate the Potential Clinical Significance of Antibodies to Red Blood Cells
Kayluz Frias Boligan, Kayluz Frias Boligan, Gurleen Sandhu, Gurleen Sandhu, Donald R. Branch, Donald R. Branch
ADCC
antibody-dependent cellular cytotoxicity
clinically significant RBC antibodies
MMA
monocyte monolayer assay
Abstract
Immune-mediated red blood cell (RBC) destruction due to antibodies is an ongoing problem in transfusion medicine for the selection of the safest blood. Serological testing often revealed incompatibility with donors’ RBCs. When this incompatible blood was transfused, destruction was due mostly to extravascular-mediated phagocytosis of the antibody-opsonized RBCs; however, intravascular hemolysis was sometimes observed without explanation. Based on serology, antibodies with potential for clinical sequalae could not be ascertained; thus, antigen-negative blood was usually selected for transfusion to avoid problems. Antibodies to antigens having very high frequency in the general population (>95%), however, made selection of antigen-negative blood difficult and sometimes impossible. Some patients, who were sensitized by previous transfusions or by pregnancy, developed multiple antibodies, again creating a problem for finding compatible blood for transfusion, without the ability to discern which of the antibodies may be clinically irrelevant and ignored. Transfusion medicine scientists began searching for an in vitro means to determine the in vivo outcome of transfusion of blood that was serologically incompatible. Methods such as chemiluminescence, monocyte-macrophage phagocytosis, and antibody-dependent cellular cytotoxicity (ADCC) were described. Over the years, the monocyte monolayer assay (MMA) has emerged as the most reliable in vitro assay for the prediction of the clinical relevance of a given antibody. ADCC has not been fully studied but has the potential to be useful for predicting which antibodies may result in intravascular hemolysis. This article captures the protocols for the implementation and readout of the MMA and ADCC assays for use in predicting the clinical significance of antibodies in a transfusion setting. © 2022 The Authors. Current Protocols published by Wiley Periodicals LLC.
Basic Protocol 1 : Monocyte monolayer assay (MMA)
Basic Protocol 2 : Antibody-dependent cellular cytotoxicity assay (ADCC)
INTRODUCTION
Since the discovery of the ABO blood group system by Landsteiner (1901), the clinical discipline of transfusion medicine has grown with leaps and bounds. Indeed, the four blood group antigens, A, B, AB, and O, described by Landsteiner in the early 1900s became the first human blood group system (ABO). Since then, additional blood group systems have been described so that there are now >250 blood group antigens and 25 blood group systems (Garratty et al., 2000). Despite the serologic and genetic methods developed to identify blood group antigens and their corresponding antibodies, the question of which antibodies are important from a clinical standpoint has never been completely answered.
Originally, in vivo assays to determine the clinical significance of recipient-produced antibodies to red blood cell (RBC) antigens were used. The so-called biological crossmatch (Mollison, 1972) and Chromium-51 (51Cr) RBC survival assays (Donohue et al., 1955; Mollison, 1984; Silvergleid et al., 1978) were the first assays for the determination of the clinical significance of RBC antibodies based on the survival of infused RBCs in the presence of recipient antibodies. More recently, a non-radioactive, biotin-labeling assay has been used with some success (Mock et al., 2014). Although, these in vivo assays were useful, involvement of radioactivity or having to transfuse incompatible blood without any preconception of outcome is dangerous; so, in the early 1980s, investigators began in earnest to try and address the clinical significance of detected RBC antibodies by designing in vitro cellular assays to mimic the in vivo environment (Branch, Gallagher, Mison, Sy Siok Hian, & Petz, 1984; Conley et al., 1982; Gallagher, Branch, Mison, & Petz, 1983; Hunt, Beck, Hardman, Tegtmeier, & Bayer, 1980; Schanfield, Schoeppner, & Stevens, 1980; Stevens, Schanfield, & Braley, 1976). Assays to assess the potential for an antibody to cause hemolysis of transfused red blood cells in patients having the corresponding alloantibody included a chemiluminescence test (CLT; Downing, Templeton, Mitchell, & Fraser, 1990; Hadley, Wilkes, Poole, Arndt, & Garratty, 1999; Lucas, Hadley, Nance, & Garratty, 1993), monocyte-macrophage assays (MMAs; Tong & Branch, 2017; Tong, Burke-Murphy, et al., 2016; Zupanska, 1985), flow cytometry (Balola, Mayer, Bartolmas, & Salama, 2021), and less characterized assays such as antibody-dependent cellular cytotoxicity (ADCC; Barcellini, 2015). Herein, we provide detailed protocols for the MMA and ADCC assays for use to determine the clinical significance of antibodies in patients requiring transfusion of serologically incompatible donor blood.
Basic Protocol 1: MONOCYTE MONOLAYER ASSAY (MMA)
MMA is an in vitro assay which is used to predict blood transfusion outcomes in patients with auto- or alloantibodies to RBCs. In this assay, anti-RBC antibodies are assessed for their Fcγ receptor (FcγR)-mediated phagocytosis. Through serological methods, compatibility testing or crossmatching is performed. Once the presence of antibodies to RBCs is detected, MMA can be further used to identify the clinical significance of the anti-RBC antibody by testing them against specific RBC antigens (e.g., opsonizing Kell positive RBCs with anti-Kell antibody). The phagocytosis results from the MMA can help reduce the risk of post-transfusion hemolysis. Cells of interest in the MMA are the peripheral blood mononuclear cells (PBMCs) due to the mononuclear phagocyte system's involvement in mediating the extravascular hemolysis of antibody-bound RBCs. Apart from predicting post-transfusion survival or clearance of RBCs, MMA can be used to study other aspects of the IgG antibody and FcγR interaction that induce phagocytosis.
This protocol is modified from the work by Tong and Branch (2017).
Materials
-
RPMI-1640 complete culture medium (see recipe)
-
Cytiva Ficoll Paque PLUS, density 1.077 g/L (Thermo Fisher Scientific, 17-1440-03)
-
Fresh whole blood collected in acid-citrate-dextrose (ACD) tubes (yellow-top tubes; minimum of two blood tubes should be collected for MMA)
-
Fresh whole blood collected by venipuncture into red-top (no additive) serum separator tubes
-
Rh positive (R2R2) red blood cells (control; Blood Collection Center, Canadian Blood Services; also commercially available)
-
Anti-Rh(D):
- Commercial source of Rh immune globulin (e.g., WinRho® SDF CDN, Saol Therapeutics, 1003092)
- Rh immune globulin (to opsonize control red blood cells; R2R2 RBCs)
-
ACK lysis buffer (see recipe)
-
1× PBS, pH 7.4, without Ca2+/Mg2+ (Wisent Bioproducts, 311-425-CL)
-
Red blood cell storage/stabilization solution: ID-CellStab (Bio-Rad, 005650 05740)
-
100% methanol
-
Anti-Human Globulin (AHG; commercial source)
-
Elvanol mounting medium (see recipe)
-
Trypan Blue solution (Thermo Fisher Scientific, 15250061)
-
Nunc® Lab-TekTM II Chamber SlideTM with Cover, RS Glass Slide Sterile (Thermo Fisher Scientific, 154534)
-
Coverslips (24 × 50 mm; VWR, 48393-081)
-
Manual cell counters
Isolate PBMCs
1.Obtain human whole blood from donor or patient in ACD tubes and red-topped serum tubes. Store whole blood in ACD tubes at room temperature (18°C to 22°C) and red-topped serum tubes at 4°C to allow separation of serum.
2.Transfer room temperature blood from whole blood ACD tubes to 50-ml Falcon tubes (approximately one 50-ml Falcon tube for every two ACD tubes). Add equal volume of the room temperature RPMI-1640 complete medium to the blood (1:1 ratio of whole blood to medium), for a final volume of 35 ml.
3.Add 15 ml room temperature Ficoll Paque Plus to a new 50-ml Falcon tube.
4.Carefully layer the 1:1 diluted whole blood on top of the Ficoll Paque Plus density gradient to minimize amount of mixing at the interface for optimal separation of blood.
5.Centrifuge the layered mixture at 700 × g for 30 min with brakes OFF.
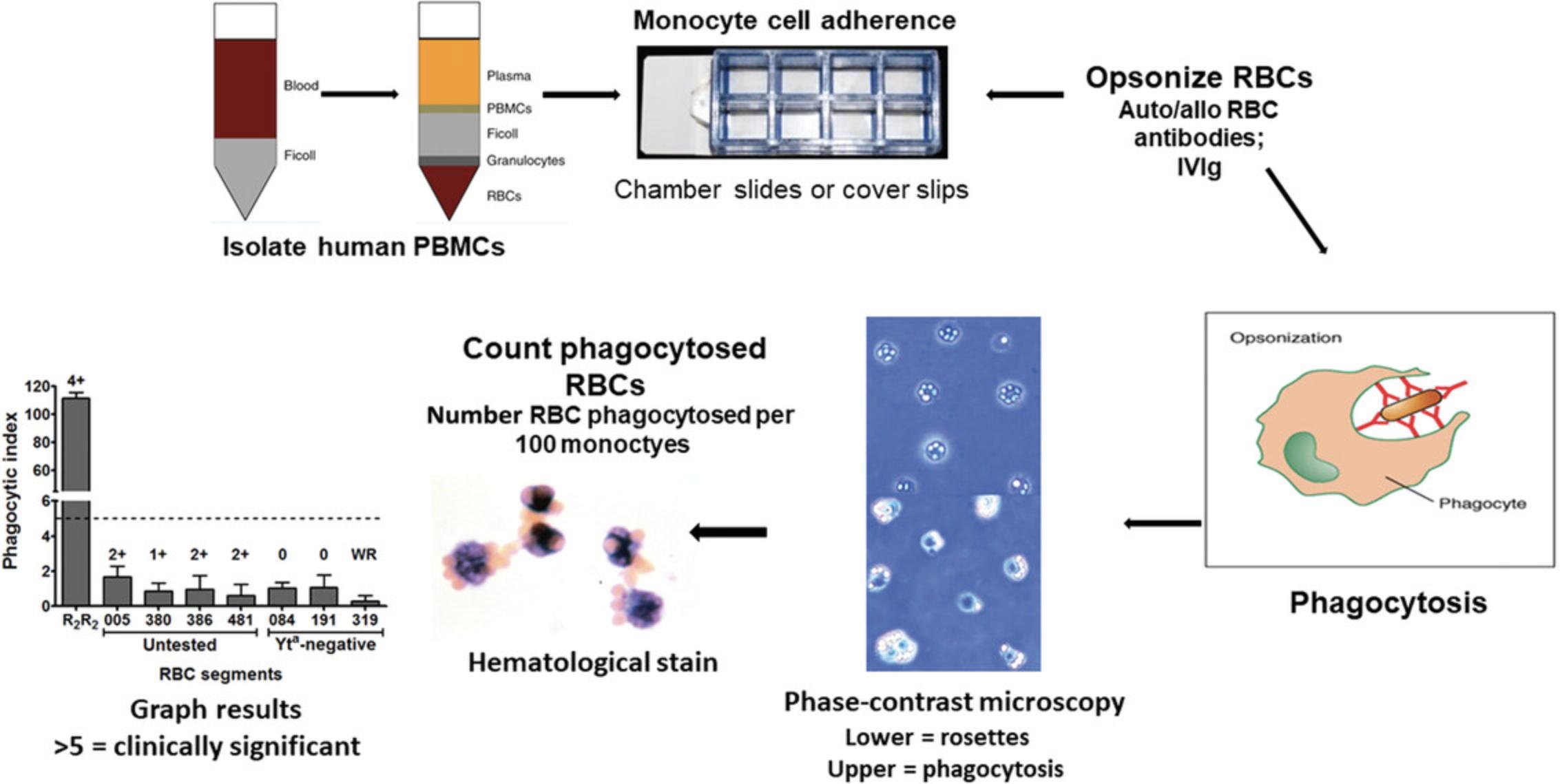
6.Aspirate and discard the majority of the topmost layer (supernatant) which is plasma, leaving 1-2 ml remaining above the buffy coat layer, and carefully retrieve the buffy coat (PBMCs) layer. (Using plastic or glass Pasteur pipets with a suction bulb while performing circular motions is recommended.) Transfer the retrieved PBMCs into a new 15-ml tube.
7.Wash the isolated buffy coat layer two times with pH 7.4 PBS solution for 10 min at 350 × g with full brakes ON in between washes.
8.Lyse any RBCs carried over with ACK lysis buffer. Add 5-10 ml ACK lysis buffer, depending on pellet size, and incubate at room temperature for 5 min. After incubation, top up with pH 7.4 PBS and centrifuge 10 min at 350 × g with full brakes ON and wash one more time.
9.Reconstitute PBMC pellet in 3-7 ml (depending on the size of the pellet) RPMI-1640 complete medium.
10.Count PBMCs using a hemocytometer in a 1:1 staining ratio with trypan blue by only counting the cells not stained with trypan blue. Reconstitute PBMCs to 1,750,000 cells/ml in RPMI-1640 complete medium.
11.Seed 400 μl (700,000 cells) using a micropipet into each well of the 8-well chamber slide and incubate at 37°C, 5% CO2 for 1 hr in a humidified tissue culture incubator.
Pre-treatment of adhered monocytes
Steps 12-13 are only necessary if trying to inhibit or enhance phagocytosis.
12.Adhered monocytes can be pre-treated with any drug(s) or compound(s) of interest. Reconstitute the test material in RPMI-1640 complete medium to the desired concentration.
13.Aspirate any non-adhered PBMCs from the wells using a micropipet and discard. Add 400 μl of test material and incubate at 37°C, 5% CO2 for 1 hr.
Opsonization of Rh(D)+ R2R2 red blood cells
R2R2 RBCs are used as a positive control for FcγR-mediated phagocytosis. R2R2 RBCs can be centrifuged at 870 × g for 15 min with brakes ON and reconstituted in ID-CellStab (RBC storage/stabilization solution) and stored at 4°C for up to a month.
Although we use D+ R2R2 RBCs for opsonization with anti-D for our positive control, any Rh+ RBCs could be used and do not have to be phenotyped.
14.Wash R2R2 (cDE/cDE) RBCs in pH 7.4 PBS three times by centrifugating at 350 × g for 5 min each time. (You may need to wash more than three times if hemolysis or red supernatant observed.)
15.Opsonize R2R2 RBCs with anti-Rh(D) at 100 ng/ml and incubate at 37°C, 5% CO2 for 1 hr. Intermittent reconstitution by mixing is recommended as RBCs settle at the bottom (e.g., vortex every 15 min). A five percent RBC suspension for opsonization is recommended (e.g., 15 μl of packed RBCs in 300 μl antibody mixture).
16.Wash opsonized R2R2 RBCs with pH 7.4 PBS three times by centrifugating at 350 × g for 5 min each time.
17.Reconstitute washed opsonized R2R2 RBCs to 1.25% (v/v) using RPMI-1640 complete medium, for example, 1,200 μl medium to 15 μl RBCs to achieve 1.25% (v/v) per well.
Fc receptor-mediated phagocytosis
18.Aspirate pre-treatment test material (if added) or non-adhered cells (if not added) and discard, following gentle aspiration and adding technique (see step 13).
19.Gently wash wells one time with pH 7.4 PBS.
20.Add 400 μl of 1.25% (v/v) opsonized R2R2 RBCs mixture to each well of the triplicate. Incubate at 37°C, 5% CO2 for 2 hr undisturbed.
21.After incubation, prepare two beakers with 150 ml pH 7.4 PBS in each beaker. Invert the 8-well chamber slides to discard non-phagocytosed RBCs into one of the beakers. Remove chambers using the manufacturer's adaptors. Dab off excess on paper towel while keeping slides moist.
22.Submerge slide into the beaker with the discarded supernatants and wash slide by moving it back and forth slowly (20-30 strokes) to remove any remaining non-phagocytosed RBCs.
23.Using the other beaker without any discarded supernatants, submerge slide and wash slowly for 20-30 strokes more.
24.Remove slide from PBS and dab off excess on paper towel. Air dry slide.
25.When almost dry, fix by submerging in 100% methanol for 45 s then air dry.
26.Mount slide using an in-house made Elvanol mounting medium and add coverslips (24 × 75 mm).
27.Allow mount to dry overnight before quantification.
Quantification of phagocytosis
28.Using a phase contrast microscope and 40× objective lens, quantify phagocytosis using a manual cell counter.
Have two manual cell counters in each hand to count phagocytosed RBCs in one and total number of monocytes in the other (300 monocytes should be counted per well).
29.Calculate average phagocytic index (PI) per test (across triplicates) by dividing the number of phagocytosed RBCs by the number of total monocytes counted and multiplying by 100:
(Number of phagocytosed RBCs/Number of total monocytes counted) × 100.
Express data as average PI ± the standard error of the mean (SEM).
Interpretation of results
We and others have found that a PI >5 is correlated with clinically significant antibodies. The MMA may not correlate with serologic results as shown in Figure 1.In this result, the antibody (anti-Cartwright (Yta)) reacts by IAT serology with the Yt(a+) RBCs but not with Yt(a-) RBCs. This would suggest that this antibody is clinically significant and only Yt(a-) blood should be selected for transfusion. Despite the IAT reactivity, the MMA assay indicates that the PI is <5 and the antibody is, thus, considered clinically insignificant and that all these serologically incompatible donor bloods can be transfused into this patient without sequalae. Indeed, this patient was transfused with Yt(a+) blood and did not have any problems.
Basic Protocol 2: ANTIBODY-DEPENDENT CELLULAR CYTOTOXICITY ASSAY (ADCC)
Natural killer (NK) cells are classical mediators of ADCC through the interaction of their low affinity Fcγ receptor CD16 with IgG antibodies present in circulation. Some of these antibodies (alloantibodies) can lead to unwanted reactions in the case of patients receiving a transfusion thus matching between recipient and blood donor is required. Therefore, as a complementary method to determine the clinical significance of antibodies in transfusion reactions, and test for compatibility between donor and recipient, we evaluate the capacity of NK cells to mediate ADCC against red blood cells, when the latter have been exposed to a specific human serum.
Materials
-
RPMI-1640 complete culture medium (see recipe)
-
Cytiva Ficoll Paque PLUS, density 1.077 g/L (Thermo Fisher Scientific, 17-1440-03)
-
Fresh whole blood collected into acid-citrate-dextrose (ACD) tubes (yellow-top tubes; minimum of nine to ten blood tubes should be collected for NK cell-mediated ADCC assays; store blood at room temperature up to 48 hr)
-
Fresh whole blood collected by venipuncture into red-top (no additive) serum separator tubes
-
Isolation buffer (see recipe)
-
1× PBS, pH 7.4, without Ca2+/Mg2+ (Wisent Bioproducts, 311-425-CL)
-
Patient's isolated serum (for opsonization), or utilize patient's plasma or a purified antibody of your interest
-
51Cr (Na2CrO4, sodium chromate), 1 mCi (37 Mbq; PerkinElmer, NEZ030S001MC), lead-protected at room temperature; use within 2 half-lives
-
1 N HCl
-
Cell purification kits:
- EasySep™ Human NK Cell Isolation Kit (Stemcell Technologies, 17955) or
- EasySep™ Human NK Cell Enrichment Kit (Stemcell Technologies, 19055)
-
Radioactivity counter (e.g., MicroBeta2 Microplate Counter for Radiometric and Luminescence Detection, PerkinElmer)
-
LumaPlate-96, shallow wells, White Opaque 96-well Microplate with Scintillant Coated on the Bottom, sample capacity:100 µl (PerkinElmer, 6006633)
-
96-well suspension culture plates, U-bottom (Greiner Bio-One, 650 185)
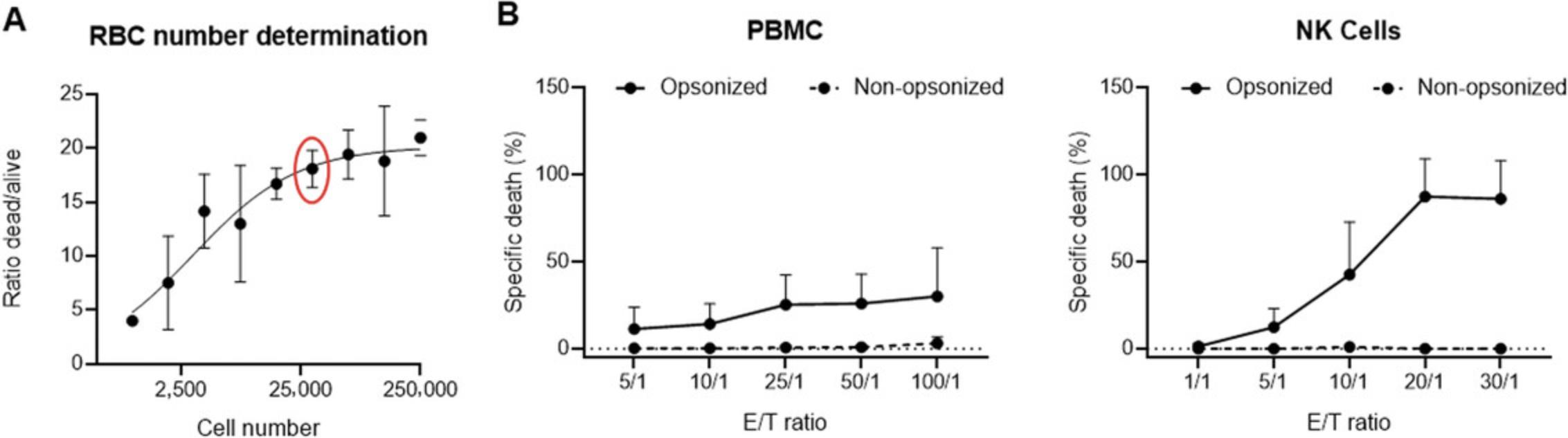
Optimization of number of RBCs to use in the ADCC assay
Optimization steps should be done just once at the beginning of the study.
1.Wash RBCs with PBS three times by centrifugating at 350 × g for 5 min at room temperature each time.
2.Count 1 × 107 RBCs, centrifuge to remove PBS, and resuspend in 50 μCi of 51Cr.
Incubate 1 hr in a 37°C incubator, 5% CO2. Resuspend RBCs every 15 min by tapping the tube on the side. Radionuclide should be used within 2 half-lives (27.71 days), adjusting the amount used based on radioactivity levels at the time of the assay.
3.Add 5 ml complete culture medium, centrifuge at room temperature (423 × g , 5 min), discard supernatant, and repeat for a total of three washes.
4.Resuspend washed RBCs in complete RPMI and adjust the cell concentration (in volume of 100 µl) to the highest number of cells that will be included in your titration curve (∼ 250,000 RBCs/well).
5.Seed at least six wells (two sets of triplicates) of the 51Cr loaded RBCs in round-bottom 96-well plates and do serial dilutions (by a factor of 2) starting from your most concentrated RBC sample down to 976 RBCs/well.
6.Lyse one set of triplicates for each dilution by adding 100 µl 1 N HCl and add 100 µl RPMI in the other set of triplicates.
7.Incubate plate at 37°C, 5% CO2 for the duration of the assay (usually 4 hr).
8.After 4 hr, take plate out of the incubator and carefully collect 50 μl cell-free supernatant from each well (with a multichannel pipet), and transfer to LumaPlates.
9.Let the LumaPlates dry overnight.
10.Count (1 min/sample) in a radioactivity counter (e.g., MicroBeta2 Microplate Counter).
11.Calculate the ratio of dead versus alive cells [divide the counts per minute (cpm) of the dead cells by the cpm of the alive cells] and plot in an xy graph against the corresponding cell number (Fig. 2A). Select the most adequate number of cells for your subsequent experiments.
Purify NK cells
12.Perform PBMC isolation as described in monocyte monolayer assay (Basic Protocol 1).
13.Count isolated PBMCs and adjust to 50 × 106 cells/ml in isolation buffer to proceed with the NK purification.
14.Purify NK cells using the NK isolation kit or the NK enrichment kit (Stemcell Technologies; EasySepTM) according to the manufacturer's instructions (Fig. 3).
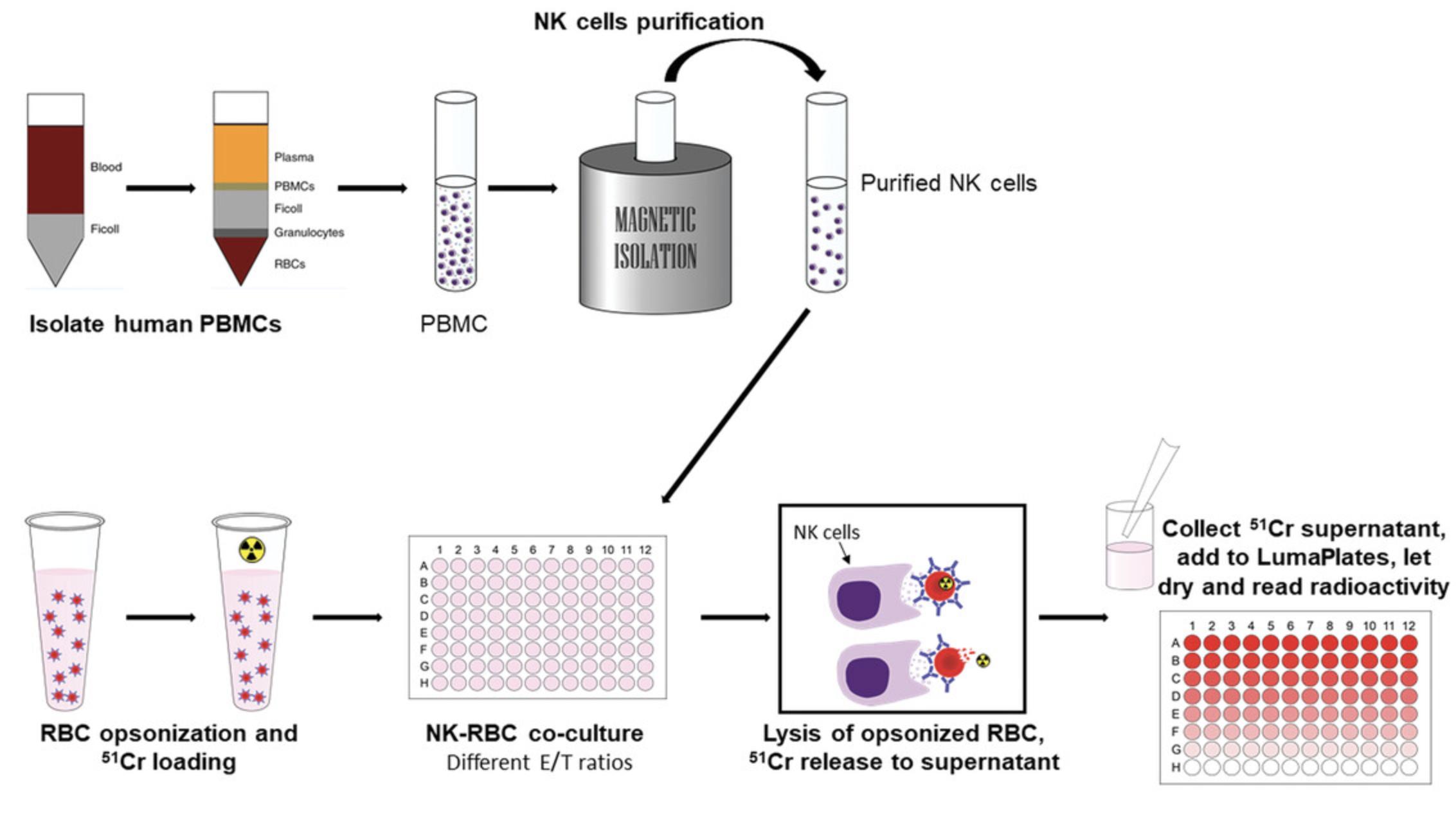
15.Count purified NK cells (ready to use) and resuspend in complete RPMI at the desired concentration.
Target cell labeling
16.Patient RBCs are washed and opsonized as described in Basic Protocol 1, steps 14-17.
17.Count RBCs and add 50 μCi of 51Cr per 1 × 107 cells.
Incubate for 1 hr in a 37°C incubator, 5% CO2. Resuspend every 15 min by tapping the tube on the side.
18.Add 5 ml complete medium, centrifuge (423 × g , 5 min), discard supernatant, and repeat for a total of three washes.
19.Resuspend washed RBCs in complete RPMI and adjust cell concentration to 30,000 cells in 100 µl.
Cytotoxicity assay
20.Plate 100 μl effector cells (at the desired concentrations) in triplicates on a 96-well round-bottom microtiter plate.
21.Add 100 μl radiolabeled target cell suspension to the wells already containing 100 μl of effector cells and to six additional wells.
22.Add 100 μl complete RPMI (spontaneous release) to three of the six additional wells and 100 μl 1 N HCl solution to the remaining three wells (maximum release).
23.Incubate 4 hr (5% CO2 at 37°C).
24.Carefully collect 50 μl cell-free supernatant (with a multichannel pipet) and transfer to LumaPlates.
25.Let the LumaPlates dry overnight.
26.Count (1 min/sample) in a radioactivity counter (e.g., MicroBeta2 microplate counter).
27.Determination of specific 51Cr release assay and calculation: The percent of specific 51Cr release (equivalent to specific lysis) is calculated as: (Experimental value-spontaneous release)/(Maximum release-spontaneous release) × 100. Each value is calculated as the mean of triplicates.
REAGENTS AND SOLUTIONS
ACK lysis buffer
- 155 mM NH4Cl, 0.1 mM EDTA, and 10 mM KHCO3.
- Store at 4°C for 1 year.
Elvanol mounting medium
- Dulbecco's PBS (D-PBS) without Ca2+/Mg2+, 15% (w/v) polyvinyl resin, and 30% (v/v) glycerine.
- Store at room temperature for 1 year.
Isolation buffer
- 1× PBS, supplemented with 2% heat-inactivated FBS, and 1 mM EDTA.
- Store at 4°C for 1 year.
RPMI-1640 complete culture medium
- RPMI-1640 (Wisent, 350-000-CL), supplemented with 10% heat-inactivated FBS, 1 mM GlutaMAX supplement, 1 mM HEPES, and 1% penicillin/streptomycin.
- Store at 4°C for 1 year.
COMMENTARY
Background Information
Evaluation of the clinical significance or insignificance of antibodies to RBC antigens when deciding on the selection of blood in anemic patients requiring RBC transfusion support has historically been based on the specificity of the antibodies using serological methods. In complicated transfusion cases, such as when patients have alloantibodies against high-prevalence antigens of uncertain clinical significance or multiple alloantibodies whereby it is difficult to find crossmatch compatible blood, the in vitro method most tested and proven to be predictive of in vivo antibody clinical significance is the monocyte monolayer assay (MMA; Hadley, 1998; Noums, Billingsley, & Moulds, 2015; Tong, Cen, & Branch, 2019; Zupanska, 1993). However, cell-mediated cytotoxicity—the direct lysis of RBCs—may be an important mechanism of antibody-dependent (ADCC) or antibody-independent RBC lysis (Flegel, 2015; Garratty, 2008). ADCC, although proposed to be a mechanism of RBC lysis (Barcellini, 2015), has not been as well studied as other methods; but due to cases of brisk intravascular lysis seen in some instances (Michelis et al., 2014), ADCC would be a method of RBC lysis that should be further evaluated (Garratty, 2008). We have described the MMA and ADCC assays used in our laboratory in great detail so that they can be used to predict the potential clinical significance of RBC auto- and alloantibodies.
Critical Parameters
For NK assays and MMA, buffy coat can be used.
Best results are obtained with a control antibody having a known concentration. Although any anti-D could be used, we use WinRho® SDF CDN (Saol Therapeutics, 1003092) Rh immune globulin as its concentration of anti-D is known. Any Rh immune globulin could be used for the positive control to ensure reproducibility.
Troubleshooting
See Tables 1 and 2 for common problems encountered when performing these protocols and suggested solutions.
| Problem | Possible cause | Solution |
|---|---|---|
| Low monocyte attachment to the chamber slide |
Blood stored at cold temperatures Blood not drawn within 48 hr of the assay |
Always make sure the ACD blood tubes are stored at room temperature Perform the assay within the 48 hr of blood collection |
| Lysis of cells on the chamber slide at the end of the assay | PBS used for washing is not adequate | Use cell culture grade PBS for washing |
| Low phagocytic index | Poor resuspension of RBCs | Frequently resuspend RBCs when adding them to the monocytes seeded on the chamber slide |
| Problem | Possible cause | Solution |
|---|---|---|
| High counts in the low control | Aspiration of RBC when transferring to the LumaPlate | Aspirate supernatant from the top of the plate. If possible, centrifuge the plate before collecting supernatant. |
| High variability among replicates | Poor resuspension of cells | Ensure frequent resuspension of NK cells and RBCs when seeding them on the 96-well plates |
Understanding Results
As a general rule, ≥5% killing is considered significant.
Time Considerations
The described assays are time consuming and it is important to have a dedicated person well trained to perform these assays. The MMA assay takes ∼4 to 5 hr to get to a stopping point, when slides are finalized for reading using phase-contrast microscopy or looking at hematological stained slides. The reading of the phagocytosis is subjective; thus, well trained individuals should be reading the slides and readings should be compared between individuals to insure similar results. The reading can take a half-day if looking at 300 monocytes per slide. The ADCC is also time consuming and purification of the NK cells can take a couple of hours as it involves isolating the PBMCs using Ficoll-Hypaque and then utilizing an NK purification kit. Opsonization of the RBCs takes about 1.5 hr and setting up the plates for reading in the scintillation counter can take a couple of hours. The plates also need to be left overnight prior to reading the radioactivity. Thus, the entire ADCC assay takes about 2 days, similar to the MMA assay.
Acknowledgments
We would like to acknowledge Dr. Christine Cserti-Gazdewich for her unwavering support for the assays described in this article. We also acknowledge the support of the University of Toronto, Quality in Utilization, Education, and Safety in Transfusion (QUEST) consortium in Toronto. The article is based on research that was partly funded by the Canadian Blood Services Postdoctoral Fellowship Award to K.F.B. This research was also supported by the Centre for Innovation of the Canadian Blood Services, using general resources provided in part by Health Canada, a department of the federal government of Canada, to D.R.B.
Author Contributions
Kayluz Frias Boligan : Data curation, methodology, supervision, writing original draft, writing review and editing; Gurleen Sandhu : Data curation, methodology, writing original draft, writing review and editing; Donald R. Branch : Conceptualization, methodology, writing original draft, writing review and editing.
Conflict of Interest
The authors declare no conflicts of interest.
Disclaimer
The views expressed herein do not necessarily represent the view of the Federal Government of Canada.
Open Research
Data Availability Statement
Data available on reasonable request from the authors.
Literature Cited
- Balola, A. H. A., Mayer, B., Bartolmas, T., & Salama, A. (2021). A fluorometric erythrophagocytosis assay using differentiated monocytic THP-1 cells to assess the clinical significance of antibodies to red blood cells. Vox Sanguinis , 116, 1106–1116. doi: 10.1111/vox.13105
- Barcellini, W. (2015). New insights in the pathogenesis of autoimmune hemolytic anemia. Transfusion Medicine and Hemotherapy , 42, 287–293. doi: 10.1159/000439002
- Branch, D. R., Gallagher, M. T., Mison, A. P., Sy Siok Hian, A. L., & Petz, L. D. (1984). In vitro determination of red cell alloantibody significance using an assay of monocyte-macrophage interaction with sensitized erythrocytes. British Journal of Haematology , 56, 19–29. doi: 10.1111/j.1365-2141.1984.tb01268.x
- Conley, C. L., Lippman, S. M., Ness, P. M., Petz, L. D., Branch, D. R., & Gallagher, M. T. (1982). Autoimmune hemolytic anemia with reticulobytopenia and erythroid marrow. The New England Journal of Medicine , 306, 281–286. doi: 10.1056/NEJM198202043060507
- Donohue, D. M., Motulsky, A. G., Giblett, E. R., Pirzio-Firoli, G., Viranuvatti, V., & Finch, C. A. (1955). The use of chromium as a red-cell tag. British Journal of Haematology , 1, 249–263. doi: 10.1111/j.1365-2141.1955.tb05508.x
- Downing, I., Templeton, J. G., Mitchell, R., & Fraser, R. H. (1990). A chemiluminescence assay for erythrophagocytosis. Journal of Bioluminescence and Chemiluminescence , 1990(5), 243–250. doi: 10.1002/bio.1170050406
- Flegel, W. A. (2015). Pathogenesis and mechanisms of antibody-mediated hemolysis. Transfusion , 55, S47–S58. doi: 10.1111/trf.13147
- Gallagher, M. T., Branch, D. R., Mison, A., & Petz, L. D. (1983). Evaluation of reticuloendothelial function in autoimmune hemolytic anemia using an in vitro assay of monocyte-macrophage interaction with erythrocytes. Experimental Hematology , 11, 82–89.
- Garratty, G., Dzik, W., Issitt, P. D., Lublin, D. M., Reid, M. E., & Zelinski, T. (2000). Terminology for blood group antigens and genes – historical origins and guidelines in the new millennium. Transfusion , 40, 477–489. doi: 10.1046/j.1537-2995.2000.40040477.x
- Garratty, G. (2008). The James Blundell Award Leture 2007: Do we really understand immune red cell destruction? Transfusion Medicine (Oxford, England) , 18, 321–334. doi: 10.1111/j.1365-3148.2008.00891.x
- Hadley, A. G. (1998). A comparison of in vitro tests for predicting the severity of hemolytic disease of the fetus and newborn. Vox Sanguinis , 74(Suppl. 2), 375–383. doi: 10.1111/j.1423-0410.1998.tb05445.x
- Hadley, A., Wilkes, A., Poole, J., Arndt, P., & Garratty, G. (1999). A chemiluminescence test for predicting the outcome of transfusing incompatible blood. Transfusion Medicine , 9, 337–342. doi: 10.1046/j.1365-3148.1999.00218.x
- Hunt, J. S., Beck, M. L., Hardman, J. T., Tegtmeier, G. E., & Bayer, W. L. (1980). Characterization of human erythrocyte alloantibodies by IgG subclass and monocyte interaction. American Journal of Clinical Pathology , 74, 259–264. doi: 10.1093/ajcp/74.3.259
- Landsteiner, K. (1901). Ueber agglutinationserscheinungen normalen menschlichen blutes [About agglutination phenomena in normal human blood]. Wiener klinische Wochenschrift , 14, 1132–1134.
- Lucas, G. F., Hadley, A. G., Nance, S. J., & Garratty, G. (1993). Predicting hemolytic disease of the newborn: A comparison of the monocyte monolayer assay and the chemiluminescence test. Transfusion , 33, 484–487. doi: 10.1046/j.1537-2995.1993.33693296810.x
- Michelis, F. V., Branch, D. R., Scovell, I., Bloch, E., Pendergrast, J., Lipton, J. H., & Cserti-Gazdewich, C. M. (2014). Acute hemolysis after intravenous immunoglobulin amid host factors of ABO-mismatched bone marrow transplantation, inflammation, and activated mononuclear phagocytes. Transfusion , 54, 681–690. doi: 10.1111/trf.12329
- Mollison, P. L. (1972). Survival in vivo as a test for red cell compatibility. Haematologia , 6, 139–145.
- Mollison, P. L. (1984). Methods of determining the posttrasfusion survival of stored red cells. Transfusion , 24, 93–96. doi: 10.1046/j.1537-2995.1984.24284173365.x
- Mock, D. M., Widness, J. A., Veng-Pedersen, P., Strauss, R. G., Cancelas, J. A., Cohen, R. M., … Franco, R. S. (2014). Measurement of post-transfusion red cell survival with the biotin label. Transfusion Medicine Reviews , 28, 114–125. doi: 10.1016/j.tmrv.2014.03.003
- Noumsi, G. T., Billingsley, K. L., & Moulds, J. M. (2015). Successful transfusion of antigen positive blood to alloimmunised patients using a monocyte monolayer assay. Transfusion Medicine , 25, 92–100. doi: 10.1111/tme.12189
- Schanfield, M. S., Schoeppner, S. L., & Stevens, J. O. (1980). New approaches to detecting clinically significant antibodies in the laboratory. Progress in Clinical and Biological Research , 43, 305–323.
- Silvergleid, A. J., Wells, R. F., Hafleigh, E. B., Korn, G., Kellner, J. J., & Grumet, F. C. (1978). Compatibility test using 51chromium-labeled red blood cells in crossmatch positive patients. Transfusion , 18, 8–14. doi: 10.1046/j.1537-2995.1978.18178118571.x
- Stevens, J. O., Schanfield, M. S., & Braley, J. F. (1976). Detection of clinically significant IgG antibodies by an in vitro human peritoneal macrophage phagocytosis assay. Transfusion , 16, 523.
- Tong, T. N., Burke-Murphy, E., Sakac, D., Pendergrast, J., Cserti-Gazdewich, C., Laroche, V., & Branch, D. R. (2016). Optimal conditions for the performance of a monocyte monolayer assay. Transfusion , 56, 2680–2690. doi: 10.1111/trf.13766
- Tong, T. K., & Branch, D. R. (2017). Use of a monocyte monolayer assay to evaluate Fcγ receptor-mediated phagocytosis. Journal of Visualized Experiments , 119, e55039. doi: 10.3791/55039
- Tong, T. N., Cen, S., & Branch, D. R. (2019). The monocyte monolayer assay: Past, present and future. Transfusion Medicine Reviews , 33, 24–28. doi: 10.1016/j.tmrv.2018.07.004
- Zupanska, B. (1985). Assays to predict the clinical significance of blood group antibodies. Current Opinion in Hematology , 5, 412–416. doi: 10.1097/00062752-199811000-00010
- Zupanska, B. (1993). Clinical application of functional assays for assessing the red cell antibody activity. Transfusion Science , 14, 371–381. doi: 10.1016/S0955-3886(05)80010-4
Citing Literature
Number of times cited according to CrossRef: 1
- Zhuolin Li, Yufeng Wang, Mao Sun, Tangdong Chen, Yuanming Wu, Kun Chen, Application of droplet digital PCR combined with TaqMan real‐time PCR in Dombrock blood group genotyping in Northwest China, Vox Sanguinis, 10.1111/vox.13428, 118 , 6, (488-496), (2023).

