Methods for Imaging Inflammation and Transendothelial Migration in Vivo and ex Vivo
Vivienne Fang, Vivienne Fang, Maureen E. Haynes, Maureen E. Haynes, Vanessa Hayashi, Vanessa Hayashi, Erika Arias, Erika Arias, Jeremy A. Lavine, Jeremy A. Lavine, David P. Sullivan, David P. Sullivan, William A. Muller, William A. Muller
Abstract
Inflammation is the body's response to injury and harmful stimuli and contributes to a range of infectious and noninfectious diseases. Inflammation occurs through a series of well-defined leukocyte-endothelial cell interactions, including rolling, activation, adhesion, transmigration, and subsequent migration through the extracellular matrix. Being able to visualize the stages of inflammation is important for a better understanding of its role in diseases processes. Detailed in this article are protocols for imaging immune cell infiltration and transendothelial migration in vascular tissue beds, including those in the mouse ear, cremaster muscle, brain, lung, and retina. Also described are protocols for inducing inflammation and quantifying leukocytes with FIJI imaging software. © 2023 The Authors. Current Protocols published by Wiley Periodicals LLC.
Basic Protocol 1 : Induction of croton oil dermatitis
Alternate Protocol 1 : Induction of croton oil dermatitis using genetically fluorescent mice
Basic Protocol 2 : Intravital microscopy of the mouse cremaster muscle
Support Protocol : Making a silicone stage
Basic Protocol 3 : Wide-field microscopy of the mouse brain
Basic Protocol 4 : Imaging the lungs (ex vivo)
Alternate Protocol 2 : Inflating the lungs without tracheostomy
Basic Protocol 5 : Inducing, imaging, and quantifying infiltration of leukocytes in mouse retina
INTRODUCTION
Inflammation is critical to responding to and resolving insults, as infiltration of immune cells known as leukocytes contributes to the expulsion and/or destruction of damaging mediators. However, while it is critical to the body's response to infection and injury, inflammation is a double-edged sword, also underlying the pathology of most diseases. As a result, it is of the utmost importance to understand and characterize inflammatory responses. Throughout history, the inflammatory response has been characterized through a number of methods, with imaging being of great importance due to its ability to reveal inflammatory processes occurring in various tissues under physiologically normal conditions and in disease. Diseases manifest their pathology in different organs, so it is useful to study inflammation in the appropriate vascular beds. To investigate the response of leukocytes to various insults in different vascular beds and to characterize the transendothelial migration step of the inflammatory cascade, we have established and calibrated several methods of inducing and imaging inflammation. Here we describe methods by which to image leukocyte-endothelial cell interactions in the murine skin, cremaster muscle, brain, lung, and eye along with techniques for image analysis and post-processing.
NOTE : All protocols involving animals must be reviewed and approved by the appropriate Animal Care and Use Committee and must follow regulations for the care and use of laboratory animals.
NOTE : These protocols require basic knowledge of mouse handling. Please see appropriate guidelines before attempting this procedure.
Basic Protocol 1: INDUCTION OF CROTON OIL DERMATITIS
This protocol is used to study acute inflammation in the skin of mice. The tissue is later stained with appropriate antibodies and imaged to determine the percentage of transendothelial migration (TEM) events. We developed this protocol to study TEM in vivo.
This protocol is best suited for experiments in which the purpose is to test drugs or treatments that are expected to block inflammation. Mice that have been exposed to such drugs or treatments are challenged with a dose of the inflammatory substance croton oil administered to the ears. Under control conditions where mice have not received any treatment, the ears become acutely inflamed; however, if the tested drug has anti-inflammatory properties, it is expected that the ears will have decreased inflammation despite croton oil application.
CAUTION : Croton oil is a stimulant of inflammation and at high concentrations can cause severe irritation to exposed skin. The active ingredient is phorbol myristate acetate (PMA), an activator of protein kinase C and a known tumor promoter. It is therefore important to wear gloves when handling the oil. We recommend that you double glove and keep the bottle of croton oil in a secondary containment of a 50-ml conical tube to prevent potential spillage.
NOTE : This protocol starts with croton oil application. Any experimental conditions to be tested must be performed on the mice prior to the steps listed in this protocol. There is no definite number of mice that should be processed per batch, but 6 mice or fewer would be reasonable for the workload.
Materials
-
Carrier solution (see recipe)
-
Croton oil mixture (see recipe)
-
Mice (we have tested FVB, C57BL/6, and 129S backgrounds)
-
NairTM hair-removal lotion
-
1× DPBS without calcium and magnesium (Corning, product no. 21-031-CV)
-
Fixing solution (see recipe)
-
Permeabilization buffer (see recipe)
-
Anti-PECAM 2H8 (Millipore Sigma, cat. no. MAB1398Z)
-
Anti-S100A9 (Abcam, cat. no. ab105472)
-
Bovine serum albumin (BSA) solution (see recipe)
-
Normal goat serum (NGS; Jackson, cat. no. 005-000-121)
-
Fluorophore-conjugated goat anti-Armenian hamster secondary antibody
-
Fluorophore-conjugated goat anti-rat secondary antibody
-
Normal mouse serum (NMS; Jackson, cat. no. 015-000-120)
-
FluorSave mounting reagent (Calbiochem, cat. no. 345789-20L)
-
1.5-ml microcentrifuge tubes
-
Isoflurane anesthetic apparatus
-
Heating pad
-
CO2 tank with mouse euthanasia apparatus
-
Cotton swabs
-
Dissection scissors and tweezers
-
15-ml conical tubes
-
12-well flat bottom plate
-
Plate shaker
-
100-mm culture dish
-
Dissection microscope
-
Fine forceps
-
24-well flat-bottom plate
-
Aluminum foil
-
Glass microscope slides
-
22 × 22-mm no. 1.5 coverslips
-
Slide holder
-
Spinning-disk confocal microscope
Day 1
1.Prepare the carrier and croton oil mixtures in two 1.5-ml microcentrifuge tubes.
2.Set the air flow rate to 2 liters/min and anesthetize the mice with isoflurane. Wait for the mouse's breathing to slow down enough to indicate a sufficient plane of anesthesia.
3.Pipet 10 μl carrier mixture onto the back of the left ear and 10 μl of the same carrier mixture onto the front side.
4.Pipet 10 μl croton oil onto the front and back of the right ear. Repeat for all mice.
5.Leave mice in cage with access to a heating pad for 5 hr.
6.Sacrifice mice by CO2 asphyxiation and cervical dislocation.
7.Rub Nair on both sides of the ears using a cotton swab, leave for a minute, and wipe off with disposable wipes to remove hair. Cut the ears off the mouse. Pour 12 ml of 1× DPBS into each of two 15-ml conical tubes, and place an ear into each tube, keeping track of which ear received which treatment. Shake the tubes to wash the Nair off the ears.
8.Place the ears into a labeled 12-well flat-bottom plate. Fill each of the wells with 1 ml fresh fixing solution. Leave the plates on a shaker for 0.5-1 hr at room temperature.
9.Add 1× DPBS to a 100-mm culture dish. Temporarily transfer one ear at a time to this 100-mm dish, place under dissection microscope, and zoom in on the cut edge of the ear. Separate the front and back sides of the ear by gripping each side at the cut edge with fine forceps and then slowly pulling them apart. Keep track of which side is the epithelial side and which is the side with interstitial tissue/endothelium.
10.Making sure the interstitial tissue/endothelium side of the ear is facing down, place each ear half individually in a 12-well plate with fresh fixing solution. Leave the plate on a shaker overnight at 4°C.
Day 2
11.Wash ears with 1× DPBS three times, 10 min each, at room temperature.
12.Place each half-ear in the same orientation as before into a well of a 24-well plate with 500 ml permeabilization buffer in each well. Leave to permeabilize on a shaker overnight at 4°C.
Day 3
13.Wash ears with 1× DPBS three times, 10 min each, at room temperature.
14.Dilute anti-PECAM 2H8 and anti-S100A9 in a solution of 2.5% bovine serum albumin (BSA) + 5% normal goat serum (NGS). Add 250 ml antibody mixture into each well of a 24-well plate. Place each half-ear in the same orientation as before in the wells. Leave the plate on a shaker overnight at 4°C.
Day 4
15.Wash ears with 1× DPBS three times, 10 min each, at room temperature.
16.Dilute goat anti-Armenian hamster secondary and goat anti-rat secondary in a solution of 2.5% BSA + 5% NGS + 5% normal mouse serum. Leave this mixture for 15 min on ice to preabsorb the secondary antibodies. Add 250 ml of the mixture into each well of a 24-well plate. Place each half-ear in the same orientation as before in the wells. Cover the plate with aluminum foil to protect the fluorophores from light. Leave the plate on a shaker for 4 hr at room temperature.
17.Wash ears with 1× DPBS three times, 10 min each, at room temperature.
18.Mount specimens on glass microscope slides. Place two drops of FluorSave mounting reagent onto the slide, and place the half-ear onto the slide with the epithelial side facing down. Add as many drops of FluorSave as needed and place coverslip on top. Allow slides to dry in a dark place for at least 1 hr at room temperature.
19.Store slides in a slide holder to maintain them protected from light and store at 4°C until imaging.
Imaging
20.Turn on the spinning-disk confocal microscope. Place the slide on the mechanical stage and add a drop of immersion oil on top of the coverslips. Adjust the position of the slide so that the objective lens (40× oil) is directly above the ear that received carrier mixture. Lower the objective until it barely touches the drop of oil.
21.Focus the image until the blood vessels are visible in the goat anti-Armenian hamster secondary channel.
22.Switch to the goat anti-rat secondary channel, and scan to verify that there are few to no leukocytes (0-2 per field).
23.Adjust the position of the slide so that the objective lens is directly above the ear that received croton oil mixture. Lower the objective and focus the image as in step 20.
24.Scan the sample for blood vessels whose cell-to-cell junctions have a cobblestone pattern, and that have a diameter between 15 μm and 50 μm, ideally between 30 μm and 50 μm.
25.Switch the laser to the goat anti-rat secondary channel. Take a z stack if there are 10-30 leukocytes in the field.
26.Repeat steps 24 and 25 for 10 or more fields in each ear that received the croton oil mixture.
27.Repeat entire imaging process for all the experimental conditions.
Counting of leukocytes
28.On each picture, look at the z plane and score how many leukocytes are inside and outside the vessels (area within 50 mm from a venule). If there are leukocytes that are neither fully inside nor outside, count them separately.
29.Calculate the percent TEM per image: Divide the number of leukocytes that were outside the vessels by the total amount of leukocytes in the given image. Repeat this for all images.
30.Calculate the average percent TEM: Take the average of the values from step 29.Compare the average percent TEM between/among experimental conditions and perform statistical tests as needed.
Alternate Protocol 1: INDUCTION OF CROTON OIL DERMATITIS USING GENETICALLY FLUORESCENT MICE
If the mice used in the experiment have fluorescent neutrophils (e.g., LysM-eGFP, Catchup), it is possible to skip the steps of the protocol that involve staining leukocytes with antibodies after fixation.
Additional Materials (also see Basic Protocol 1)
- Mice with fluorescent neutrophils (e.g., LysM-eGFP, Catchup)
- Non-blocking CD31 (PECAM) monoclonal antibody (390) conjugated with fluorophore of choice
Day 1
Perform Basic Protocol 1, steps 1-10, but between steps 5 and 6 in the original protocol, perform the following steps, if applicable.
1.Anesthetize the mice as in step 2.
2.Take one mouse and inject 100 μl fluorescent 390 antibody retro-orbitally. Wait 5 min.
3.Skip the steps for Day 2 and 3 (steps 11-14), and continue Basic Protocol 1 from Day 4 (step 15) onward.
COMMENTARY: BASIC AND ALTERNATE PROTOCOLS 1
Background Information
Transendothelial migration (TEM) is a step in the inflammatory cascade that involves leukocytes crossing the walls of blood vessels. It involves homophilic binding of neutrophil PECAM to PECAM on the endothelial junctions, and sequential binding of CD99L2 and CD99 (Rutledge et al., 2022). Studying TEM is a promising endeavor because it opens up more possibilities for anti-inflammatory drug development, given that many of the molecules involved are specifically expressed in endothelial junctions.
The current protocol for croton oil dermatitis is one that aids in the study of TEM. The advantage of this protocol is that it allows in vivo experimentation without requiring complex equipment or a high level of skill. The disadvantage is that it is an end-point assay, unlike protocols involving intravital fluorescence microscopy (IVM), which can record the movement of leukocytes in real time. Even so, it is a useful assay to complement the findings from IVM in a different tissue bed.
The current protocol can be used on any mouse; thus, the researcher is at liberty to study the effect of pharmacological or genetic manipulations on TEM. Croton oil is an irritant that induces acute inflammation at low concentration when applied to the skin. This inflammation will be visualized at the end of the protocol, and the effect on TEM quantified by counting how many leukocytes are inside and outside the blood vessels. If TEM is blocked, it is expected that there will be more leukocytes inside the vessels, unable to transmigrate, than in wild-type mice or other mice used as control.
Critical Parameters
Though the protocol has been used in C57BL/6 and FVB mice strains with success, before applying it to another strain, it would be best to conduct a croton oil dose-response experiment on mice of that strain, as too little croton oil will recruit too few leukocytes and too much will severely hurt the mice. In response to severe irritation, the mice will scratch their ears, resulting in excoriation and local trauma that can hinder interpretation of the results. The best croton oil concentration will be one at which, when the ears are imaged with a 40× objective lens, there are several fields of view containing 10-20 leukocytes.
If the fluorophore-conjugated antibodies used are sensitive to light, it is advisable to shield the ears from light using aluminum foil from the antibody incubation step onward.
Troubleshooting
Table 1 lists problems that may arise with the croton oil dermatitis procedures, along with their possible causes and solutions.
| Problem | Possible cause | Solution |
|---|---|---|
| The front and back sides of the ear tear when trying to separate them | Fixation time was insufficient | Leave in fixing solution for 10 min longer. |
| Poor signal | Ears were over-fixed | Fix for 1-2 hr instead of overnight. |
| The ear was mounted with the epithelial side facing the lens | Remove the ear from the slide, wash with 1× DPBS, and mount on a new slide with the epithelial side facing down. | |
| Microscope settings are not ideal | Adjust sensitivity and/or auto-contrast settings. | |
| Too little or too much inflammation (<10 or >30 leukocytes per 170 × 170-μm field) | Croton oil concentration is too low/too high | Increase/decrease croton oil concentration to 1% or 2% (v/v). |
Understanding Results
If there is no block in transmigration, it is expected that most neutrophils will be located outside blood vessels (Fig. 1A); if there is a block in transmigration, it is expected that most neutrophils will be inside the vessels (Fig. 1B). One will likely encounter imaging fields that have leukocytes inside and outside the vessels, depending on how responsive the mouse strain is to inflammation (Fig. 1C).
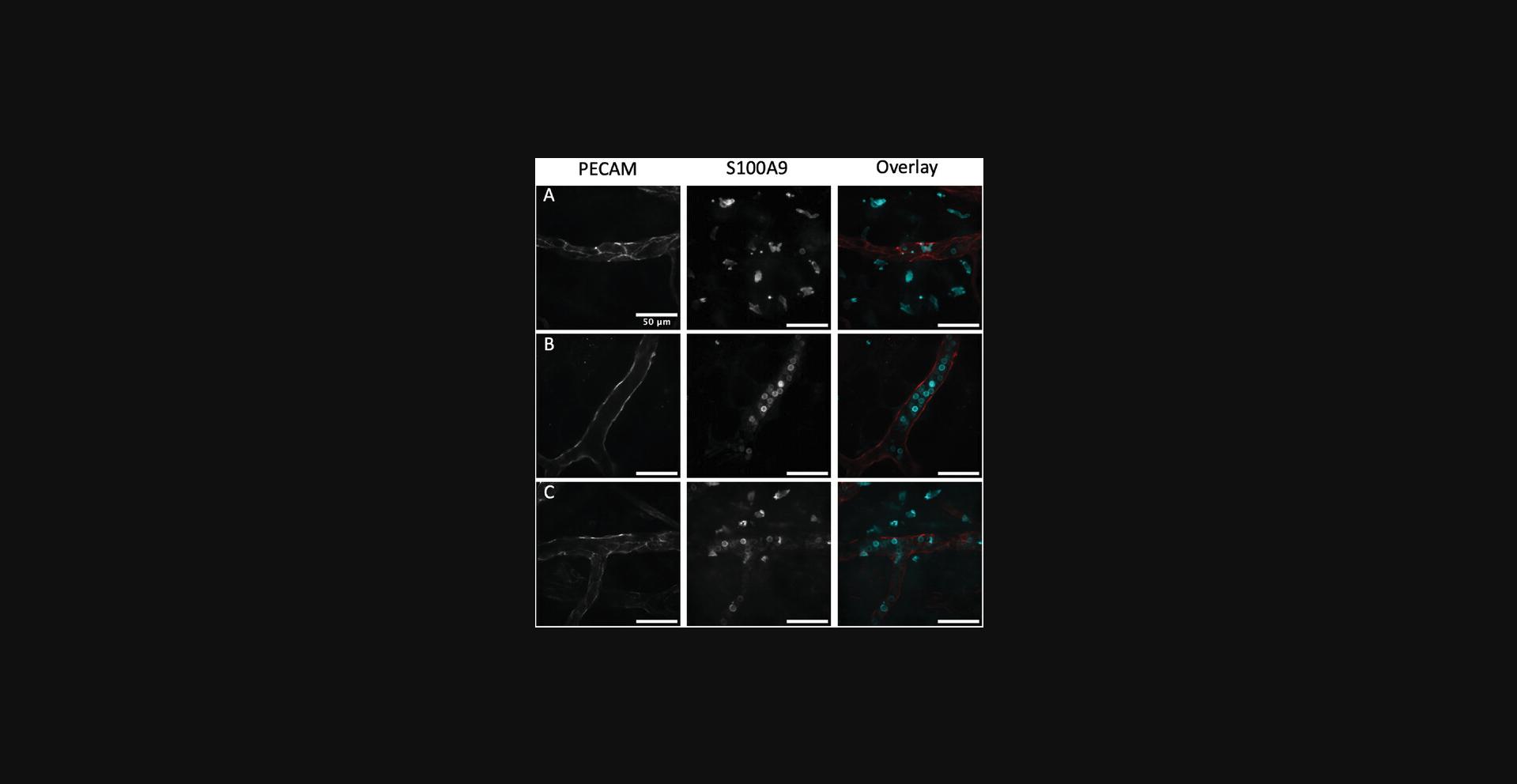
Time Considerations
This protocol takes 4 days up to the point when samples are loaded on microscope slides. The imaging and counting processes take a variable number of hours, depending on how many samples are being imaged and what microscope settings are used.
Basic Protocol 2: INTRAVITAL MICROSCOPY OF THE MOUSE CREMASTER MUSCLE
The cremaster muscle is an extremely thin, muscular sheath that surrounds the testes in male mice. This protocol describes methods to surgically expose, mount, and image the vasculature of the cremaster muscle to study the infiltration of inflammatory cells in vivo in real time. IVM of the cremaster muscle is used to visualize the movement of inflammatory cells like leukocytes. This protocol is useful for visualizing the response to localized inflammatory stimuli. IL-1β is routinely used as a generic stimulus, particularly when studying general leukocyte adhesion and diapedesis. Other inflammatory stimuli could be used to interrogate specific leukocyte subsets and/or other components of the inflammatory cascade.
Materials
-
1× DPBS without calcium and magnesium (Corning, product no. 21-031-CV)
-
Recombinant mouse interleukin-1β (IL-1β; R&D Systems, 401-ML-005/CF)
-
Mice with genetically encoded fluorescent leukocytes, wild-type or knockouts, with or without prior treatment of interest (see paragraph on Fluorescent Mice in the Critical Parameters section below)
-
Ketamine hydrochloride (Covetrus, NDC no. 11695-0703-1)
-
Xylazine (Akorn Inc., NDC no. 59399-110-20)
-
Anti-PECAM antibody conjugated to a fluorophore (EMD Millipore, CBL1337 clone 390, rat anti-mouse, non-blocking)
-
NairTM hair-removal lotion
-
Perfusion buffer (see recipe)
-
Plexiglas platform with glass window
-
Heating pad
-
Sutures
-
Threaded needle
-
Modeling clay
-
Silicone stage with quartz pedestal
-
Dissecting microscope (Leica or equivalent)
-
Suture needles bent into an “L” shape
-
Fine forceps and scissors
-
Dissecting pins
-
Buffer warmer
-
Cotton tip applicators
-
Syringe pump
-
Confocal microscope (Nikon or equivalent) with 20× water-immersion objective
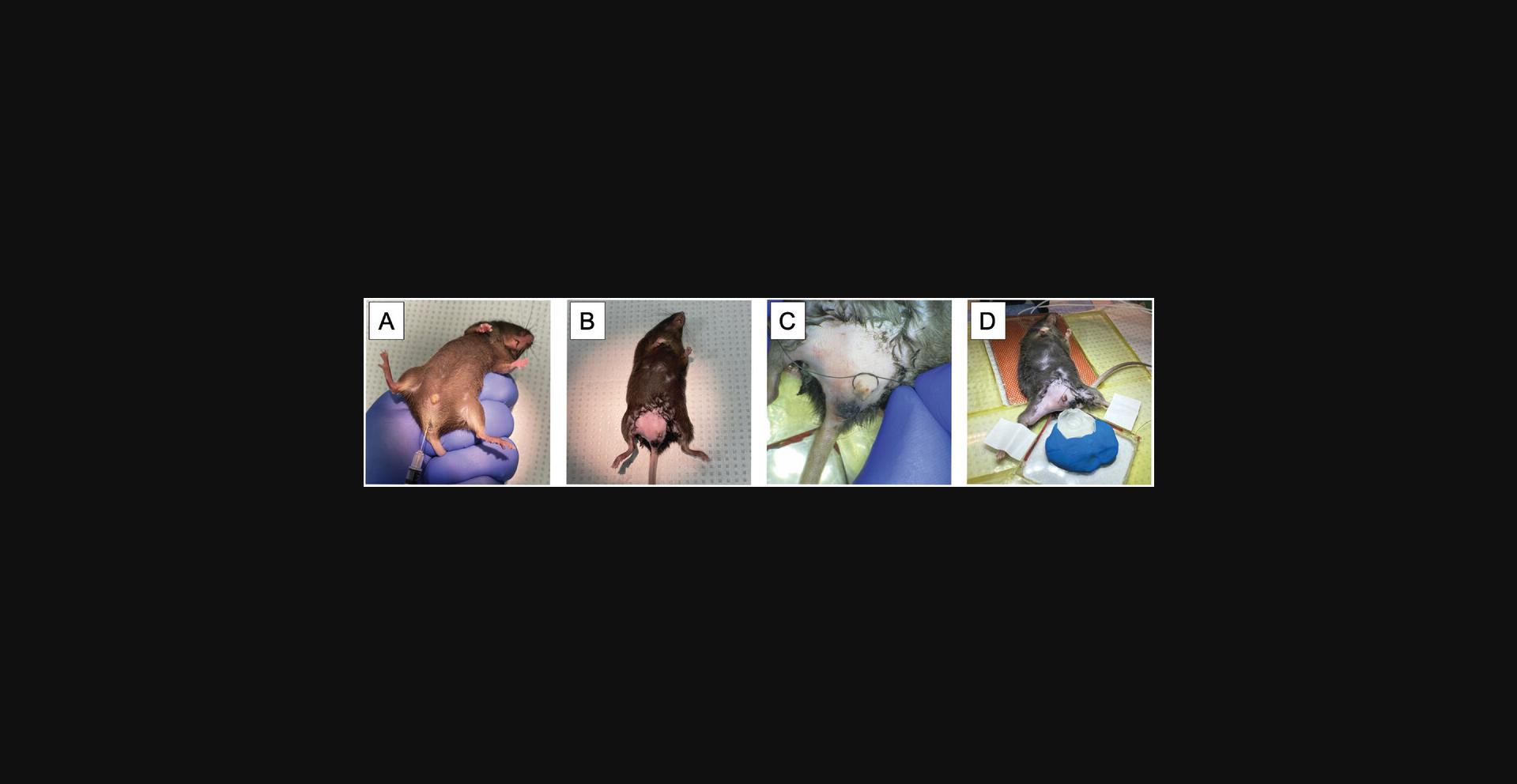
1.Inject 150 µl DPBS containing 50 ng mouse IL-1β intrascrotally (Fig. 2A).
2.Wait 3.5 hr.
3.Anesthetize the mouse by intraperitoneal injection of ketamine and xylazine (100 mg/kg and 10 mg/kg body weight, respectively).
4.Inject 100 mg of fluorescently conjugated non-blocking anti-PECAM antibody (clone 390) and/or any additional labeling antibodies of interest intravenously or retro-orbitally.
5.Remove hair on the scrotum and surrounding area with depilatory gel (Nair®; Fig. 2B). The total area of exposed skin should be 1 cm × 1 cm.
6.Secure the mouse in a supine position on a Plexiglas platform containing an internal heating pad set to ∼37°C. Use suture to securely tie the urethra to prevent urination during the procedure. Tape each lower leg such that it straddles the edge of a glass window in the platform (Fig. 2C and D).
7.Position the silicone stage (see Support Protocol) in modeling clay on the Plexiglas platform (Fig. 2D).
8.Grab the outer skin of the scrotum and secure it with a needle to the silicone mount. Make a ∼1 cm incision along the left ventral side of the scrotum (Fig. 3A and B).
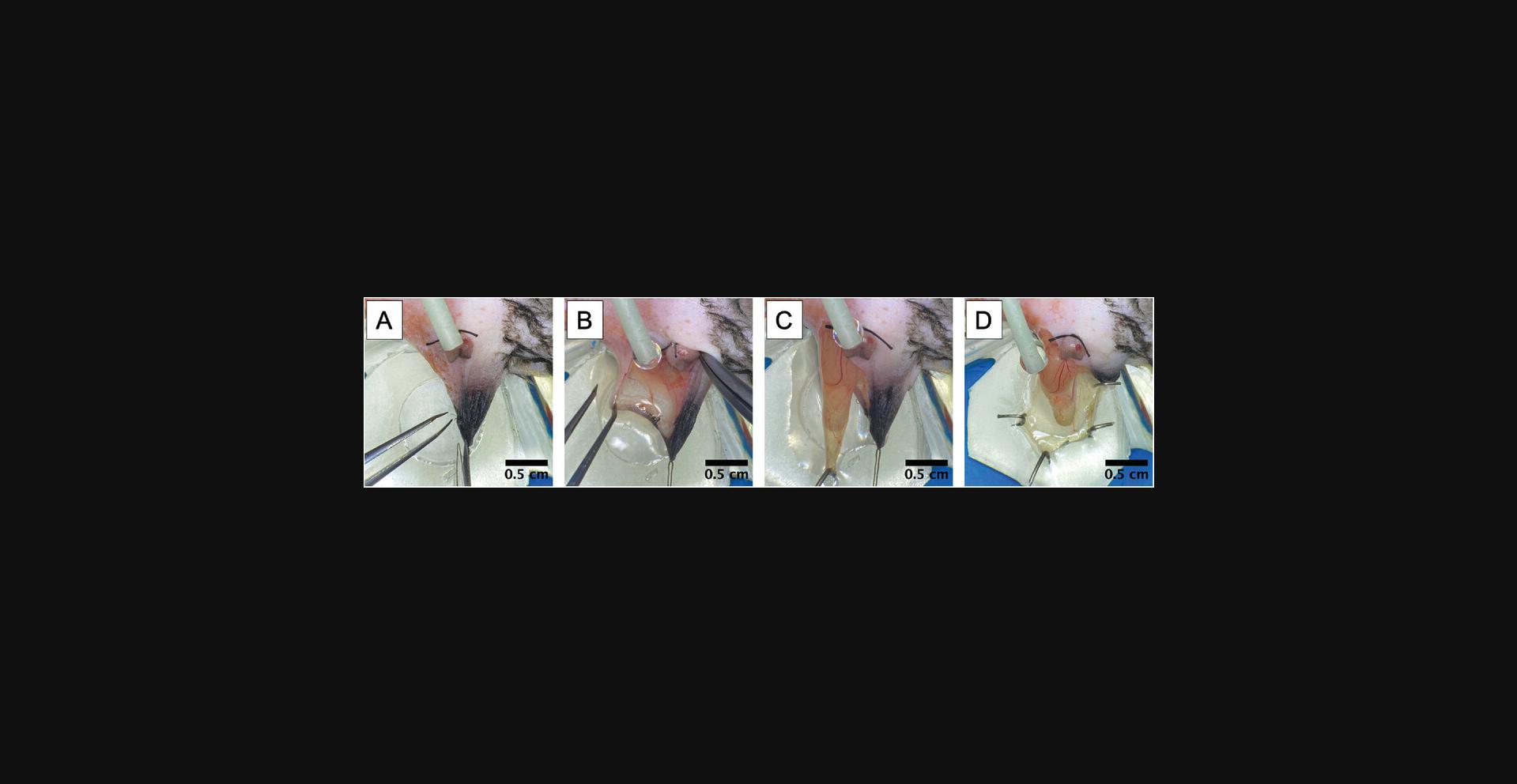
9.Grasp the base of the cremaster muscle and gently pull the tissue and testicle it encompasses out of the body cavity (Fig. 3C).
10.Secure the tissue across the quartz pedestal using bent suture needle(s) (Fig. 3D).
11.Begin perfusion with warmed perfusion buffer. Allow buffer to drain directly onto the objective by bringing the perfusion tube directly next to it. The buffer can then drain down onto the tissue in a way that does not induce motion artifacts throughout the rest of the protocol. Using a syringe pump helps provide for a smooth, steady flow and is preferable to a peristaltic pump.
12.Carefully remove connective tissue from the cremaster muscle using forceps by grabbing small pieces of the overlying sheath and pull them down and away from the body.
13.Open the muscle using microsurgery scissors by making a single distal-to-proximal incision that avoids severing as many large vessels as possible.
14.Cut the small tissue connection and vessel running from the epididymis (and associated testicle) to the cremaster muscle using microsurgery scissors and replace them in the inguinal canal (Fig. 4A).
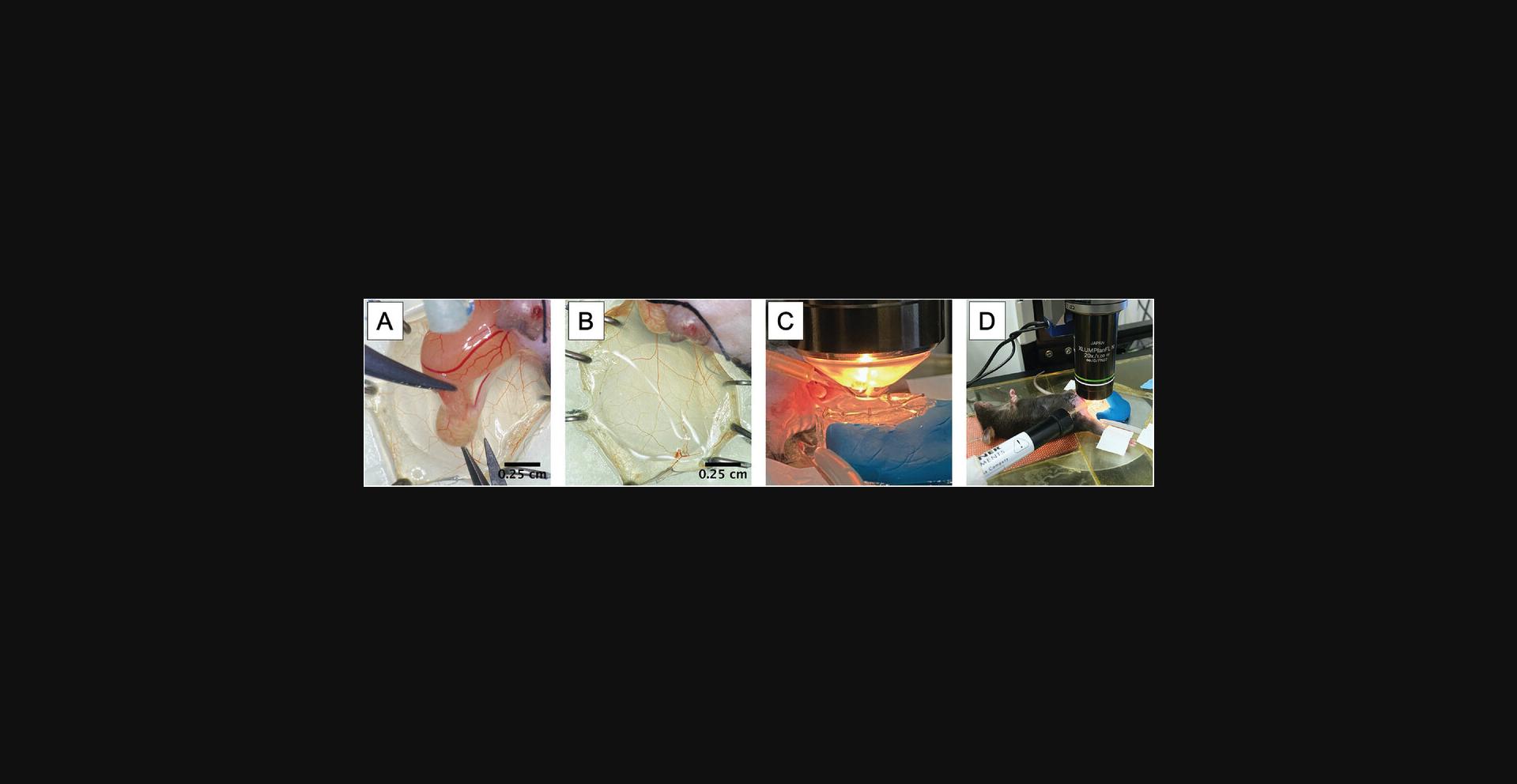
15.Spread the muscle across the silicone stage centered over the quartz pedestal and pin it along its periphery (Fig. 4B).
16.Transfer the platform and secured mouse to the microscope for visualization (Fig. 4C and D).
17.Identify target fields containing post-capillary venules that have normal flow using bright-field illumination.
18.Record desired image sequences.
Image acquisition
19.Collect the following image series as needed for the experiment:
-
In bright-field, acquire one plane for 1 min at maximum speed to confirm flow.
-
In fluorescence, acquire azstack through the entire field with a 1-µm step size to document the entire field.
Unlike for z stacks of fixed objects, where the entire stack is first acquired in one color, then a second stack in a second color, etc., to best visualize the temporal and spatial relationships of cells to vessels, it is best to take an image in each color at each z plane. This results in slower acquisition of the entire stack, but truer anatomic relationships.
20.Turn the contrast settings up while shortening the acquisition time to the shortest reasonable time (60-80 ms works well). Find a viewing plane with the vessel both centered on your screen and near the top of the viewing plane where adherent leukocytes can be seen. Record one plane in the leukocyte channel at maximum speed for 1 min to document rolling flux and adhesion.
21.Restore the settings back to those reasonable for long-term acquisition. Ideal settings will minimize both intensity and exposure time, which will help reduce photobleaching while allowing a maximum acquisition rate (frames per minute).
22.Define the upper and lower boundaries for the vessel that is to be imaged longitudinally.
23.Check the mouse to determine if it needs a booster dose of anesthesia and administer if necessary.
24.Check perfusion buffer to make sure there is enough to complete the recording (typically 15 min).
25.Begin the longitudinal acquisition. Monitor the images in real time in order to adjust the focal plane as needed so that the desired vessel is always collected. Example acquisition of transendothelial migration in the cremaster is shown in Figure 5.
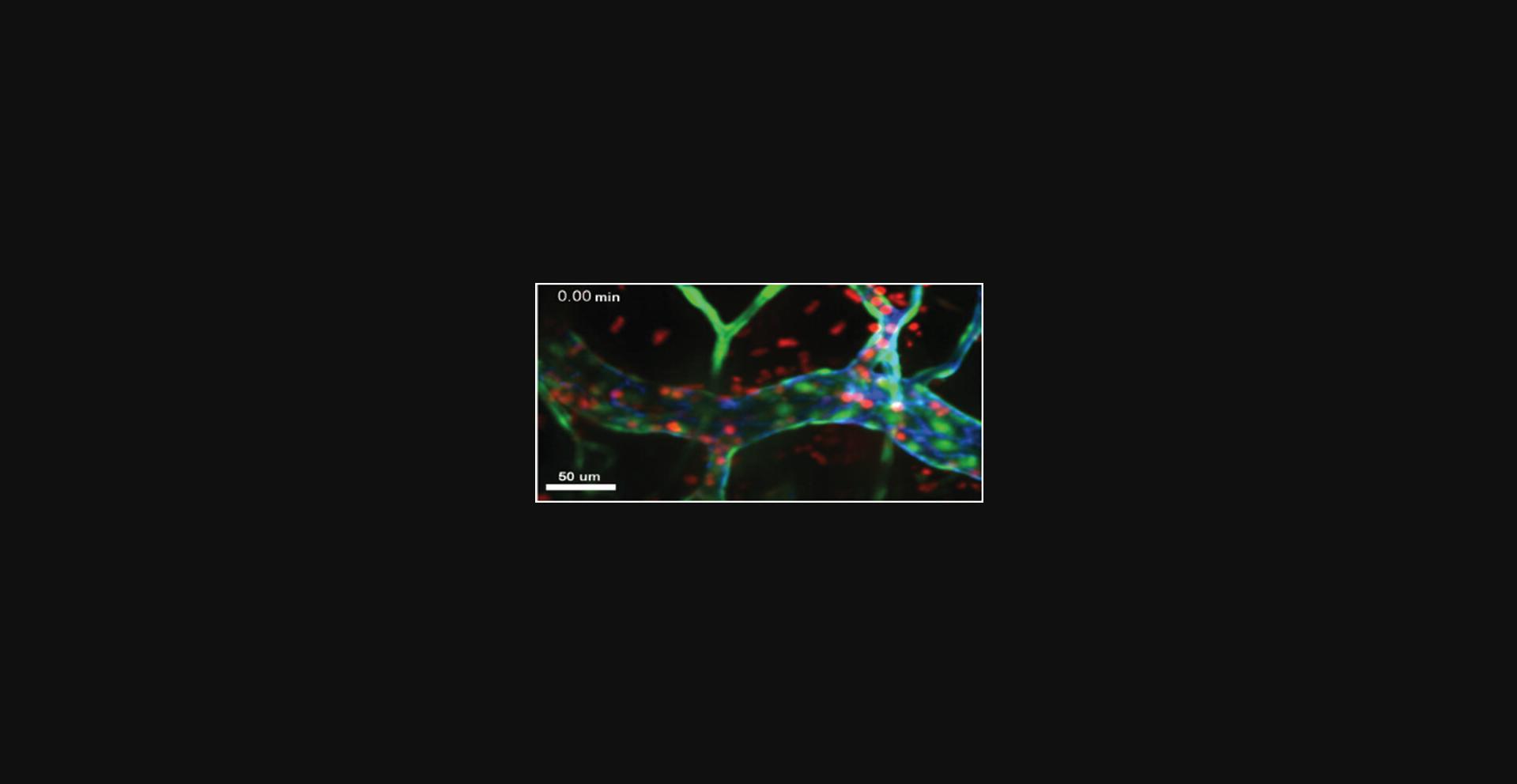
26.In fluorescence, acquire a z stack through the entire field with a 1-µm step size to document the entire field at the end of the imaging.
27.After recording, examine the field again using bright-field illumination and acquire one plane for 1 min at maximum speed to check for any undesired changes in flow rate.
28.If desired, find another field and repeat these steps.
29.Sacrifice the mouse after the last images have been acquired.
Image analysis
30.Open the image in the desired image analysis program.
31.Confirm that the scale and time has been transferred or retained faithfully.
32.Adherent cells may be counted by examining individual leukocytes along the vessel and assessing their movement over the 60-s recording. Adherent cells are defined as remaining attached and moving less than one cell diameter over 30 s. Vessel walls may be confirmed and measured with the thick stack acquired above. The vessel length is measured along the middle and average diameter is averaged from multiple measurements along the length. Surface area (SA) can by calculated by modeling the vessel as half a cylinder with the equation SA = π rl + π r 2, where l is the length and r is the average radius.
33.Determine the rolling flux by identifying a position in the vessel where rolling and free-flowing leukocytes can be easily seen both in and out of focus, and count the number of leukocytes that roll past that point over the recording. Data are expressed either as number of rollers (past this point) per second or per minute.
34.Calculate rolling flux fraction by counting the number of leukocytes that pass the same point in the blood stream. Leukocytes in the blood stream will appear as streaks and should be able to be tracked in sequential frames.
35.Quantify transendothelial migration from the long recording by tracking individual cells as the move over time. Typically, cells will adhere and/or roll into the view and spend a few minutes crawling around, usually along endothelial junctions (identified by staining with the anti-PECAM 390 antibody). When they undergo TEM, they will stop moving and “ball up.” Shortly thereafter, they will be observed dramatically flattened and on the outside of the vessel.
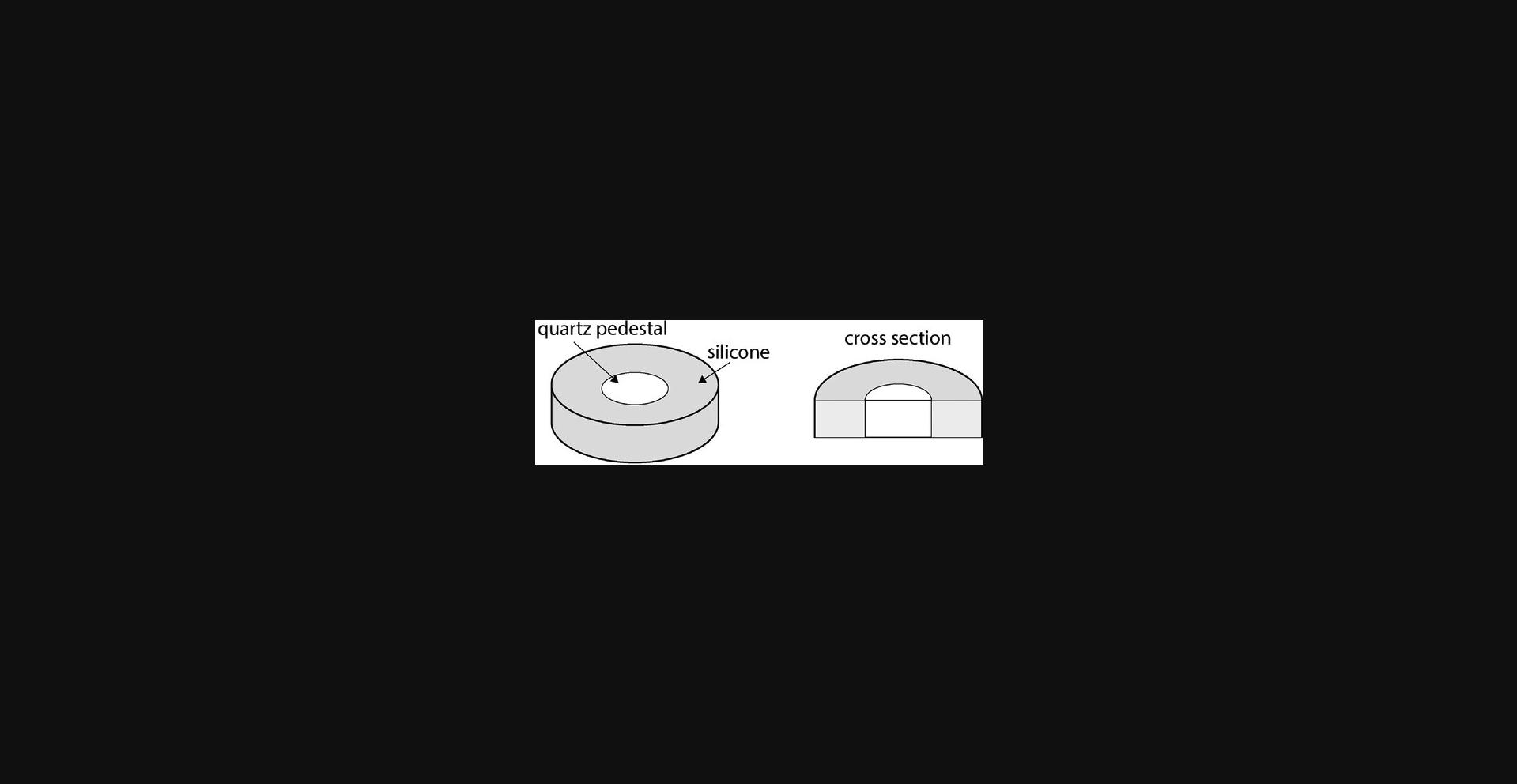
Support Protocol: MAKING A SILICONE STAGE
The cremaster muscle is uniquely useful for imaging inflammation and extravasation from systemic vasculature due to its thin, vascularized structure. To effectively image this membranous muscle with confocal and/or bright-field microscopy, the muscle must be exteriorized onto an imaging stage that is conducive to microscopy and capable of keeping the structure exposed and still. The use of a quartz viewing stage surrounded by silicone allows the tacking of the cremaster muscle and exposure of vasculature of interest. Here we describe the use of quartz, which is a stable, optically clear, and predictable viewing platform critical for the use of microscopy.
Materials
- SYLGARD 184 Silicone Elastomer Kit (Dow Corning, no. 4019862)
- 1 × 0.25-inch clear fused quartz ground and polished disc (Technical Glass Products)
- 6-well tissue culture dish
- Vacuum chamber
- Dissecting light
1.Mix SYLGARD silicone reagents A and B according to manufacturer's directions in 1:10 proportions.
2.Place the quartz disc with a flat side facing down and up in one well of a 6-well tissue culture dish.
3.Pour the silicone reagents into the well, surrounding the quartz, until the meniscus of the silicone is flush with the top surface of the quartz (Fig. 6).
4.Place the tissue culture dish in a vacuum chamber.
5.Leave under direct light overnight at room temperature.
6.Remove the silicone-embedded quartz stage from the dish.
7.Silicone-embedded quartz stages last for approximately 20 surgeries before the silicone loses its structural integrity. Rinsing with water is adequate to clean the stage. The quartz can be removed from the silicone using a blade and reused.
COMMENTARY: BASIC PROTOCOL 2 AND SUPPORT PROTOCOL
Background Information
Intravital microscopy of the cremaster muscle has been integral in the discovery and further study of the inflammatory cascade, the process by which inflammatory cells migrate to sites of inflammation. It is a site in which the mechanisms of leukocytes migration in the systemic vasculature have been interrogated and is an ideal place to study inflammation. This protocol is advantageous in that the cremaster muscle is easily accessible with minor surgery and is transparent enough to be imaged with ease. The results of this protocol can be used to interrogate the response of leukocytes to inflammatory stimuli, blockades and treatments, and various interventions relevant to inflammation in the systemic circulation.
Critical Parameters
Ideal fields
One of the best ways to ensure consistent results and striking movies is to pick consistent fields to observe and record. The ideal venule will:
- 1.be largely straight and not tortuous for a length of at least 150 µm;
- 2.be 30-50 µm in diameter;
- 3.be near the surface/relatively close to the objective;
- 4.have consistent flow, without any pulsating or occasional pausing; and
- 5.show signs of inflammation, including adherent and rolling leukocytes and some leukocytes already out in tissue.
Although it is common to have to compromise on some these details, it is important to understand how they may affect the results. It is difficult to measure velocities in tortuous vessels, and they often have abnormally high adhesion. Narrower vessels and vessels with inconsistent flow usually have slower flow rates and often dramatically increased adhesion and slower rolling. In addition, vessels with intermittent flow will often cease flowing by the end of the recording, making the entire effort fruitless. Signs of active inflammation are often a good indicator of ongoing recruitment and TEM. However, one must be careful not to bias the results by only choosing “hot” areas. In almost every mouse, there are regions of hyper- and hypoinflammation, possibly due to damage during tissue preparation or exposure to the stimulus. Where rolling and adhesion are to be examined, it is more important to compare vessels of similar size and tortuosity. Where TEM is to be examined, extra attention should be given to finding venules that have a modest amount of active inflammation, to avoid hyperactive areas (obviously little TEM can occur where there is no rolling or adhesion).
Drifting field and tenting
It is somewhat common for the field to drift (move in the x and y dimensions) slightly over the course of the recording due to changes in the muscle tension and relaxation. Small drifts can be corrected in the image-processing software to align the images together. If given the option, align the time points according to the vessel fluorescence channel, because the structures are more consistent than the leukocyte channel. If the field moves too much, longer recordings may become impossible. Typically, fields are more stable at the periphery of the tissue, near where it has been pinned. Unfortunately, these regions are less likely to contain appropriate venules to record. Drifting can be limited somewhat by using more pins to secure the tissue, especially proximal to the body. Take care when adding pins in this region, though, as it is easy to apply too much tension to the major vessels that feed and drain tissue, thus affecting the flow rates to the entire tissue. Another common issue is tenting, or drift in the z direction. This occurs when fluid slowly accumulates under the tissue, pushing it up and moving it out of focus. Slow changes can be corrected for by adjusting the z focus manually (if the scope allows). Tenting can typically be avoided by making sure that the tissue exits the body parallel to the silicone mount and pedestal, and that it is well secured. Keeping the flow rate of the perfusion buffer slow but steady will help keep it from accumulating beneath the tissue.
Tissue drying out
Typical recordings of TEM can go for 15 min or longer. During this time, the perfusion buffer running down the objective will hydrate the field of view. However, depending on subtleties of the tissue and silicone support, the distal regions of the tissue may not receive adequate flow to remain viable. After finding a view of interest, take note of the hydration state of distal regions of the tissue to make sure they are receiving enough buffer to remain hydrated. Ideally, much of the tissue will be clearly covered with buffer. To prevent the tissue from drying out and ruining potentially ideal fields, the flow rate of the perfusion buffer can be increased. This issue is more common for regions visualized near the periphery and less so nearer the center of the tissue. If this is not possible alter the flow rate or change positions, the tissue may be hydrated manually by gently pipetting a small amount of warm perfusion buffer directly onto it. Take care to apply it between images, and avoid touching the lens or tissue.
Fluorescent mice
To avoid the use of labeling antibodies that might interfere with leukocyte or endothelial cell function, it is often easiest to use mice that express fluorescent proteins in the specific subset of leukocyte one wishes to observe. For example, GFP inserted into the LysM locus adequately labels all myelomonocytic cells (neutrophils, monocytes, and macrophages), although neutrophils are typically brighter than monocytes, whereas macrophages are somewhat dim, more ramified, and within the tissue rather than intravascular. Along these lines, it is possible (and convenient) to use mice whose leukocytes have been rendered fluorescent. Mice with genetically encoded fluorescent leukocytes are available with virtually any leukocyte subtype labeled. These can be bred to your mouse of interest or, assuming histocompatibility, provided via a standard adoptive bone marrow transfer.
Troubleshooting
Table 2 lists problems that may arise with the procedures for intravital microscopy of mouse cremaster muscle, along with their possible causes and solutions.
| Problem | Possible cause | Solution |
|---|---|---|
| Poor signal | Focal plane is too deep | Find a field closer to the surface. |
| The labeling method was not effective | Inject more labeling antibody intravascularly | |
| Poor flow | Vessel is constricted or clotted | Follow the venule back to a rapidly flowing region and the examine the vascular tree to find a region with adequate flow. |
| Vascular bed/area has been compromised | Check a distal region. | |
| Tissue has dried out | Not enough perfusion buffer is reaching the region | Increase the perfusion buffer flow rate and/or manually hydrate the tissue. |
| Field of view is drifting in x or y dimension | Tissue relaxation or constriction | Image areas closer to the periphery. |
| Field of view is drifting in z dimension (tenting) | Perfusion buffer is accumulating under the tissue | Secure the tissue with more pins proximal to the body and/or image closer to the periphery. |
Understanding Results
An image series collected as described here will allow the investigator to carefully compare various components of the leukocyte adhesion and diapedesis pathway. Because of the sequential nature of this process, it is common that one component will be affected while others remain relatively unchanged. For example, disrupting the diapedesis step will typically not alter the ability of the leukocytes to roll or adhere to the vessel wall and thus the measured rolling velocity and the number leukocytes per field would remain similar controls. However, disrupting diapedesis will likely lead to more leukocytes found inside the vessel lumen (because they are unable to migrate out). Likewise, disrupting adhesion would reduce the total number of leukocytes observed on the vessel wall and, because it is downstream, the total number of leukocytes found in tissue. In this case, however, the diapedesis of those leukocytes that do adhere would likely be rapid and robust. Only by recording the multiple components of the pathway is it possible to identify which processes are fundamentally affected. Upon successful acquisition of the suggested image series, the investigator will have high-quality images detailing leukocyte rolling, adhesion, diapedesis, and migration through the tissue.
Time Considerations
When deciding on timing, take into consideration the experience level of the surgeon and the microscopist. An experienced technician can perform the entire surgery (from anesthetizing the mouse to tissue ready to image) in ∼30 min. Likewise, setting up the microscope, finding a field, and beginning the TEM recording need not take more than 15 min. Less experienced personnel may take two or three times as long. Ideally, one would plan to begin recording just before the peak inflammation time, just at the start of the biologically relevant window (i.e., at 4 hr post stimulus injection for neutrophils.)
Care must be taken when determining when to exteriorize the tissue to ensure one is within the correct biological window. Neutrophils are typically the early responders, with their egress peaking 4-8 hr after the injection of stimulus, whereas monocytes typical peak around 24 hr. The ideal time point for a particular mouse strain and stimulus combination can be determined by using a time course and simply harvesting the tissue to check the progress.
Basic Protocol 3: WIDE-FIELD MICROSCOPY OF THE MOUSE BRAIN
Wide-field imaging allows the rapid acquisition of images over an entire field. Unlike confocal imaging, wide-field imaging has limited resolution in the z dimension, making it difficult to delineate whether a particular leukocyte has extravasated outside of a vessel. However, in contrast to confocal imaging, wide-field imaging allows the visualization of global leukocyte migration patterns. In this protocol, wide-field microscopy is used to visualize leukocytes in relation to the vasculature across an entire fluorescently labeled brain slice. This protocol includes advice on how to remove the brain and image with wide-field microscopy to quantify leukocyte extravasation. Here we provide an outline to quantify transmigration in the brain vasculature.
Materials
-
Genetically modified mice with fluorescent leukocytes, such as LysMGFP, CatchupIVM , or Ccr2RFP mice
-
Anti-PECAM antibody conjugated to a fluorophore (EMD Millipore, CBL1337 clone 390, rat anti-mouse, non-blocking)
-
1× DPBS without calcium and magnesium (Corning, product no. 21-031-CV)
-
4% paraformaldehyde solution (see recipe)
-
CO2 source
-
30-ml syringes
-
Fine forceps and scissors
-
Dissecting pins
-
Razors
-
Murine Brain Matrix (RWD; device for holding brain in place for precise slicing)
-
Coverslip dishes (Mattek; with no. 1.5 coverslips)
-
Wide-field microscope with 4× objective: e.g., Nikon Eclipse Ti2 wide-field microscope with a Nikon 4× objective (0.20 NA) equipped with a Nikon DS-Qi2 camera
1.Inject 100 μg of fluorescently conjugated anti-PECAM antibody (clone 390) into the mouse intravascularly to label the vasculature.
2.Wait 30 min.
3.Sacrifice the mouse with CO2 based on the euthanasia guidelines for rodents.
4.Conduct thoracotomy and transcardial perfusion through the left ventricle with 30 ml DPBS until the blood is cleared as previously described (Wu et al., 2021).
5.Decapitate mice, cut the skin, and secure the head with dissecting pins (Fig. 7).
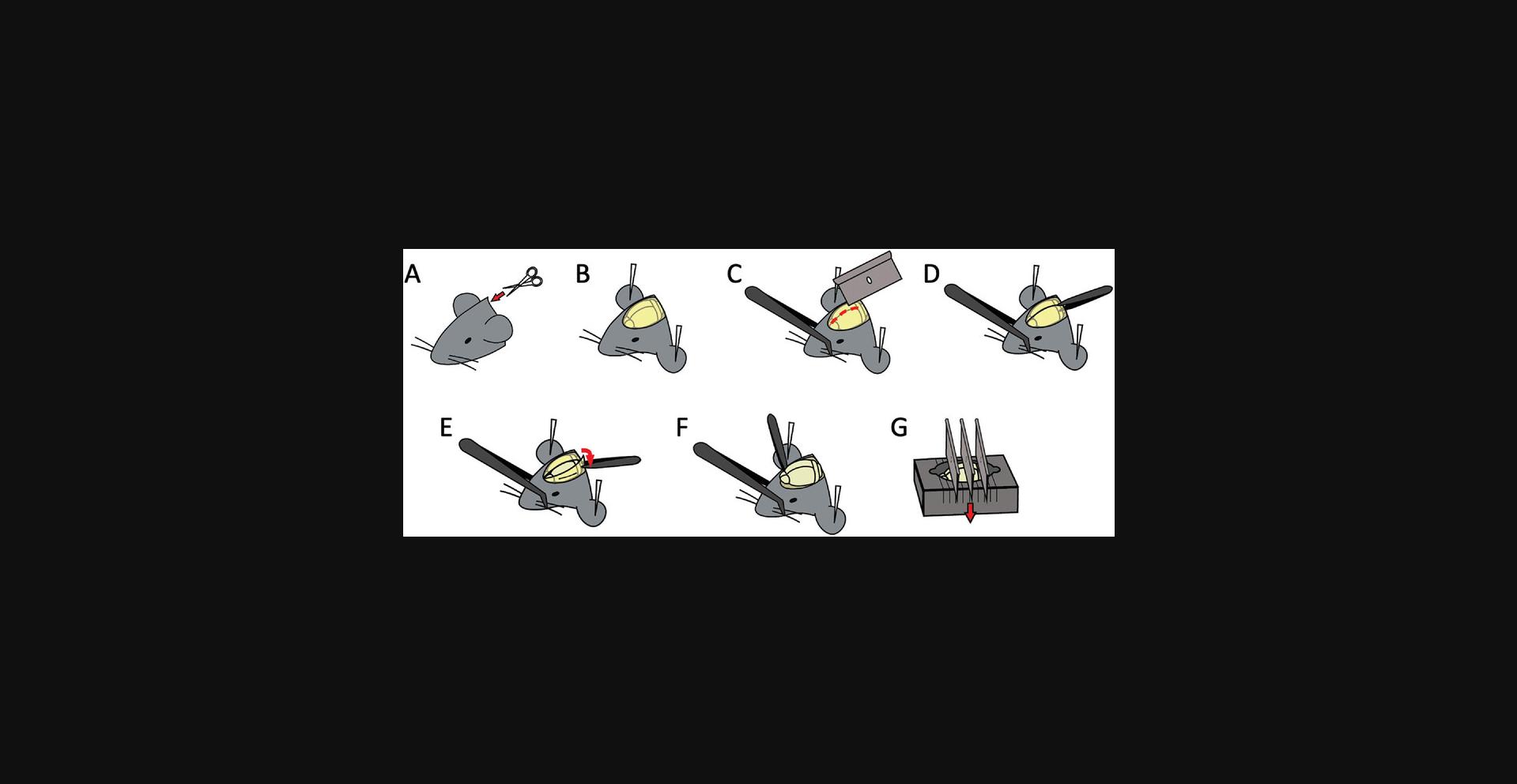
6.Score the skull along the brain midline with a razor.
7.Gently remove the skull with forceps.
8.Remove the brain, place the brain into the murine brain matrix (RWD), and cut the brain into 2-mm sections using single-edged razor blades.
9.Incubate the brain slices in 4% paraformaldehyde for at least 2 hr.
10.Place the brain slices on coverslip dishes (e.g., Mattek with no. 1.5 coverslips).
11.Image the brain from below using a Nikon Eclipse Ti2 wide-field microscope with a Nikon 4× objective (0.20 NA) equipped with a Nikon DS-Qi2 camera.
12.Collect images over the entire slice with a 15% overlap. Stitch the images together using the embedded stitching algorithm of NIS elements (Nikon, version 5.11.01) with the Optimal Path option.
COMMENTARY: BASIC PROTOCOL 3
Background Information
This protocol is useful for visualizing the position and density of infiltrating leukocytes and can be used to generate a cohesive, unbiased spatiotemporal image of leukocyte infiltration in the brain. Generating a spatiotemporal image of infiltrating leukocytes is helpful for understanding leukocyte migration patterns in various diseases and the global effects of therapeutic agents on leukocyte distribution in the brain.
Critical Parameters
It is important to prepare the samples for imaging in a consistent fashion. Because blood contains leukocytes that have not adhered to endothelial cells, inconsistent perfusion may affect the number of leukocytes quantified by this method. Additionally, paraformaldehyde has been shown to quench some fluorescent molecules. Therefore, to preserve fluorescence, samples should not be over-fixed.
Troubleshooting
Table 3 lists problems that may arise with the procedure for wide-field microscopy of the murine brain, along with their possible causes and solutions.
| Problem | Possible cause | Solution |
|---|---|---|
| High background | Brain slice may be too thick, increasing background, particularly in the GFP channel | Cut the brain into 1-mm intervals to decrease the background signal. |
| Debris in the section | Debris in the murine brain matrix | Clean the murine brain matrix after every use to limit debris. Debris can also be removed from the sections under a dissecting microscope. |
| Weak fluorescence | Cells are over-fixed | Reduce the time of incubation in 4% paraformaldehyde. |
Understanding Results
Wide-field imaging is useful due to the ability to acquire images over a large field. Processing and segmentation of the image allows quantification of the total number of leukocytes that have migrated into a field and visualization of leukocyte migration patterns in relation to areas of injury and major structures. The manner one would use to present the data depends on the research question, as wide-field imaging is useful to characterize leukocyte migration patterns. For example, one could use this technique to quantify the number of leukocytes that have migrated into an ischemic region surrounding an infarct in comparison to the area that surrounds the infarct.
Time Considerations
Preparation for imaging will take ∼2.5 hr. Imaging may take 10-30 min, depending on the size of the field, the number of channels used, and the microscope objective (we recommend a 4× objective, but other objectives may be used).
Basic Protocol 4: IMAGING THE LUNGS (EX VIVO)
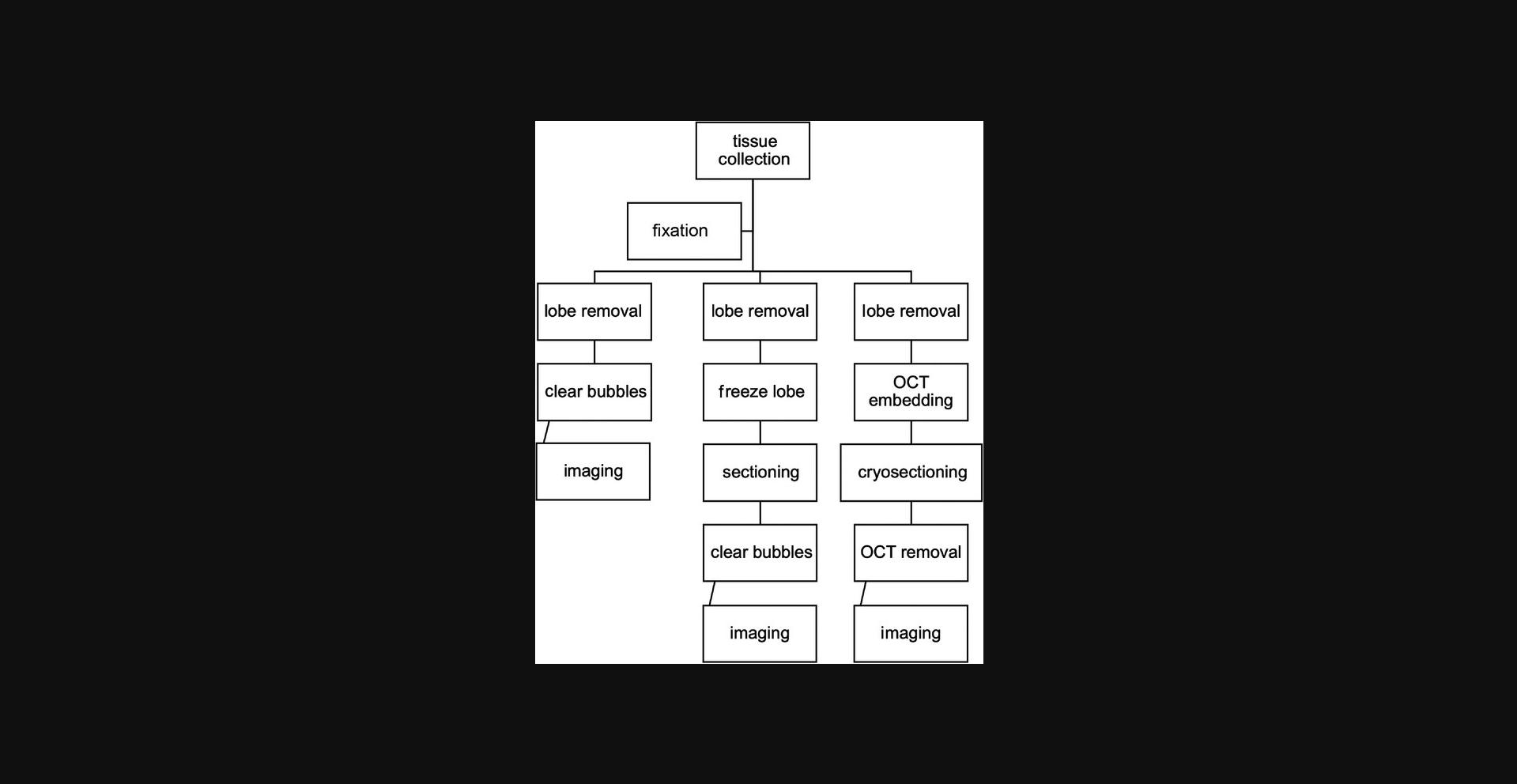
Imaging the vasculature of the lungs can be challenging because of the dense capillary network surrounding the alveolar sacs of the lungs. The vasculature of the lungs is unique compared to that of any other tissue, and inflammation in the lungs underlies many pathologies. These methods are used for imaging inflammation in the lungs after induction of injury or at baseline physiological conditions. These protocols can be used to prepare lungs to image leukocyte extravasation by wide-field microscopy or any type of fluorescence microscopy. Here we provide an outline of how to image inflammation in the murine lung. This protocol includes two experimental techniques for inflating the lungs. One protocol involves inflation via tracheostomy, whereas the other utilizes a long, blunted gavage needle to inflate the lungs. The experimenter can choose whichever method they feel most comfortable executing, as the end result will be identical. Figure 8 show a general workflow for imaging the lungs ex vivo, the details of which are described below.
Materials
-
Optional : Labeling antibodies (e.g., 390 to label endothelial cell junctions)
-
Avertin (see recipe)
-
70% ethanol
-
4% paraformaldehyde fixing solution (see recipe)
-
1× DPBS, without calcium and magnesium (Corning, product no. 21-031-CV)
-
OCT Compound (Sakura, cat. no. 4583)
-
Dissecting board
-
Dissecting tools
-
Dissecting board pins
-
Blunt forceps
-
Blunt-tipped scissors
-
Sutures
-
Sharp scissors
-
Luer stubs (Instech Labs, cat. no. LS20)
-
1-ml slip-tip syringe
-
Dissecting scope
-
MatTek dishes (MatTek, cat. no. P35G-1.5-14-C)
-
22 × 22-mm cover slips (Fisher Scientific, cat. no. 50-143-780)
-
15 × 15 × 5-mm Tissue-Tek Cryomold (Fisher Scientific, cat. no. NC9511236)
-
Dry ice
-
Razor blades
-
Cryostat for tissue sectioning
-
Cryostat blades
-
Optional : Brain matrix (RWD)
-
Slides
-
Wide-field microscope
-
FIJI software
Removing the lungs
1.Induce inflammation or perform the procedure necessitated by the experiment along with any appropriate controls.
2.Prior to euthanasia, inject any needed live antibody labels. Make sure that these are fixable.
3.Inject the mouse with an overdose injection of Avertin (800 µl-1 ml of Avertin, prepared as noted previously) to achieve deep anesthesia.
4.Once the mouse is in a deep plane of anesthesia (unresponsive to toe pinch), you can proceed with the lung removal.
5.To remove the lungs, place the mouse flat on a dissecting board on its back, pinning all four limbs to the board using pins.
6.Use 70% ethanol to wet the hair on the top of the mouse, focusing on the abdomen and the head. This is not to sanitize or clean the mouse but rather for fur control (to avoid getting fur in the sample).
7.Gross-dissect the skin away from the abdomen by gently pulling up on the skin on the abdomen with blunt forceps and making a small incision with blunt-tipped scissors. Insert the scissors in the cut, below the skin but above the chest plate. Open and close the scissors, gently moving them up the abdomen until you have bluntly dissected the skin away from the chest plate up to the jaw.
8.Using the blunt-tipped scissors, cut away the loose skin covering the chest and bottom of the jaw.
9.Underneath the jaw of the mouse, there will be two pieces of white-pink tissue (salivary glands). Pull these two apart or remove them completely, exposing the trachea.
10.Remove the chest wall by cutting away the diaphragm and up the left and right side of the ribs. This will expose the lungs.
11.Carefully dissect away the protective tissue around the trachea, exposing the horizontal rings of cartilage. You should dissect away the tissue both ventrally and dorsally.
12.Thread a 10-cm piece of suture under the trachea, leaving equal visible trachea space above and below the suture.
13.Using small, sharp scissors, make a small (∼1 mm) cut in between the rings of cartilage of the trachea.
14.Insert the Luer stub into the incision, and tightly tie the suture around the Luer stub and trachea with a single square knot that can be undone.
15.Slowly instill the lung with up to 700 µl of 4% paraformaldehyde fixative using a 1-ml slip-tip syringe.
16.Remove the syringe and Luer stub while simultaneously tightening the suture to keep the fixative inside the lungs. Tie off the suture with a box knot.
17.Holding the trachea up with the suture (gently pulling away from the body), cut at the top of the trachea, then dissect the lungs and heart away from the body.
18.Remove the lungs/heart together with the suture. Place in a container filled with fixative.
19.At this point, you can either quick-fix or fully fix. If you are quick-fixing, leave the lungs in the fixative for 30 min and then move on to imaging. If you are fully fixing, leave the lungs in fixative overnight. The next day, transfer the lung to 15% sucrose in PBS. Once the lungs have sunk to the bottom in the 15% sucrose (usually 1-2 days), transfer them to 30% sucrose until they sink (1-2 days).
20.Once the lungs are ready for imaging, you can image whole lungs, thick sections, or thin sections. You can also split the lungs up into left and right lungs or individual lobes for different imaging methods.
For whole lung imaging
21a. Remove the lungs from the fixative. Dissect away any blood clots or excess tissue from the outside of the lungs.
22a. Using sharp scissors, cut off the whole lobe that you plan to image.
23a. Under the dissecting microscope, pop any surface bubbles and clear the top of the lobe of blood and tissue as much as possible.
24a. Place the lobe, viewing side down, in a MatTek dish. Hydrate with DPBS and place a cover slip on top.
25a. The lung is now prepared for imaging. Proceed to step 27.
For thick sectioning
21b. Remove the lungs from the fixative. Dissect away any blood clots or excess tissue from the outside of the lungs.
22b. Using sharp scissors, cut off the whole lobe that you plan to image.
23b. Place the lobe in a Tissue-Tek Cryomold and either quick or slow freeze the lobe. To quick-freeze the lobe, place the cryomold on dry ice (5-10 min). To slowly freeze the lobe, place the cryomold in a −20°C freezer until frozen (30-60 min).
24b. Remove the lobe and section in 1-mm slices with a razor blade. This can be done by eye or in a brain matrix for sectioning.
25b. Place the section in a MatTek dish in 3-4 drops of PBS. Under dissecting microscope, pop any surface bubbles and place a cover slip on top.
26b. The lung is now prepared for imaging. Continue with step 27.
For thin sectioning
27.Remove the lungs from fixative. Dissect away any blood clots or excess tissue from the outside of the lungs.
28.Using sharp scissors, cut off the whole lobe that you plan to image.
29.Place a dot of OCT compound in a 15 × 15 × 5-mm Tissue-Tek Cryomold. Wait until the OCT has settled at the bottom and all bubbles have popped. The OCT should fully coat the bottom of the cryomold.
30.Place the lung lobe you plan to image on the OCT in the cryomold. Cover the lung with more OCT until it is fully submerged. Allow the OCT to settle and all the bubbles to pop (2-3 min).
31.Place the cryomold on dry ice.
32.Once frozen, remove the frozen block from the mold and place inside a cryostat set to 18°C.
33.With a razor blade, cut off the edges of the OCT that do not contain lung tissue.
34.Using the cryostat, section the lung tissue into 10-15 µm sections and place on a slide. Keep the slides uncovered on dry ice.
35.Rinse the OCT off by placing 2-3 drops of PBS on the slide for 5 min before placing a cover slip on the slide.361.Image the slides.
Counting cells in FIJI
36.Import your image into FIJI using the BioFormats importer.
37.Split any color channels into their own separate image tabs by going to Image, Color, Split Channels.
38.For each color channel with cells, perform background subtraction based on pixel size. Select Process, Subtract Background, and input the approximate pixel size of the cell type for the background subtraction.
39.To count the cells in a given channel, convert the channel to a binary. Select Process, Binary, Make Binary. Select Analyze, Analyze Particles…, and input your criteria. The Analyze Particles function can discriminate on the basis of size and circularity. This will differ depending on your cell type (Fig. 9).
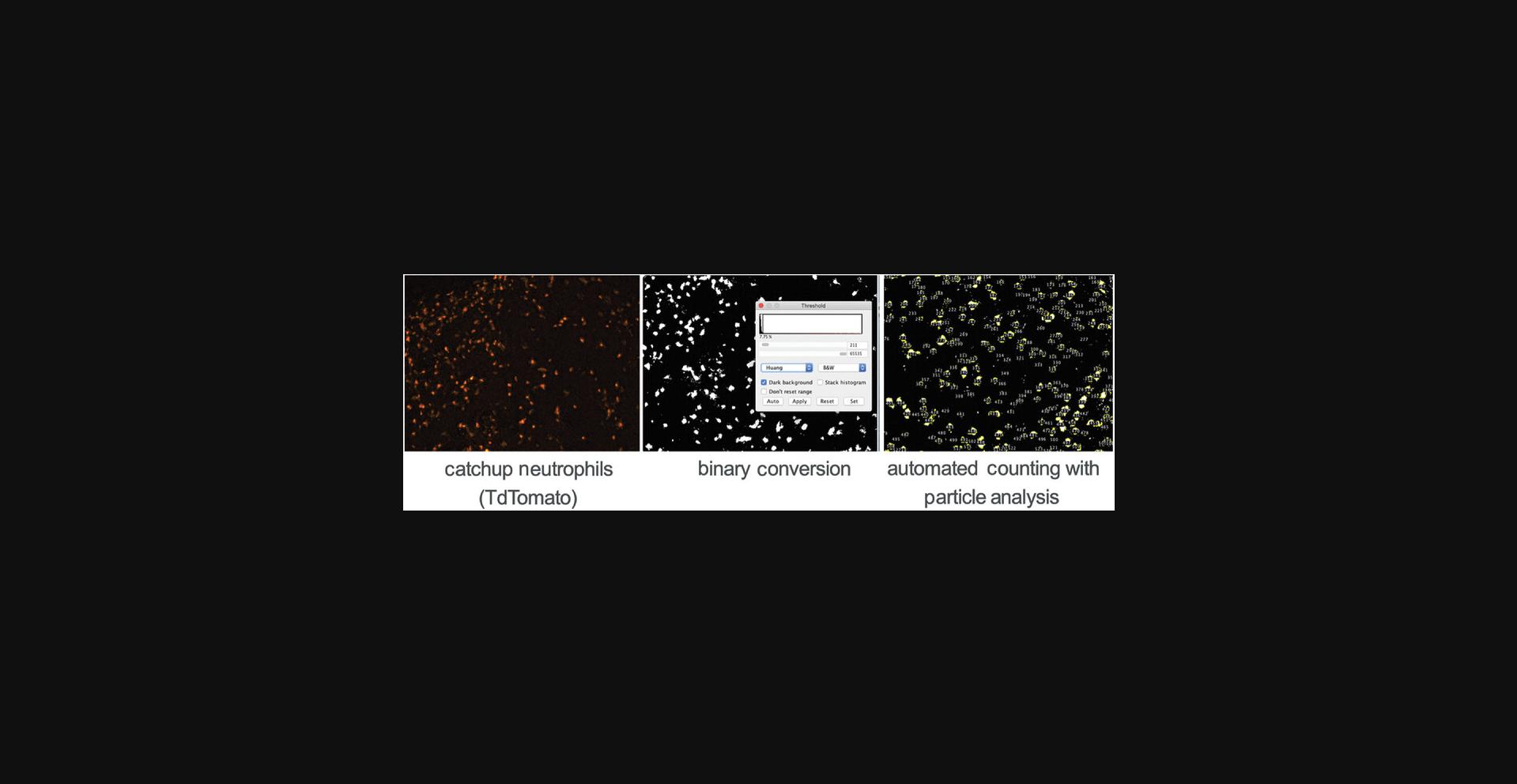
40.This will open the ROI Manager, which will contain the results of your count. This can be used as your final count.
Alternate Protocol 2: INFLATING THE LUNGS WITHOUT TRACHEOSTOMY
The gavage device is reusable and runs less risk of fully tearing the trachea and being unable to tie the suture. This alternative protocol may also be easier for someone newer to the technique.
Additional Materials (also see Basic Protocol 4)
- Straight stainless steel olive tip cannula with Luer-Lok base
- Perform Basic Protocol 4, replacing steps 12-16 with the following.
12a. Attach a straight stainless steel olive tip cannula with a Luer-Lok base to a 1-5 ml syringe containing fixative.
13a. Gently insert the cannula through the oropharynx into the trachea. It is helpful to pull up on the mouse's lower jaw with forceps to help guide it. The trachea is anterior to the esophagus, so guide the cannula, pushing it slightly anteriorly and caudally to avoid insertion into the esophagus.
14a. You will see the tip of the cannula enter the trachea through the cartilage rings. When it is more than halfway to the lungs, thread a piece of suture behind the esophagus and tie the first half of a square knot around the trachea, holding the cannula in place.
15a. Slowly instill the lung with up to 700 µl of 4% paraformaldehyde fixative using a 1-ml slip-tip syringe.
16a. Remove the cannula from the trachea while simultaneously tying the second half of the square knot, not too tightly, around the trachea to prevent the fixative from leaking out.
COMMENTARY: BASIC PROTOCOL 4 AND ALTERNATE PROTOCOL 2
Background Information
This technique is advantageous in that the preparation of the lungs for imaging is flexible. If necessary, the protocol can be done quickly, and the tissue can be prepared and serially sectioned. This is particularly useful if the fluorophore being used is easily quenchable. If a longer fixation is more beneficial to the final imaging product, this can also be done. This method also allows mixed use of protocols, i.e., you can use different methods for the different lobes of the murine lung. These protocols are useful for determining whether your stimulus or intervention results in the induction of inflammatory cells in the pulmonary vasculature. They are also useful for determining the degree to which varying stimuli induce inflammatory cell infiltration.
Critical Parameters
In this protocol, it is important that you remove the lungs carefully. Mishandling during removal can cause damage to the tissue. Additionally, it is important that the section you are imaging is the section that you are interested in. Because the physiology of the lungs can vary from the surface to the deepest parts of the organ, it can be useful to serially section (either thin or thick) and image a number of different sections to validate results as being consistent throughout the tissue.
Troubleshooting
Table 4 lists problems that may arise with the procedures for imaging inflammation in the murine lungs ex vivo, along with their possible causes and solutions.
| Problem | Possible cause | Solution |
|---|---|---|
| High background | Lung slice may be too thick | Opt to use thin sectioning rather than thick sectioning. |
| Bubbles | Bubbles in the image during microscopy | After placing the lung section in a MatTek dish, place under dissecting microscope. Gently disperse as many bubbles as possible and image as soon as possible. |
| Weak fluorescence | Cells are over-fixed | Reduce the time of incubation in 4% paraformaldehyde. |
| Inflammation in controls | Euthanasia method is inducing inflammation | Opt for an overdose of a drug such as Avertin as opposed to CO2 asphyxiation. |
Understanding Results
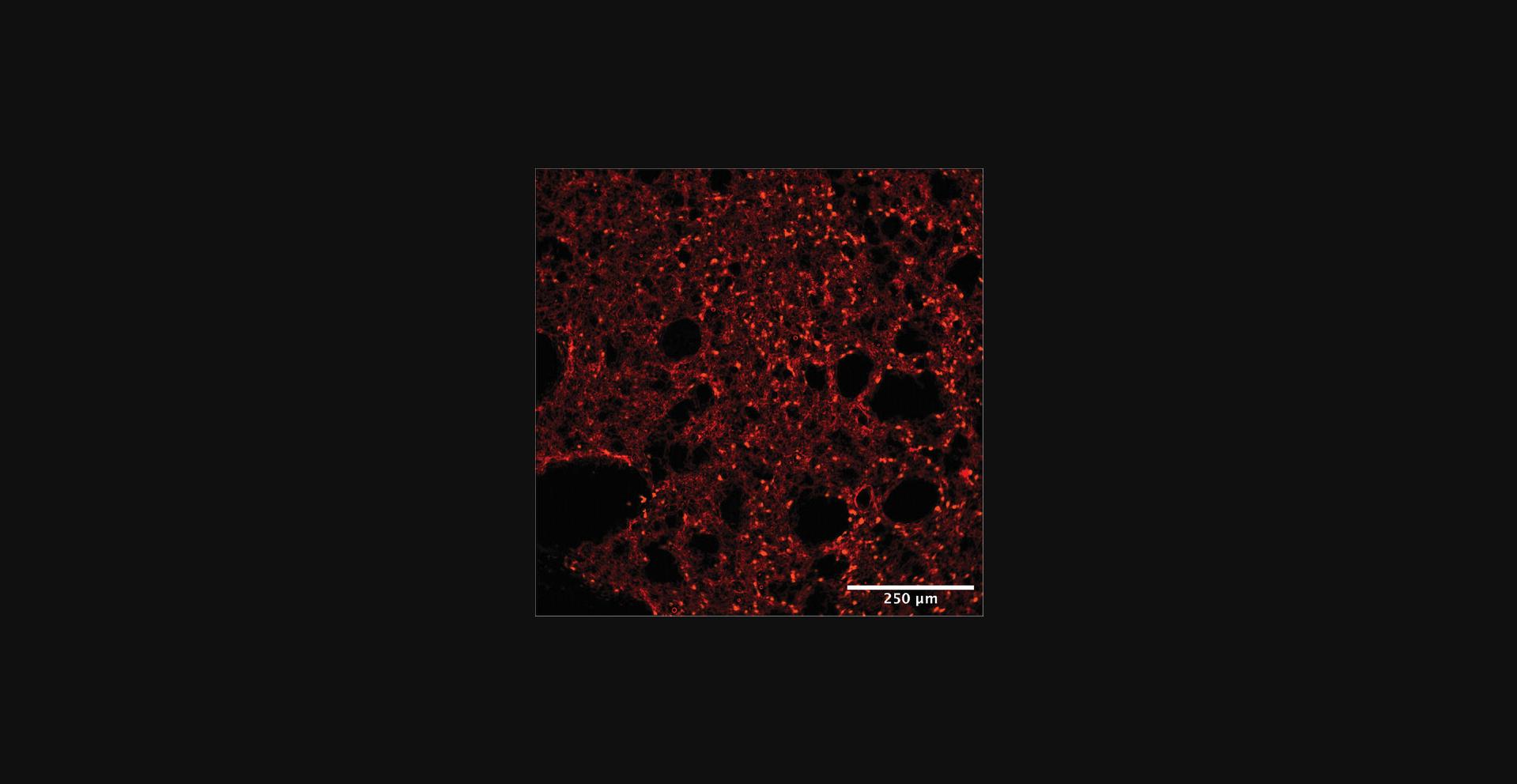
Analyzing and interpreting the results of this protocol will vary based on the type of label used for the fluorescent tagging of leukocytes and/or blood vessels. After completion of imaging, you should have labeled vasculature with leukocytes present in the appropriate fluorescent channel that corresponds to the fluorescent tag used. As shown in Figure 10, you should see small, round leukocytes in the vasculature of the lungs (marked by its alveolar shaping).
Time Considerations
The time from induction of inflammation to imaging depends on the model. After the euthanasia of the mouse, the protocol should take 2-3 hr if quick-fixing or 24-72 hr if using a lengthier fixation process. The length of the fixation is the main determinant of time.
Basic Protocol 5: INDUCING, IMAGING, AND QUANTIFYING INFILTRATION OF LEUKOCYTES IN MOUSE RETINA
The objective of Basic Protocol 5 is to induce, image, and quantify leukocyte extravasation from vasculature in the mouse retina. The eye is an immune-privileged organ and has few leukocytes at baseline; thus, to visualize leukocyte infiltration, the experimenter will need to use a stimulus to induce inflammation in the eye. This can be done by intravitreal injection of an inflammatory stimulus prior to sacrifice. The experimenter then conducts immunofluorescence staining on whole-mount retina and acquires fluorescent images to visualize the leukocytes and retina vasculature. The experimenter can use FIJI to process the immunofluorescence images and quantify leukocyte extravasation. If the protocol is conducted properly, the experimenter will induce inflammation, visualize, and quantify leukocyte infiltration in the mouse retina.
Materials
-
Mice of interest: e.g., C57BL/6 or Ccr2RFPCx3cr1GFP mice
-
Ketamine/xylazine anesthetic (see recipe)
-
Meloxicam
-
1× DPBS without calcium and magnesium (Corning, product no. 21-031-CV)
-
Proparacaine hydrochloride eye drops
-
Phenylephrine hydrochloride eye drops
-
Tropicamide eyedrops
-
4% paraformaldehyde (PFA) fixing solution (see recipe)
-
Blocking solution (see recipe)
-
Washing solution (see recipe)
-
Recombinant Mouse CCL2/JE/MCP-1 Protein (R&D Systems Inc., cat. no. 479-JE-050/CF)
-
Primary antibody, such as:
- Rabbit Anti-S100A9 antibody [2B10] (Abcam, ab105472) to detect neutrophils
- Rabbit Anti-IBA1 antibody (Wako, 019-19741) to detect macrophages
- Goat Anti-Collagen IV antibody (Abcam, ab19808) to detect vessels
- Goat Anti-CD31 antibody (R&D Systems, AF3628) to detect vessels
-
Secondary antibody, such as:
- AffiniPure Goat Anti-Rat IgG (Jackson, cat. no. 112-005-167)
- Alexa Fluor® 647 AffiniPure Goat Anti-Rabbit IgG (Jackson, cat. no. 111-605-144)
-
ProLong™ Gold Antifade Mountant (Thermo Fisher, cat. no. P36930)
-
Ear punch tool
-
Scale
-
30-G needle
-
and 5-ml syringes
-
Scissors
-
Model 901 Removable Needle Syringe, 10-µl capacity (Hamilton, cat. no. 7648-01)
-
Small Hub Removable Needle, 32 G, 0.4 inch, point style 3 (Hamilton, cat. no. 7803-04)
-
Light microscope
-
CO2 tank with mouse euthanasia apparatus
-
Carcass disposal bag
-
Curved forceps (Fine Science Tools, cat. no. 91117-10)
-
Light dissecting microscope
-
Ring forceps (Fine Science Tools, cat. no. 11103-09)
-
Spring scissors (Fine Science Tools, cat. no. 15003-08)
-
Straight forceps (two; Fine Science Tools, cat no. 11254-20)
-
Weigh boats
-
12- and 24-well plates (Thermo Scientific, cat. nos. 130185 and 130186)
-
3-ml transfer pipets (VWR cat. no. 52947-948)
-
Orbital shaker
-
3-inch × 1-inch × 1.2-mm microscope slides (VWR, cat. no. 16004-370)
-
20 × 60-mm no. 1.5 microscope cover glasses (VWR, cat. no. 16004-312)
-
FIJI software
-
Stirring hotplate (Thermo Fisher, cat. no. SP88854100)
Tagging and sedating mice
1.Obtain mice of interest, such as C57BL/6 or Ccr2RFPCx3cr1GFP mice.
2.If using multiple mice, tag to differentiate one from another. Suggestions for tagging include ear punch or ear tag.
3.Weigh mice.
4.Calculate dose of ketamine/xylazine to administer to mice based on weight. See recipe for more information on ketamine/xylazine anesthetic.
5.Scruff mice and inject correct dose of ketamine/xylazine intraperitoneally. Wait a few minutes until mouse is sedated.
6.Once mouse is sedated, inject 0.4 ml of meloxicam (1:100 meloxicam/1× DPBS mixture) subcutaneously with 30-G needle attached to a 5-ml syringe. To perform a subcutaneous injection, place mouse on a flat surface, pinch and lift the skin on its back, and inject into the lifted skin.
Intravitreal injection of inflammatory stimulus CCL2
7.Trim whiskers to prevent disruption of microscope examination.
8.Scruff mouse and position its head with one eye facing up. Administer one drop of proparacaine hydrochloride to eye. Balance the drop for 2-3 s and then shake off. Repeat in other eye.
9.Administer phenylephrine hydrochloride eye drops (same handling technique as in step 8).
10.Administer tropicamide eye drops (same handling technique as in step 8).
11.Place mouse under light microscope on a paper towel. Position the head with the eye of interest facing up.
12.With a 30-G needle attached to an empty 1-ml syringe, make an incision just posterior to the sclerocorneal junction on the lateral side of the eye (Fig. 11). The incision should be bevel deep and no more.
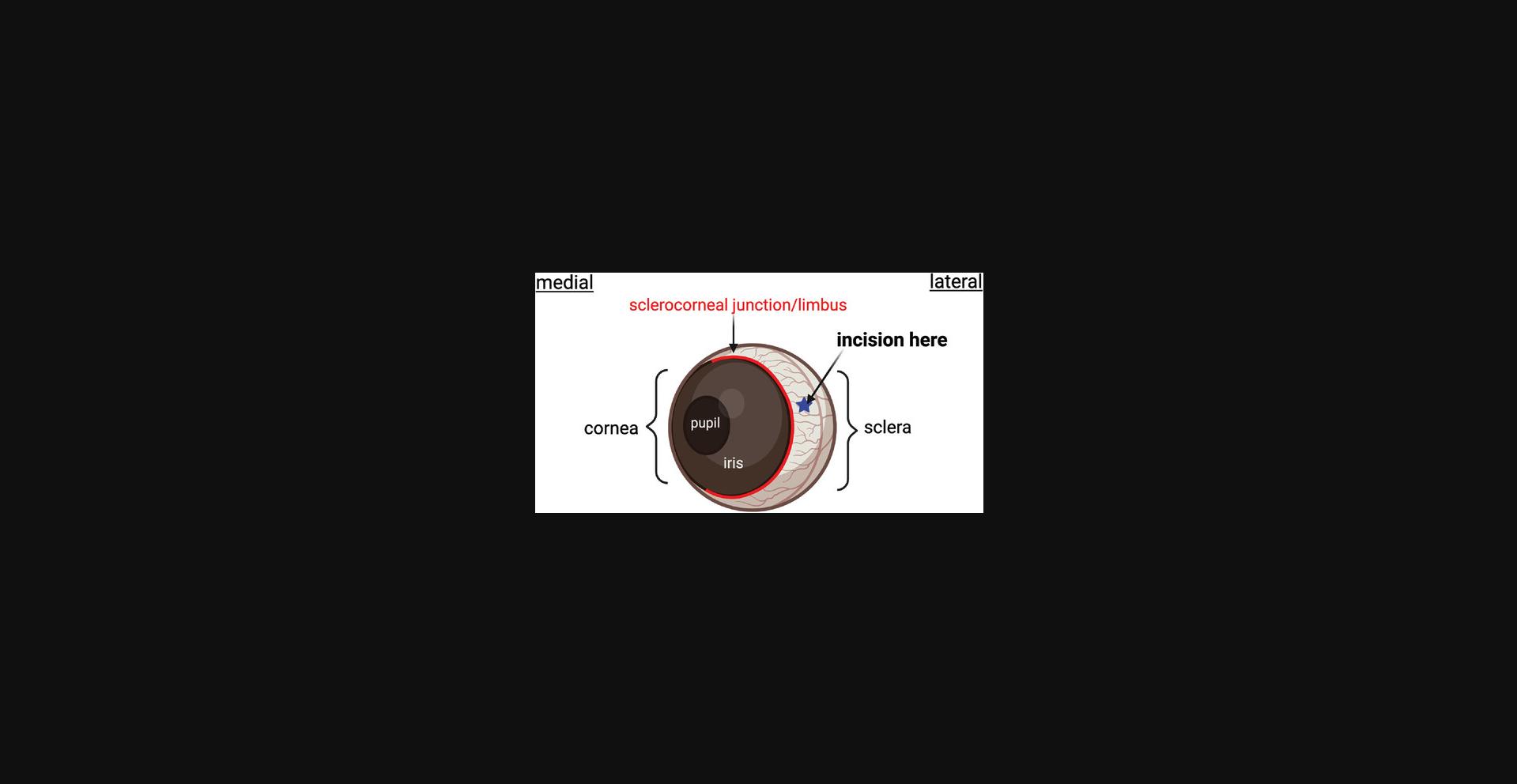
13.With a Hamilton syringe loaded with your inflammatory stimulus (CCL2, 5 ng/µl), insert the tip of the needle 2 mm deep into the incision you created in step 12.Have a second person help push the plunger to inject 1 µl of CCL2.
14.Flip the mouse to its other side and repeat steps 11-13 to inject the other eye. Omit this step if you are only injecting one eye.
15.Place mouse in an empty cage on a cage warmer until the mouse wakes up.
16.Return mouse to its original cage and provide food and water as usual. Sacrifice mice to harvest eyes 24 hr after intravitreal injection.
Enucleation and dissection of retina
17.At the desired time after intravitreal injection, euthanize mice per IACUC protocol.
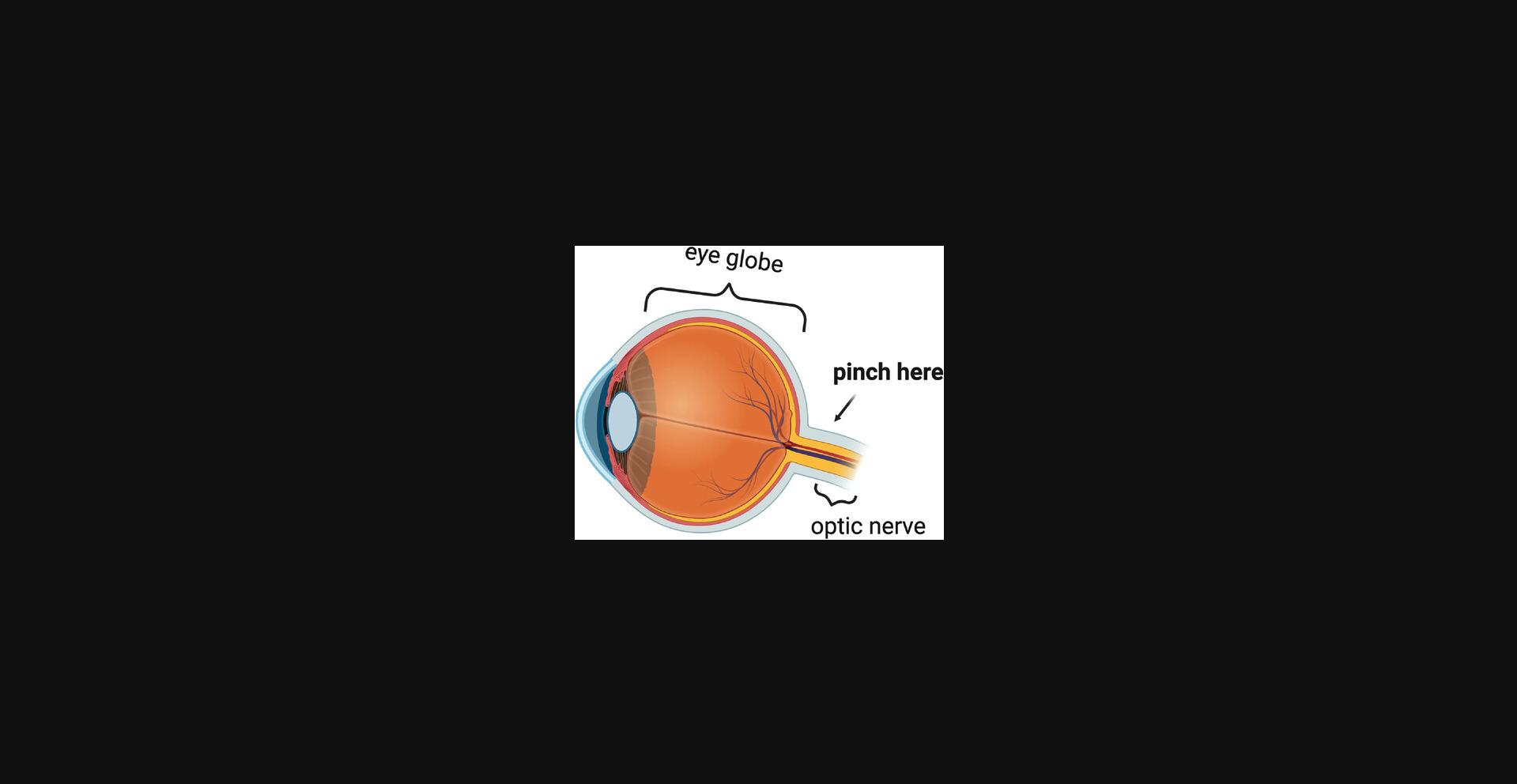
18.Using curved forceps, enucleate the eyes: maneuver the forcep tips around and under the eye globe, pinch the optic nerve, and gently pull the eye free from the socket (Fig. 12). Place enucleated eyes in a dish with 1× DPBS and place under a dissecting light microscope.
19.Dissect globe to remove conjunctiva, extraocular muscles, and orbital fat. Stabilize globe with ring forceps and make a small incision (1 mm) with spring scissors at the limbus of the eye.
20.Substitute ring forceps with straight forceps. Insert straight forcep tip into the incision from step 19 and grip the anterior portion of the eye.
21.Cut along the limbus around the globe using spring scissors in one hand, while gripping the anterior portion of the globe using straight forceps with the other hand.
22.Remove the anterior portion of the eye.
23.Separate the lens from the posterior globe.
24.Pinch the choroid-sclera complex with straight forceps in both hands and tear. Continue tearing at different parts of the globe until the cup is easily separable from the retina.
25.Pinch the retina at the optic nerve to separate it from the cup.
Immunofluorescence staining of whole-mount retina
Day 1
26.Place dissected retinas immediately into 4% PFA and fix for 1 hr at room temperature.
27.Manually aspirate PFA with a transfer pipet and wash retina with washing solution (PBS + 0.1% Tween) for 10 min on an orbital shaker at room temperature. Repeat this step 4 times.
28.Block retinas by submerging in blocking solution overnight at 4°C or for 1 hr at room temperature on an orbital shaker.
Day 2
29.Move retinas to a 24-well plate and incubate in primary antibodies overnight at 4°C on an orbital shaker.
Day 3
30.Move retinas back to the 12-well plate and wash with washing buffer for 10 min at room temperature on an orbital shaker. Repeat four times.
31.Move retinas back to 24-well plate and add secondary antibody in blocking solution for 1 hr at room temperature on an orbital shaker.
32.Move retinas back to the 12-well plate and wash with washing buffer for 10 min at room temperature on an orbital shaker. Repeat four times.
Mounting retina onto microscope glass slide
Day 3 continued
33.Using a transfer pipet, place stained retina into 1× DPBS in a plastic weigh boat or similar-sized vessel under a dissecting light microscope.
34.Cut retina into four leaflets using cuts at 90° (Fig. 13).
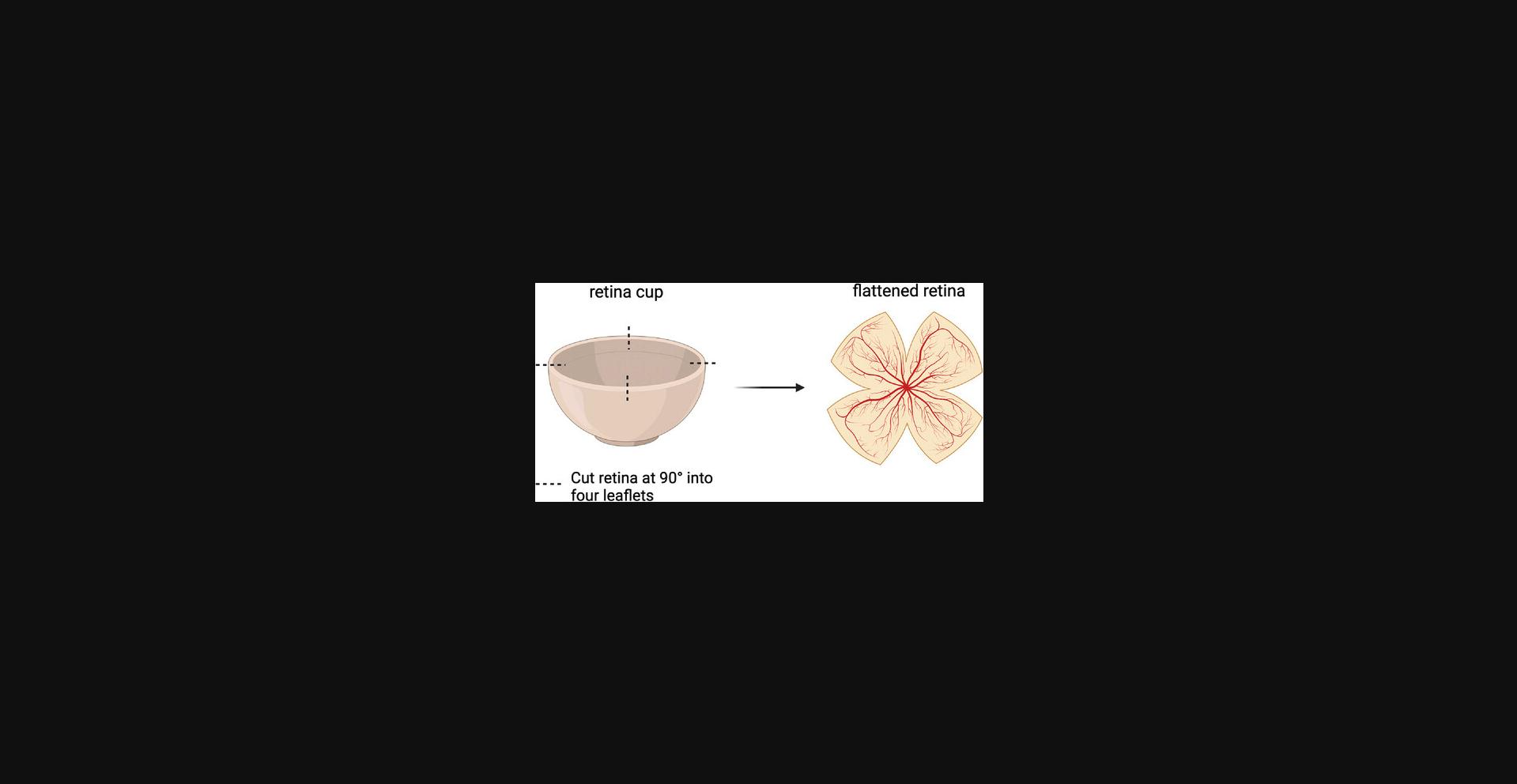
35.Place retina onto a glass microscope slide using a transfer pipet.
36.Wick away DPBS with a Kimwipe, and flatten the retina by maneuvering the leaflets with straight forceps.
37.Place 1-2 drops of ProLong Gold on retina and cover with a coverslip. Seal coverslip with nail polish and allow it to cure at 4°C overnight or until ready to image.
Image acquisition
Acquire fluorescence images of stained retina as desired. Confocal images are necessary to investigate the superficial and deep capillary plexuses independently. For quantification in FIJI, it is advisable to acquire images that encompass the entire retina.
Quantification of leukocytes with FIJI
38.Open .nd2 file in FIJI.
39.Use horizontal scroll bar to scroll to the channel of interest in the .nd2 file.
40.Adjust the brightness/contrast settings until your cells of interest are clear. To do this, go to Image > Adjust > Brightness/Contrast > Auto. If the “auto” option does not work well, you can manually adjust the settings in the pop-up window.
41.Next, duplicate your image, as it is good practice to work on duplicates and not the original file. To do this, go to Image > Duplicate > Unclick “Duplicate hyperstack” and type in the number of the channel you want to duplicate (i.e., 1, 2, or 3). Duplicate your channel of interest twice. One duplicate will be your “original” and the other will be for “thresholding.”
“Threshold” duplicate
42.In your “threshold” duplicate, choose a threshold strategy for your image that highlights your cells of interest. To do this, go to Image > Adjust > Threshold. Make sure “dark background” is ticked and “B&W” is selected under the second drop-down menu. In the first drop-down menu, scroll through the different threshold options until you see one that automatically highlights your cells of interest. Click “Apply” and close the window.
43.Next, segment out debris. Go to Analyze > Analyze Particles > 300-infinity. Make sure “add manager” is ticked in the ROI pop-up window.
“Original” duplicate
44.Now we will overlay the debris selections from step 43 over your “original” duplicate. Go to your “original” duplicate, and click “show all” in the ROI manager until you see your debris selection on your original image. Click “More” in the ROI manager > Fill.
45.Next, measure the total area of the debris selections, as you will need to subtract this area from your selection of the whole retina later to calculate a “total retina area” in which to count cells. The ROI manager contains a list of selections that correspond to the segmented debris. Select all points in the ROI manager, and then Analyze > Measure in the menu at the top of your screen. The output is the area (μm2) of the debris. Write this number down.
46.Next, measure the area of retina to be analyzed. This will require you to calculate the area of the entire retina (including the debris). Go to your “original” duplicate that now has the debris filled out, select the “polygon” selection tool in the FIJI toolbar, and manually outline the entire retina. By hand drawing, you can get a count of “cells per area” and compare this in a standardized way. Once you have made your selection, go to your ROI manager and click “Add” to add your selection to the manager. In the manager, click on the selection you just added, and Analyze > Measure to calculate the area of your selection in μm2. Note that this is not the final area. To find the total area of retina to count cells in, subtract “debris area” from the area you just measured. Write this number down.
47.Next, find and calculate the number of leukocytes in the retina. To do this, you must tell FIJI what you consider to be a leukocyte. While making sure the retina area you selected is still highlighted, zoom in to your tissue and click the “hand/scrolling tool” button in the FIJI toolbar to move to an area of the retina with leukocytes. Go to Process > Find maxima in the menu at the top of the screen. The number of maxima is your leukocyte count.
48.Finally, quantify the number of leukocytes per retina by dividing your maxima calculated in step 47 by the final retina area calculated in step 46.This will give you the number of leukocytes per μm2 of retina.
COMMENTARY: BASIC PROTOCOL 5
Background Information
This protocol is for visualizing extravasation of leukocytes in the retina and could be useful for testing anti-inflammatory agents in the eye. Inflammation may play a role in the pathogenesis of retinal diseases, such as diabetic retinopathy and uveitis. The ability to visualize the course of inflammation (induction, extravasation, and resolution) can be helpful in studying disease pathology.
Critical Parameters and Troubleshooting
The mouse eye has a diameter of only 3 mm, and the retina is particularly delicate. Therefore, special attention should be paid to the intravitreal injection, as well as the retinal dissection and mounting steps. For the intravitreal injection, the experimenter should be careful to inject the inflammatory stimulus into the vitreous and not the lens. One way to check for successful injection is to practice injections using Evans blue dye and using a slit-lamp to visualize the retina after injection. If the retina is visible, the user likely injected the stimulus correctly; if the retina is opacified or blocked by blue dye, the user injected the dye into the lens. The dissection and mounting processes are also sensitive to user manipulation. For both, the experimenter needs to be careful not to tear the retina. Specific troubleshooting recommendations are outlined in Table 5.
| Problem | Possible cause | Solution |
|---|---|---|
| Oversaturated points and nonspecific binding in fluorescence channel for leukocytes | Debris in retinal dissection (choroid tissue, vitreous) | Clean retina with straight forceps as much as possible prior to mounting; however, need to balance removing debris and not creating tears in retina tissue. |
| Low fluorescence signal | Over-fixation of retinal tissue | Fix tissue for a shorter duration. |
| Tears and holes in retinal tissue | Puncturing the retina with forceps during dissection and/or mounting steps | Practice handling the retina more gently; have more retinas than needed for an experiment in case one retina becomes unusable. |
| No inflammatory cells 24 hr after intravitreal injection of CCL2 | Inflammatory stimulus was injected into the lens of the eye instead of into the vitreous, or injection was not deep enough and inflammatory stimulus leaked out of the initial incision | Practice injections with Evans blue dye. Use a slit-lamp post dye injection to see if the retina is visible. If clearly visible, injection was done correctly; if retina is not visible, dye was injected into the lens. Ensure injection needle is 1-2 mm into the initial incision. |
Understanding Results
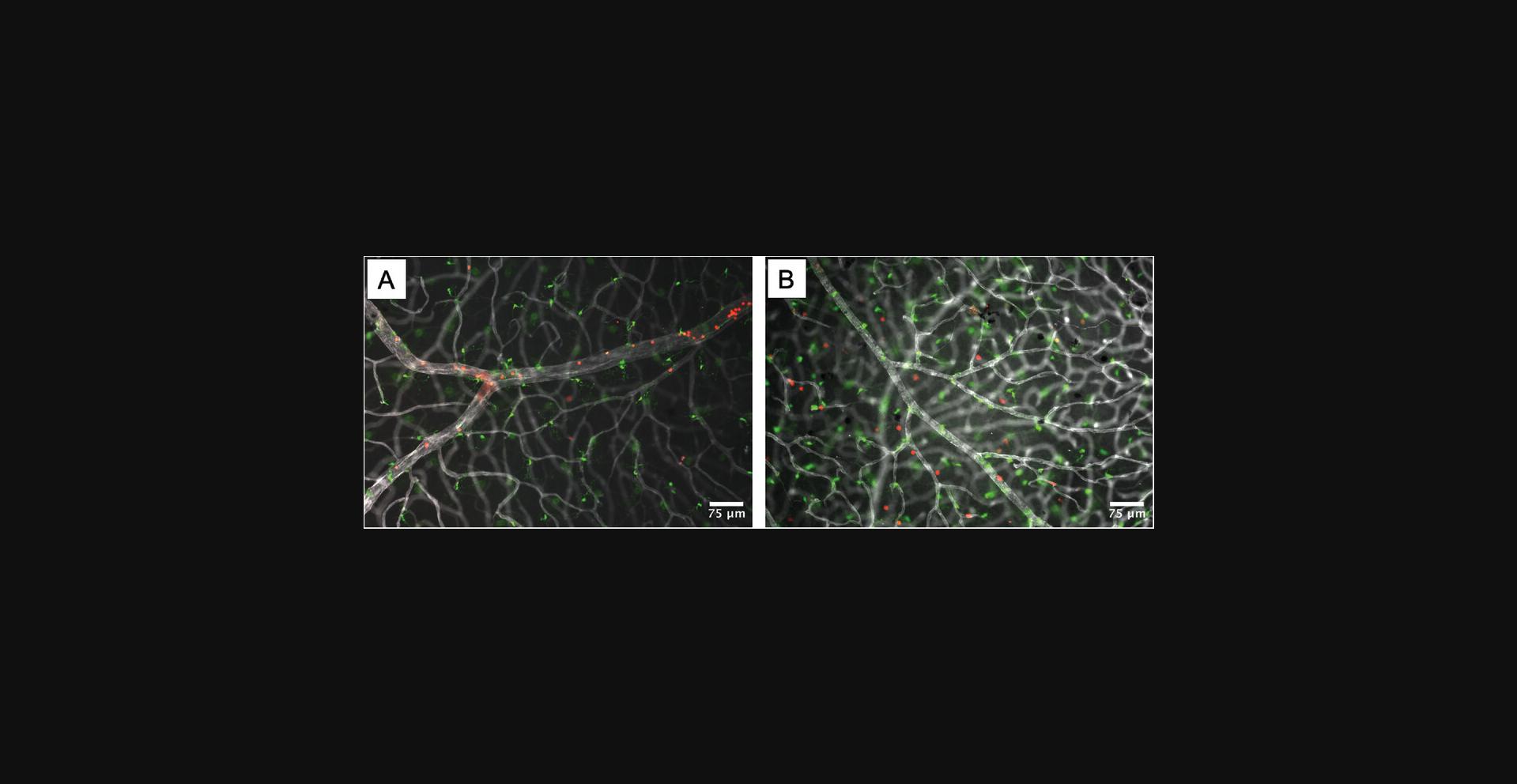
After following the steps of this procedure and acquiring fluorescent images of whole-mount retina, the experimenter should be able to visualize leukocytes within the vasculature at earlier time points after CCL2 injection (e.g., 8 hr) and leukocyte extravasation into the retinal tissue at later time points (e.g., 48 hr), as demonstrated in Figure 14. Using FIJI, the experimenter can quantify the number of leukocytes that have extravasated per mm2 area of retina for a standardized value.
Time Considerations
Several steps in the protocol require technical expertise, and the timing for these steps will vary widely depending on the experience of the experimenter. For an experienced surgeon, ∼5 min is needed per eye for injection of inflammatory stimulus, 5 min per eye for retinal dissection, and 5-10 min per eye for mounting. Time from intravitreal injection to sacrifice of the mouse will depend on what the experimenter is studying. Sacrifice 24 hr post CCL2 injection is appropriate if looking for both neutrophil and monocyte extravasation. A minimum of 2 days is required for immunofluorescence staining of whole-mount retina. In summary, it will take ∼5 days from the first step of the protocol (anesthetizing mice for injection of inflammatory stimulus) to the last step of acquiring fluorescent images of stained retinas.
REAGENTS AND SOLUTIONS
General
Avertin anesthetic
- 1.Dissolve 2.5 g of 2,2,2-tribromoethanol (99%) in 5 ml of 2-methyl-2-butanol (98%) by heating to 40°C while stirring on a stir plate.
- 2.Add sterile 1× PBS (without calcium and magnesium) to a final solution volume of 200 ml.
- 3.Using a 50-ml syringe and a 0.45-μm syringe filter, filter the Avertin into 50-ml aliquots.
- 4.Keep the aliquots at 4°C protected from light for up to 2 weeks.
- 5.Make fresh Avertin every 2 weeks or for every experiment.
Ketamine/xylazine anesthetic
For 20 ml of solution :
- 4 ml ketamine (from a 100 mg/ml stock; final concentration will be 20 mg/ml)
- 0.4 ml xylazine (from a 20 mg/ml stock; final concentration will be 0.4 mg/ml)
- 2 ml 10× DPBS
- 13.6 ml sterile water
- Mix and store at room temperature until drug expiration date
Ketamine hydrochloride should be used at a final dosage of 100 mg/kg of body weight (bwt) (e.g., 2 mg per 20 g bwt) and xylazine at a final dosage of 2 mg/kg bwt (0.04 mg per 20 g bwt).
Quick dosage guideline: Use 100 µl for a 20-g mouse. If the mouse has not entered the surgical plane of anesthesia by 10 min, boost with an additional 25%-50% of initial dose.
Basic Protocol Basic Protocol 1
Bovine serum albumin (BSA) solution
- 1.25 g BSA
- 50 ml 1× DPBS
- Mix, and filter through a 0.2-μm filter
- Store up to 6 months at 4°C
Composition: 2.5% (w/v) BSA.
Carrier solution
- 200 μl olive oil
- 800 μl acetone
- Always make fresh
Croton oil mixture
- 7.5 μl croton oil
- 492.5 μl carrier mixture
- Always make fresh
Shake the bottle of croton oil well before adding to carrier mixture.
Composition: 1.5% (v/v) croton oil.
Fixing solution
Stir 4 g paraformaldehyde and 6 drops of 1 N sodium hydroxide in 90 ml double-distilled water over a hotplate. Once the paraformaldehyde is dissolved, let cool to a temperature that allows safe handling, and add 10 ml of 10× DPBS. Maintain on ice until use. Store up to 1 month at –20°C.
Permeabilization buffer
- 200 ml normal goat serum (Jackson, cat. no. 005-000-121)
- 12 ml Triton X-100
- 3.8 ml BSA solution
- Always make fresh
Composition: 5% normal goat serum and 0.3% Triton X-100.
Basic Protocol Basic Protocol 2
Booster dose ketamine/xylazine initial anesthesia cocktail
For 20 ml of solution :
- 5 ml ketamine (from a 100 mg/ml stock; final concentration will be 25 mg/ml)
- 1.25 ml xylazine (from a 20 mg/ml stock; final concentration will be 1.25 mg/ml)
- 2 ml 10× DPBS
- 11.75 ml sterile water
- Mix and store at room temperature until drug expiration date
Supplemental dose guideline: 25 µl delivered intramuscularly.
Perfusion buffer (pH 7.4, 37°C)
- 1 liter sterile deionized H2O (lipopolysaccharide free)
- 1 packet Tyrode's salts (Millipore Sigma, T2145-10 × 1 L)
- 1 g sodium bicarbonate (Millipore Sigma, T2145)
- Store up to 5 days at 4°C
Measure the pH of the solution when it is at 37°C, as this can change with temperature. pH should be checked and readjusted before each experiment.
Basic Protocol Basic Protocol 5
Blocking solution
- For 50 ml of solution:
- 150 μl Triton X-100 (Sigma Aldrich, CAS no. 9036-19-5)
- 25 μl Tween 20 (Sigma Aldrich, product no. P9416)
- 1.5 ml normal goat serum (Jackson, cat. no. 005-000-121) or other serum
- Bring volume to 50 ml with HBSS (Corning, cat. no. 21-021-CV)
Which serum you use depends on the species your secondary antibodies are raised in. Example: use normal goat serum if you are using goat secondary antibodies, normal donkey serum if using donkey secondary antibodies.
Paraformaldehyde (PFA) solution (pH 7-7.5), 4%
- For 100 ml of solution :
- Heat 90 ml distilled, deionized water to 55-60°C on a stirring hotplate in a fume hood
- Add 4 g PFA powder, stirring with magnetic stir bar
- Add 1-3 drops of NaOH; solution should clear
- Add 10 ml of 10× DPBS without calcium and magnesium
Double check that pH is 7-7.5 (add NaOH dropwise if pH is too low and HCl if pH is too high until pH is within desired range)
Washing solution (1× PBS ± 0.1% Tween 20)
For 500 ml of washing solution : Add 500 µl Tween 20 to 500 ml of 1× DPBS.
Author Contributions
Vivienne Fang : Conceptualization, data curation, formal analysis, methodology, visualization, writing—original draft, writing—review and editing; Maureen E. Haynes : Conceptualization, data curation, formal analysis, methodology, visualization, writing—original draft, writing—review and editing; Vanessa Hayashi : Data curation, formal analysis, methodology, visualization, writing—original draft; Erika Arias : Data curation, formal analysis, methodology, visualization, writing—original draft; Jeremy A. Lavine : Methodology, supervision, writing—review and editing; David P. Sullivan : Conceptualization, data curation, formal analysis, methodology, visualization, writing—original draft; William A. Muller : Conceptualization, funding acquisition, methodology, project administration, resources, supervision, writing—review and editing.
Conflict of Interest
The authors declare no conflict of interest.
Open Research
Data Availability Statement
The data, tools, and materials (or their source) that support the protocol are available from the corresponding author upon reasonable request.
Literature Cited
- Rutledge, N. S., Ogungbe, F. T., Watson, R. L., Sullivan, D. P., & Muller, W. A. (2022). Human CD99L2 regulates a unique step in leukocyte transmigration. Journal of Immunology (Baltimore, Md.: 1950) , 209(5), 1001–1012. https://doi.org/10.4049/jimmunol.2101091
- Wu, J., Cai, Y., Wu, X., Ying, Y., Tai, Y., & He, M. (2021). Transcardiac perfusion of the mouse for brain tissue dissection and fixation. Bio-Protocol , 11(5), e3988. https://doi.org/10.21769/BioProtoc.3988

