Measuring Changes in Keap1-Nrf2 Protein Complex Conformation in Individual Cells by FLIM-FRET
Dina Dikovskaya, Dina Dikovskaya, Albena T. Dinkova-Kostova, Albena T. Dinkova-Kostova
Abstract
The nuclear factor−erythroid 2 p45-related factor 2 (Nrf2)−mediated stress response is a major cellular defense mechanism against endogenous and exogenous oxidants, electrophiles, and pro-inflammatory agents. A number of Nrf2 inducers are being developed to therapeutically stimulate this pathway. Inducers are typically sensed by Kelch-like ECH-associated protein 1 (Keap1), a negative regulator and a binding partner of Nrf2. Modifications of Keap1 by oxidants or electrophiles, or its targeting by compounds that disrupt its interaction with Nrf2, alter the conformation of the Keap1-Nrf2 protein complex, which initiates the accumulation of Nrf2 required for mounting a stress response. To detect conformational changes in the Keap1-Nrf2 complex in live cells, we have developed a procedure based on Fluorescence Lifetime Imaging−Förster Resonance Energy Transfer (FLIM-FRET). The procedure includes a FLIM time course in cells expressing fluorescently-tagged Nrf2 and Keap1, followed by an extended analysis pipeline that quantifies changes in fluorescence lifetime of labeled Nrf2. The analysis visualizes and removes intensity-dependent bias in fluorescence lifetime measured with the Time-Correlated Single Photon Counting (TCSPC) approach, thereby improving the accuracy of quantification. The throughput is increased by the whole-experiment analysis within the newly developed FLIM dataset tool (FLIMDAST) and by the time-lapse FLIM described here. This pipeline is also suitable for applications beyond the Nrf2 field that assess small changes in fluorescence lifetime of objects with variable fluorescence intensities measured using TCSPC-based FLIM. © 2020 The Authors.
Basic Protocol 1 : Lipofectamine 2000 transfection
Alternate Protocol 1 : Calcium phosphate transfection
Basic Protocol 2 : Time course with individual FLIM
Alternate Protocol 2 : Time course with time-lapse FLIM
Support Protocol : Measuring Instrument Response Function (IRF)
Basic Protocol 3 : Data analysis in SPCImage
Basic Protocol 4 : Data processing in ImageJ/FIJI
Basic Protocol 5 : Experiment analysis in FLIMDAST
INTRODUCTION
The purpose of this article is to provide a detailed protocol for measuring changes in the protein-protein interactions between Nrf2 and Keap1 in live cells using time-resolved Fluorescence Lifetime Imaging (FLIM).
This procedure monitors a crucial early step in the initiation of the oxidative stress response transcriptional program mediated by transcription factor nuclear factor−erythroid 2 p45-related factor 2 (Nrf2). In unstressed conditions, Nrf2 is bound by Kelch-like ECH-associated protein 1 (Keap1), which recruits the transcription factor to an E3 ligase complex for ubiquitination and subsequent proteasomal degradation (Dinkova-Kostova, Kostov, & Canning, 2017). Modification of Keap1 by endogenous or pharmacologic oxidants and electrophiles (termed inducers) alters the Keap1-Nrf2 complex conformation, leading to inhibition of Nrf2 degradation, and consequently accumulation of Nrf2 and enhanced transcription of Nrf2 target genes (Dinkova-Kostova et al., 2017). The state of the Keap1-Nrf2 complex can be observed in live cells co-transfected with GFP-labeled Nrf2 and mCherry-labeled Keap1 by assessing Förster Resonance Energy Transfer (FRET) between the fluorescent tags (Baird, Swift, Lleres, & Dinkova-Kostova, 2014). Fluorescence Lifetime Imaging (FLIM) provides a way to measure FRET, since FRET reduces fluorescence lifetime, that is, the time from excitation to emission of the FRET donor in the proximity of the FRET acceptor (Suhling et al., 2015) (in our case, GFP-Nrf2 and Keap1-mCherry, respectively). This effect is independent of the expression level of the FRET donor, making FLIM the method of choice for measuring FRET in systems where the level of FRET donor is highly variable, such as GFP-Nrf2 protein.
However, while the fluorescence lifetime of the FRET donor is not affected by its expression level, the equipment used for fluorescence lifetime measurement has a degree of sensitivity to its fluorescence intensity. A common Time-Correlated Single Photon Counting (TSCPC) approach for FLIM detection suffers from the so-called pile-up effect, which skews the measurements in an intensity-dependent manner (Becker, 2015). This effect is diminished but still present even at the low-intensity illumination typically used for TSCPC-based FLIM, and it can be particularly detrimental for analysis of subtle changes in fluorescence lifetimes.
Here, we describe a workflow that overcomes this limitation and allows precise quantification of small treatment-induced changes in fluorescence lifetime within live cells, in the context of variable fluorescence intensity (Fig. 1). In essence, this workflow generates the fluorescence lifetime profile of each measurement across the span of its pixel intensities and compares this profile with a profile of a reference measurement, within the overlapping range of pixel intensities (Fig. 1A and B).
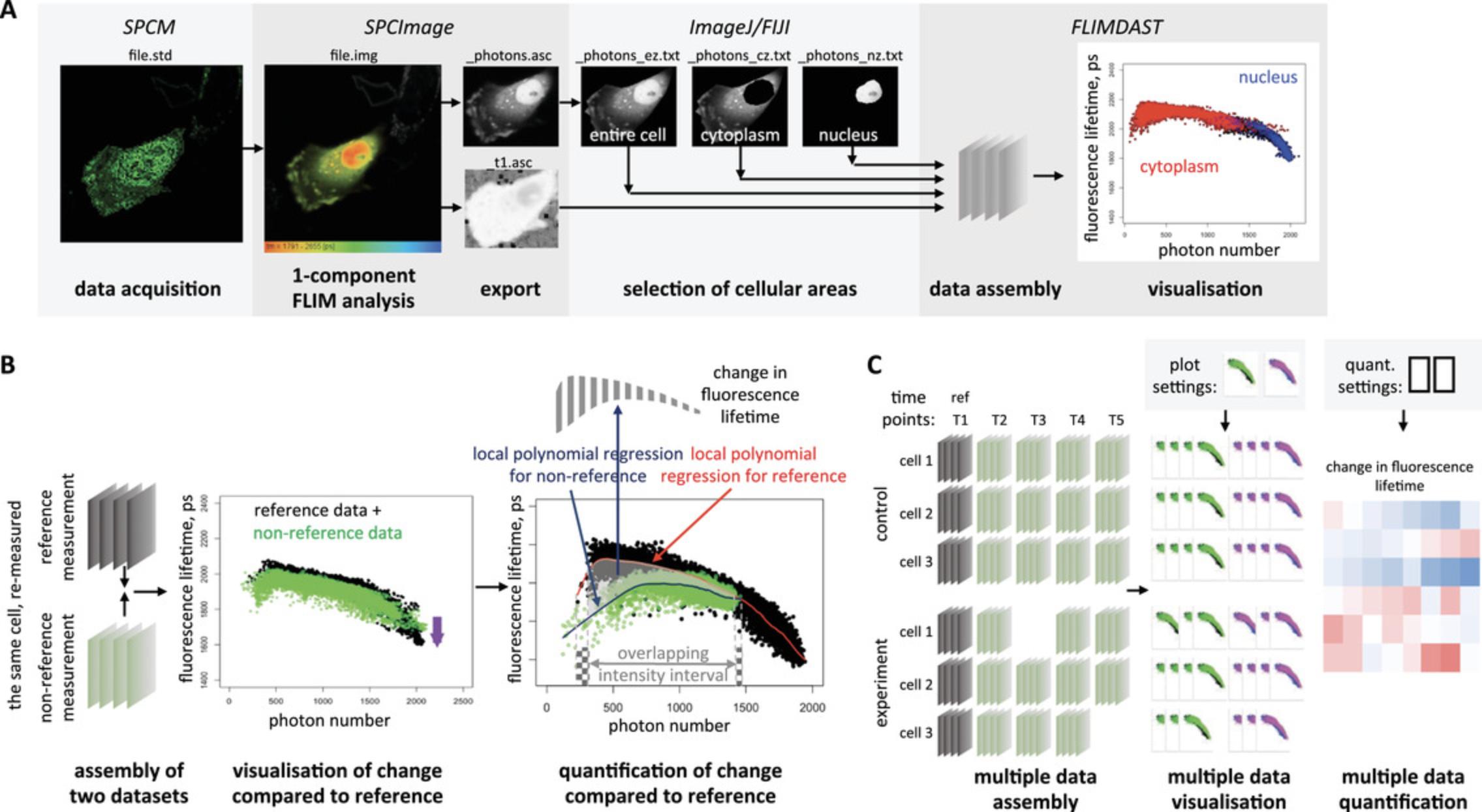
Another limitation of the TSCPC-based FLIM approach is its extremely low throughput, which ultimately limits statistical power of obtained data. Apart from very few custom-made systems, FLIM instruments and analysis software are set up for acquiring, recording, and processing one measurement at a time. To increase throughput, we exploited the inbuilt capability of one such commonly used commercial instrument assembly, the Zeiss LSM 710 microscope equipped with the Becker & Hickl TCSPC module, to establish a procedure for automatic time-lapse FLIM across multiple stage positions, which we describe here. Furthermore, we developed an R-based analytical tool for unbiased quantification of treatment-induced fluorescence lifetime changes in the entire time-course experiment (Fig. 1C) using the algorithm described above (see Fig. 1B).
In this article, we first describe the co-introduction of Nrf2 fluorescently labeled at its N-terminus with super-folder GFP (sfGFP) along with Keap1 labeled at its C-terminus with mCherry fluorophore, or control protein constructs, into cells using standard transfection procedures optimized for microscopy (Basic Protocol 1 for Lipofectamine 2000 transfection and Alternate Protocol 1 for calcium phosphate transfection). We then give step-by-step instructions for the time-course measurement, either using individual FLIM acquisitions (Basic Protocol 2) or multi-position time-lapse FLIM (Alternate Protocol 2) for the LSM 710 (Zeiss)/TCSPC module (Becker & Hickl) combination. Both protocols can be used to measure FLIM in the same cells before and after treatment, including a reference measurement for each cell. The optional Support Protocol describes a procedure for measuring the Instrument Response Function, which increases the accuracy of FLIM measurements. This protocol is only applicable for FLIM acquired with a 2-photon laser. Basic Protocol 3 gives instructions for initial processing of the acquired data in the SPCImage software (Becker & Hickl GmbH) that is compatible with the overall workflow and produces files with fluorescence lifetime and photon number matrices. Further processing of these files in ImageJ/FIJI to select cellular area(s) of interest is described in Basic Protocol 4. The entire experimental dataset, composed of fluorescence lifetime matrices exported from SPCImage and all files generated in ImageJ/FIJI for each measurement, is then analyzed using the R-based app FLIM DAtaSet Tool (FLIMDAST), which we developed for this purpose, as described in Basic Protocol 5. FLIMDAST visualizes changes in fluorescence lifetime in the context of fluorescence intensity, compared to the reference data, and calculates the intensity-independent changes in fluorescence lifetime, based on the user-defined experimental layout, settings, and file locations.
STRATEGIC PLANNING
Choice of Controls
In addition to experimental samples co-expressing sfGFP (super-folder GFP)−Nrf2 and Keap1-mCherry (Fig. 2A-E), one needs to prepare cells expressing control proteins (described below) and/or their combination (Fig. 2F-N). Measurement of the maximal fluorescence lifetime of the FRET donor sfGFP, unquenched by the FRET effect, is the most important control in this procedure. It can be done in cells expressing free sfGFP, sfGFP-labeled Nrf2, or sfGFP-Nrf2 co-expressed with either free mCherry or unlabeled Keap1. This value is essential to estimate the reduction in fluorescence lifetime produced by FRET in the experimental samples, which is the readout of the protein-protein interaction that is ultimately being measured. In addition, if a multi-exponential decay model is used to determine fluorescence lifetimes (see Basic Protocol 3), the fluorescence lifetime of the unquenched FRET donor is often incorporated into the model. It also provides a control for any biological or technological artifacts in treated and/or repeatedly imaged cells that could change fluorescence lifetime independently of the Keap1-Nrf2 complex (such as intrinsic fluorescence of the inducer or vehicle, laser-induced photobleaching, treatment-induced change in cellular autofluorescence, and so on). At least one of the abovementioned controls must be included. Among them, free sfGFP (Fig. 2N) is the least suitable: not only does it not account for the molecular context of the fluorophore known to affect fluorescence lifetime, but it is also usually too bright for FLIM measurements. In contrast, an individually expressed sfGFP-Nrf2 control (Fig. 2I-K) recapitulates both the molecular context and the expression level of the fusion protein used in the experiment. However, its predominantly nuclear location, which can influence fluorescence lifetime, makes it a poor control for the cytoplasmic sfGFP-Nrf2 co-expressed with Keap1-mCherry in experimental samples (Fig. 2A-E). Co-expression of sfGFP-Nrf2 with unlabeled Keap1 (Fig. 2L and M) is arguably the best control, since the sfGFP-Nrf2 environment in this case is identical to that in experimental samples. However, since unlabeled Keap1 is more efficient than Keap1-mCherry in degrading Nrf2 (our unpublished data), the fluorescence of sfGFP-Nrf2 in such control cells can be very low, and most bright cells found in the sample will be cells with suboptimal level of Keap1 (apparent by nuclear localization of sfGFP-Nrf2), which makes it technically challenging to select the correct cells for imaging. Another commonly used control is sfGFP-Nrf2 co-expressed with mCherry (Fig. 2F-H), which accounts for a “background” FRET from the diffuse unbound FRET acceptor. However, in our experience, the expression of free mCherry tends to be too high, which disproportionally reduces fluorescence lifetime.
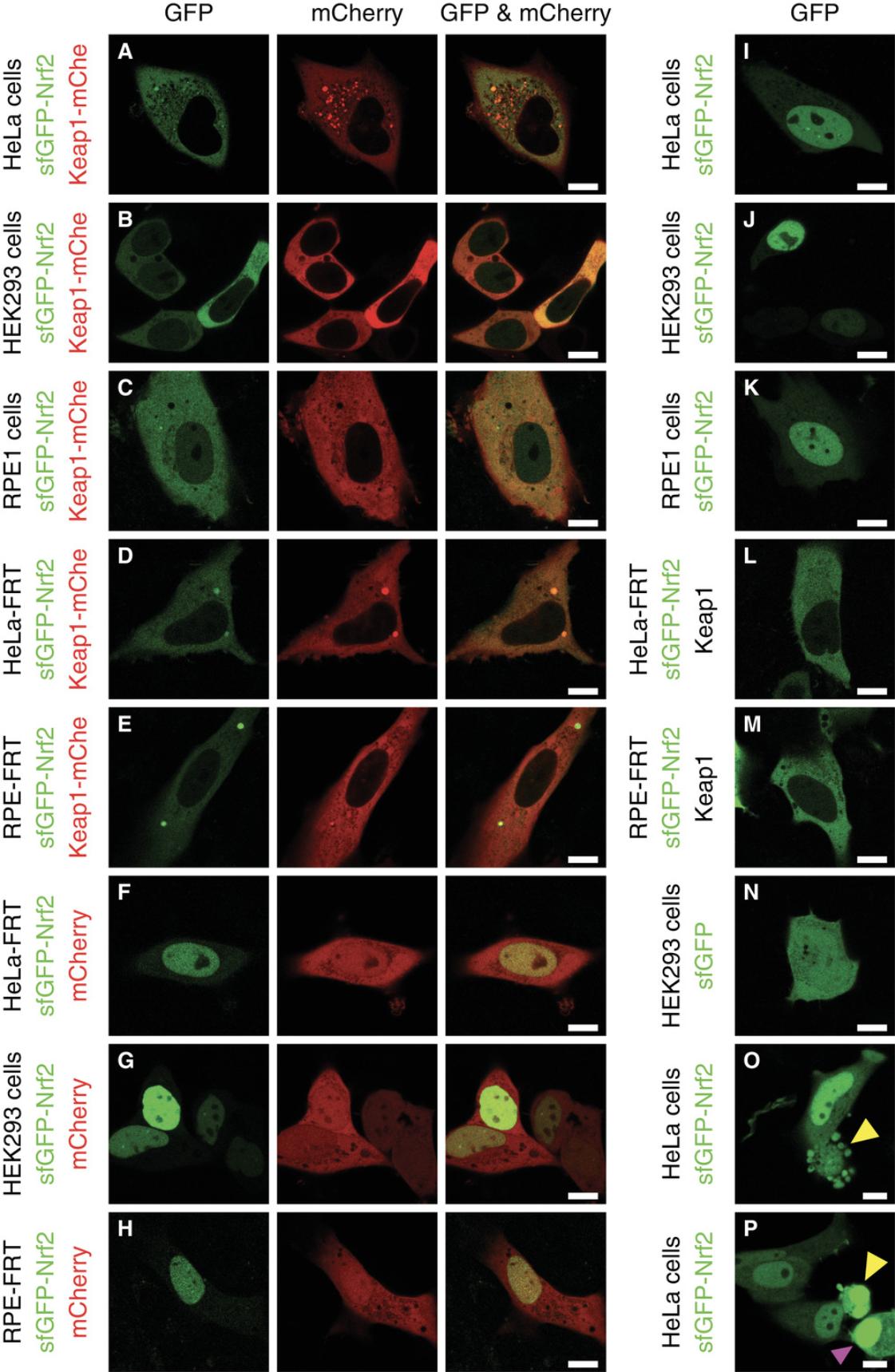
An additional technical control that establishes the sensitivity of the instrumental setup to the FRET-mediated reduction in FLIM is a fusion between sfGFP and mCherry that demonstrates maximal FRET level.
Separate consideration needs to be given to treatment controls. Using vehicle or an inactive structural analog instead of inducer under otherwise identical conditions is the best practice. Other options include inducer treatment of cells co-expressing a control plasmid combination. Specific to our system, cells co-expressing Keap1-mCherry with an sfGFP-labeled Nrf2 mutant that cannot interact with Keap1 provide a good control for inducer effects outside of the Keap1-Nrf2 interaction.
Selecting a Reference Measurement
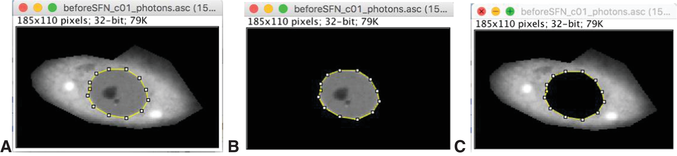
Due to the relative nature of the quantification approach described here, each measurement in the time course requires a reference measurement of the same cell for calculating the change in fluorescence lifetime. One possibility is to use FLIM data taken prior to treatment as a reference for the measurement taken after treatment (see example in Fig. 7A-D later in the article). Please keep in mind that this will generate only one experimental value. In this case, it is essential to have a negative treatment control in a different subset of cells. Another possibility is to specifically dedicate one additional measurement in each cell as a reference, to which all other measurements (before and after treatment) will be compared (see example in Fig. 7E and F later in the article). In this case, one reference could be used for multiple measurements of the same cell. The measurement and its reference should ideally be acquired and analyzed in the same way: in particular, the image resolution, duration of acquisition (Basic Protocol 2 and Alternate Protocol 2), and level of binning during analysis (Basic Protocol 3) must be identical (see Critical Parameters and Troubleshooting for details).
Sample Size
There is a considerable degree of cell-to-cell variability in this assay, partly due to differences in expression levels of Keap1-mCherry (Dikovskaya, Appleton, Bento-Pereira, & Dinkova-Kostova, 2019). Such variability requires large sample sizes to adequately power any statistical analysis of the obtained data. Therefore, within the constraints of the time-course experiment imposed by slow FLIM acquisition and limited duration of treatment, larger samples (e.g., more cells measured for each experimental condition) should be prioritized over additional experimental conditions. Some sample attrition due to cell death or migration during the experiment also has to be expected. In practice, we usually make 8-10 measurements in each condition, some of which contain more than one cell.
Basic Protocol 1: LIPOFECTAMINE 2000 TRANSFECTION
This is an adaptation of a standard cell transfection protocol using the commercially available Lipofectamine 2000 transfection reagent, which is suitable for the majority of cell lines and produces a high level of transfection. Since TCSPC-based FLIM acquisition requires moderate fluorescence intensities, the resulting transfection level might be too high for some cell types. In that case, we suggest further optimization of this protocol, or using Alternate Protocol 1.
Materials
-
HeLa cells (Ohio, ECACC 84121901; https://www.phe-culturecollections.org.uk/products/celllines/generalcell/detail.jsp?refId=84121901&collection=ecacc_gc; other cell types might require additional optimization of the procedure)
-
Complete cell growth medium: DMEM containing 10% fetal bovine serum (for HeLa cells), or a cell-type-specific medium composition (for other cell lines)
-
Purified plasmid DNA in nuclease-free water, 1 µg/µl or similar concentration: sfGFP-Nrf2, Keap1-12 fl-mCherry [referred to here as Keap1-mCherry (Dikovskaya, Appleton, Bento-Pereira, & Dinkova-Kostova, 2019), and optional controls pmCherry-N1 (Clontech 632523), sfGFP-C1 (Addgene 54579), unlabeled Keap1 (Baird, Lleres, Swift, & Dinkova-Kostova, 2013), and/or sfGFP-mCherry fusion]
-
OPTI-MEM I Reduced Serum Medium (ThermoFisher Scientific, cat. no. 3198570)
-
Lipofectamine 2000 Transfection Reagent (ThermoFisher Scientific, cat. no. 11668027)
-
Sterile phosphate-buffered saline (PBS), Ca++/Mg++-free
-
0.05% trypsin/EDTA solution
-
DMEM high-glucose with HEPES, no phenol red (ThermoFisher Scientific, cat. no 21063029) or similar phenol red-free medium
-
Phenol red−free complete growth medium: phenol red−free DMEM (see above) containing 10% fetal bovine serum (for HeLa cells), or a cell-type-specific medium composition (for other cell lines)
-
Cell culture grade 6-well plates
-
0.5- and 1.5-ml microcentrifuge tubes
-
Vortex
-
Glass-bottom dishes: e.g., 35-mm FluoroDish (World Precision Instruments, cat. no. FD35-100) or 35-mm four-compartment CELLview dish (Greiner Bio-One, cat. no. 627975); the dimensions of the glass-bottom dishes must be compatible with the microscope dish/specimen holder
-
Additional reagents and equipment for cell culture, including trypsinization and cell counting (see Current Protocols article: Phelan & May, 2015)
Day 1
1.Seed adherent cells (also see Current Protocols article: Phelan & May, 2015) onto a 6-well plate using 4−5 × 105 cells/well in 2 ml of growth medium per well.
Day 2
2.Co-transfect cells with sfGFP-Nrf2 and Keap1-mCherry plasmids, or a chosen control combination of plasmids (see Strategic Planning and Table 1) using Lipofectamine 2000 according to the following substeps.
- Replace medium in wells with 1.2 ml of fresh full medium no more than an hour before transfection, and leave plates with cells in the cell culture incubator.
The medium should not contain antibiotics.
-
For each transfection, prepare a 1.5-ml tube with 200 µl of OPTI-MEM, and mix in the required amount of plasmid DNA (see Table1).
-
For each transfection, prepare a 1.5-ml tube with 200 µl of OPTI-MEM, and add the required amount of vortexed Lipofectamine 2000 (see Table1). Resuspend.
The volume of Lipofectamine 2000 in microliters should be between 2.5 and 5 times the amount of DNA in micrograms.
Use high-purity DNA (such as produced with Qiagen Plasmid Maxi kit, but not by Qiagen plasmid Mini kit).
-
Leave for 5 min at room temperature.
-
For each transfection, mix the contents of the two tubes (from step b and c) together, and resuspend by gently pipetting up and down three to four times.
-
Leave the mixture for 25 min at room temperature.
-
Transfer each mixture into the corresponding well with cells (step a). Rock the plate gently.
-
Place cells in the cell culture incubator.
-
Optional: replace medium 4-6 hr after transfection. This step is cell-type dependent. For HeLa cells, it is acceptable to leave the transfection reagent in the medium until the day after transfection.
| Plasmid 1 | Amount of plasmid 1 | Plasmid 2 | Amount of plasmid 2 | Lipofectamine 2000 | |
|---|---|---|---|---|---|
| Experiment | sfGFP-Nrf2 | 1.2 µg | Keap1-mCherry | 0.8 µg | 6 µl |
| Choice of controls | sfGFP-Nrf2 | 1.2 µg | Keap1 | 0.8 µg | 6 µl |
| sfGFP-Nrf2 | 1 µl | 3 µl | |||
| sfGFP-Nrf2 | 1 µl | pmCherry | 0.7 µg | 5 µl | |
| sfGFP | 0.8 µl | 2.4 µl | |||
| sfGFP-mCherry | 0.8 µl | 2.4 µl | |||
Day 3
3.Wash cells twice with PBS and trypsinize (also see Current Protocols article: Phelan & May, 2015) with 0.4 ml per well of 0.05% trypsin/EDTA. Collect cells in complete growth medium, count (any method of cell counting is suitable; see Current Protocols article: Phelan & May, 2015), and seed onto a glass-bottom dish at 2−4 × 105 cells per 3.5-cm dish or 8 × 104 cells per compartment of a four-compartment 3.5-cm dish.
Day 4
4.At least 2 hr prior to imaging, wash cells one to two times with phenol red−free DMEM and replace medium on cells with 1.8 ml of phenol red−free complete medium. Inspect cells under fluorescence microscope as described in Basic Protocol 2 or Alternate Protocol 2.
Alternate Protocol 1: CALCIUM PHOSPHATE TRANSFECTION
Calcium phosphate−mediated transfection is less efficient than other transfection methods, but in easy-to-transfect cells such as HeLa or HEK293, it provides moderate transfection levels sufficient (and in fact preferable) for FLIM experiments. The advantages of this method over Lipofectamine 2000 transfection are: (i) lower cytotoxicity; (ii) intermediate level of fluorescence intensity better compatible with TSCPC-based FLIM; and (iii) significantly lower cost. Additionally, it can be performed at low cell confluency, eliminating the need to split cells after transfection to achieve the low cell density required for imaging, and therefore reducing the time needed for cell preparation from 4 to 3 days.
Materials
-
HeLa cells (Ohio, ECACC 84121901; https://www.phe-culturecollections.org.uk/products/celllines/generalcell/detail.jsp?refId=84121901&collection=ecacc_gc; other cell types might require additional optimization of the procedure)
-
Complete cell growth medium: DMEM containing 10% fetal bovine serum (for HeLa cells), or a cell-type-specific medium composition (for other cell lines)
-
Plasmid DNA in nuclease-free water, 1 µg/µl or similar concentration: sfGFP-Nrf2, Keap1-mCherry (Dikovskaya, Appleton, Bento-Pereira, & Dinkova-Kostova, 2019), and optional controls [pmCherry-N1 (Clontech 632523), sfGFP-C1 (Addgene 54579), unlabeled Keap1 (Baird, Lleres, Swift, & Dinkova-Kostova, 2013) and/or sfGFP-mCherry fusion]
-
DMEM high-glucose with HEPES, no phenol red (ThermoFisher Scientific, cat. no 21063029) or similar phenol-free medium
-
Phenol red−free complete growth medium: phenol red−free DMEM (see above) containing 10% fetal bovine serum (for HeLa cells), or a cell-type-specific medium composition (for other cell lines)
-
2× HSB (see recipe)
-
Sterile water
-
2 M CaCl2
-
Glass-bottom dishes, such as 35-mm FluoroDish (World Precision Instruments, cat. no. FD35-100); the dimensions of the glass-bottom dishes must be compatible with the microscope dish holder
-
0.5- and 1.5-ml microcentrifuge tubes
-
15-ml conical tubes (e.g., Corning Falcon)
-
Vortex
-
Additional reagents and equipment for cell culture, including trypsinization and cell counting (see Current Protocols article: Phelan & May, 2015)
Day 1
1.Seed adherent cells onto 3.5-cm glass-bottom dishes at 2 × 105 cells/dish in 2 ml of complete cell growth medium.
Day 2
2.Co-transfect cells with sfGFP-Nrf2 and Keap1-mCherry or a control combination (see Strategic Planning and Table 2) using calcium phosphate transfection according to the following substeps.
-
Within 1 hr before transfection wash cells once with phenol red−free DMEM and replace medium with 1.5 ml of fresh phenol red−free complete medium. Leave the dishes with cells in the incubator.
-
For each transfection, prepare a 15-ml Falcon tube with 200 µl 2× HSB (Solution I, Table2).
-
For each transfection, combine water (up to 200 µl final volume), 25 µl of 2 M CaCl2, 3 µg sfGFP-Nrf2 plasmid DNA, and 2 µg Keap1-mCherry plasmid DNA, or a control plasmid combination (Solution II, Table2and see note below), in a 1.5-ml microcentrifuge tube.
Considerations for DNA amount used for transfection:
Use ∼3:2 ratio of Nrf2:Keap1Use 2 µg of labeled Nrf2 per 3.5-cm dish if expressing without Keap1If co-expressing with labeled or unlabeled Keap1, increase the amount of labeled Nrf2 to 3 µg per 3.5-cm dishUse 1 µg or less of pmCherry-N1 or sfGFP-C1 per 3.5cm dish.
-
Slowly add the above DNA/CaCl2solution to the 2× HBS with continuous vortexing (1 drop/s into the middle of the tube). Continue to vortex for another 10 s.
-
Transfer the entire mixture onto the cells, evenly distributing drops throughout the dish. Put the dish back into the incubator.
| Solution I | Solution. II | ||||||
|---|---|---|---|---|---|---|---|
| 2× HBS, µl | Plasmid 1 | Amount of plasmid 1 | Plasmid 2 | Amount of plasmid 2 | H2O, µl | CaCl2 µl | |
| Experiment | 200 | sfGFP-Nrf2 | 3 µl | Keap1-mCherry | 2 µl | 170 | 25 |
| Choice of controls | 200 | sfGFP-Nrf2 | 3 µl | Keap1 | 2 µl | 170 | 25 |
| 200 | sfGFP-Nrf2 | 2 µl | 173 | 25 | |||
| 200 | sfGFP-Nrf2 | 2 µl | pmCherry | 1 µl | 172 | 25 | |
| 200 | sfGFP | 1 µl | 174 | 25 | |||
| 200 | sfGFP-mCherry | 1 µl | 174 | 25 | |||
Day 3
3.Unless fresh medium interferes with the measurements, replace medium with 1.8 ml fresh phenol red−free complete medium at least 2 hr prior to imaging. Inspect cells under fluorescence microscope as described in Basic Protocol 2 or Alternate Protocol 2.
Basic Protocol 2: TIME COURSE WITH INDIVIDUAL FLIM
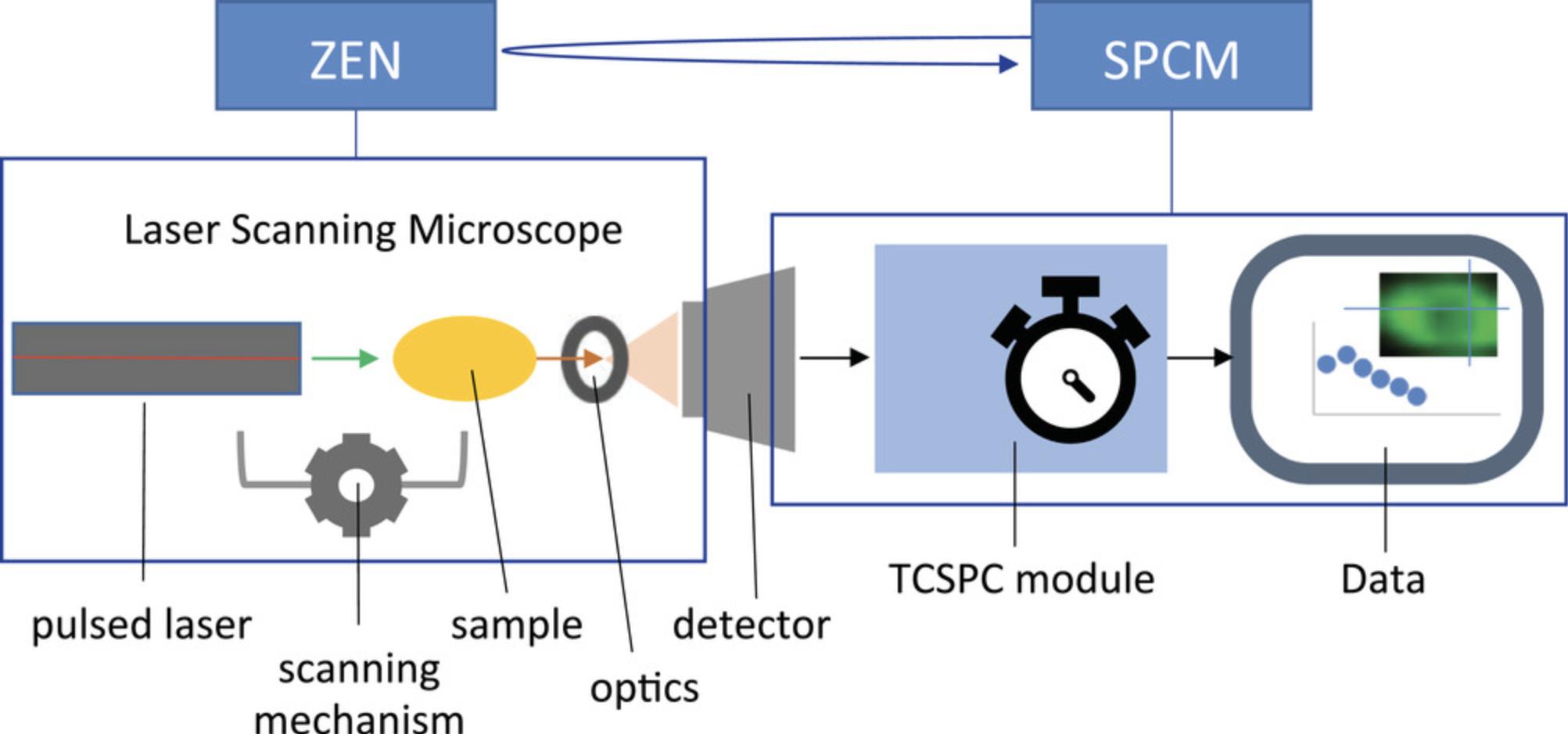
This protocol describes measurements of fluorescence lifetime using a TCSPC-based approach that works by recording the exact time of emission of individual photons after fluorophores are excited by pulsed laser illumination (Becker, 2015). The sample is scanned by a high-frequency pulsed laser, and the distribution of times between each photon emission and the respective laser pulse (so-called micro-time) accumulated during a few minutes long FLIM acquisition is generated for each pixel of the image. The measurements are performed by an assembly of a laser scanning microscope and a TCSPC module operated by two separate software applications that run simultaneously. The laser operation, light path and optics, sample position, and detector are controlled by Zen software (Zeiss), while the processing of detector outputs to generate the time-stamped photon map is controlled by the SPCM software (Becker & Hickl GmbH) (Fig. 3). The two software applications are not automatically coordinated, and the user must manually set, start, and stop the measurement process in each application. The measurement produces a three-dimensional image (X, Y, and micro-time) in a proprietary format, with an .sdt extension, readable by the SPCImage analysis software. Here we describe, from the user perspective, sample handling and setting up and operating both the Zen and SPCM software for repeated measurements of fluorescence lifetime in live cells. The protocol requires prior knowledge of standard fluorescence imaging techniques with the laser scanning microscope and does not include instrument assembly and initial software setup normally performed by the manufacturers.
Materials
-
Transfected cells in glass-bottom dishes, produced in Basic Protocol 1 or Alternate Protocol 1
-
Immersion oil with a refractive index 1.520 (if using oil-immersion lens)
-
Lens tissues
-
Phenol red−free complete medium (as described in Basic Protocol 1)
-
Compound (inducer) to be tested
-
Vehicle (such as DMSO or acetonitrile, in which the compound is dissolved)
-
This protocol is designed for the Zeiss LSM 710 confocal microscope operated by Zen software, equipped with a pulsed laser illumination source (either single photon−based, such as InTune laser with 40 MHz pulse frequency and wavelength tunable between 488 and 640 nm, or a two-photon, such as Chameleon laser (Coherent) with 80 MHz pulse frequency and wavelength tunable between 690 and 1064 nm), and HPM-100-40 GaAsP hybrid detector(s) installed at the LSM Binary GaAsP module (BiG) NDD port of the microscope or directly coupled to the rear of the scanhead. Fluorescence Lifetime measurements are performed by an attached Simple-Tau module (Becker & Hickl GmbH) for TCSPC processing, operated via the SPCM software installed on a separate computer. For other FLIM and microscope assemblies, the procedure needs to be modified according to the manufacturer's instructions. For live-cell FLIM, the microscope must also be equipped with an environmental chamber, which maintains temperature at 37°C, CO2 source that provides a humidified gas supply with 5% CO2 content, and a heating Petri-dish holder insert or other specimen holder suitable for cell culture dishes. The CO2 source is not necessary for short-term experiments with cells growing in CO2-independent media. For more details, please see The bh TCSPC handbook (Becker, 2017).
1.Switch on the microscope and the main computer; initiate Zen software.
2.Pre-warm environmental chamber and heated dish holder insert to 37°C.
3.Set humidified CO2 source to 5% CO2.
4.If the instrument contains beam splitter-detector assembly that houses fluorescence filters, make sure the correct filters are installed. For measuring fluorescence lifetime of sfGFP-Nrf2, a GFP emission bandpass filter is used (e.g., 500-550 nm or similar). For a 2-photon FLIM, an additional short-pass filter (e.g., 690 nm) is required that blocks far-red and infrared light above the specified cut-off.
5.Switch on the Simple-Tau module and the dedicated computer.
6.Place the dish with cells in to a specimen holder and move it to the imaging position, supply with CO2, and leave to equilibrate at 37°C for at least 20 min.
7.Using standard confocal optical path, select a cell with intermediate green (sfGFP-Nrf2) and red (Keap1-mCherry) intensities, and normal cell morphology—see examples in Figure 2.
8.Make a snapshot of the selected cell in green and red channels, using standard confocal lasers, with laser power and gain adjusted to avoid saturation of intensities.
9.In Zen, define the beam path for FLIM in the Light Path window. Depending on the microscope setup, choose the main dichroic beam splitter (MBS) in the “Visible light” or “Invisible light” group used for the pulsed laser (such as f-MBS 405/490c/640c for InTune laser or MBS 760+ for two-photon laser), and select “plate” for the other group. For the FLIM detectors at NDD port, guide the light into “Fiberout.”
10.In Zen, set up the following acquisition parameters in the Acquisition Mode window:
1.Frame size: 512 × 512 (pixels) 2.Scan Mode: Frame 3.Direction to —-> (unidirectional) 4.Acquisition speed: 9 or similar 5.Averaging: Number − 1
11.In Zen, select a suitable pulsed laser. If the wavelength is tunable, set up excitation to 490 nm for InTune, or to 920 nm for 2-photon laser.
12.Reduce laser power to minimal (e.g., 0.2).
13.Set pinhole to 1 AU.
14.Select zoom suitable for the entire range of samples that minimizes the empty spaces around cells.
15.Open the SPCM software on the computer that operates the TCSPC module.
16.If the Simple-Tau box is switched on, the SPCM will start in hardware mode—select OK. If, instead, the SPCM starts in simulation mode, check that all instruments are connected and switched on, and re-start the SPCM software.
17.In SPCM, set up the following parameters (or check that they are such in the used Predefined Setup):
In the main panel:
- select “StopT”
- under “Time,” select “Collection” and type “120”
This will set up the duration of FLIM acquisition that will automatically stops after 2 min.
In the System Parameters window:
- StopT: enabled
- under “Data format,” “ADC resolution” should be “256”
This is the time resolution of the fluorescence lifetime measurements, represented by the number of discrete “time bins” in the time axis for the fluorescence decay curve. Higher values could be used if desired; however, this dramatically increases the file size and, in our experience, does not significantly improve the precision of measurements in our system.
- under “Page Control,” “Image pixels X” and “Image pixels Y” should be “512” and “512,” to correspond to image size in Zen software.
Keep the rest of the parameters in System Parameters window as set during installation.
18.Click “continuous” in Zen. This will start the laser.
19.In the DCC-100 Detector Control window in SPCM, click “Enable Output.”
20.Optional : If necessary, refocus, guided by the low-level signal display in Zen (Fig. 4A) or CFD rate in SPCM (the highest when in focus). If the focus offset between standard and FLIM detector is known, and the sample is well equilibrated, refocusing could be done by stepping through Z-position within the Focus panel in Zen, after typing the value of the focus offset into the Step Size window.
21.Increase laser power to achieve CFD rates between 5 × 104 and 1 × 106, ideally 1−5 × 105 events per second.
22.In SPCM, click “Start.” The display window will become active to show the accumulation of photons.
23.When acquisition is finished (in 2 min), the total event count and the file size will be displayed. In Zen, manually stop the laser. In the DCC-100 Detector Control panel, click “Disable Outputs.”
24.In SPCM, save the file (in the Save panel, write or choose file name, and click “Save”).
25.Repeat steps 18-24 for all cells of interest.
26.Prepare an inducer or vehicle solution at a 10-fold higher concentration than the desired final concentration, in phenol red−free complete medium, in a final volume in excess of 200 µl per dish. Warm up to 37°C immediately before use.
27.Add 200 µl of the inducer or vehicle solution to 1800 µl medium (for 3.5 cm dish) or 60 µl to 540 µl medium (for a chamber of a 4-chamber dish), being careful not to move the sample and to return the instrument to the same position as before treatment.
28.Leave for 3-10 min to re-equilibrate.
29.Refocus on the same cells, using the standard lasers, and update the Position List.
30.Repeat steps 18-24 for all cells, after the desired incubation times.
31.Re-image cells with standard lasers and settings identical to those used in step 8.
Alternate Protocol 2: TIME COURSE WITH TIME-LAPSE FLIM
The procedure is similar to Basic Protocol 2, but sequential acquisition of FLIM measurements for several positions and time points is done automatically. This is an adaptation of a microscope-controlled time series routine in SPCM that accepts an external trigger to start each new measurement and allows automatic data collection and file recording. The time course details (such as positions and time intervals) are set in Zen, which sends a trigger signal to SPCM at the beginning of each imaging step as it runs the time-lapse experiment. In this scenario, unlike in Basic Protocol 2, acquisition in Zen is not continuous and, therefore, its duration time has to be specified to be slightly longer than required for each FLIM measurement within SPCM. This is achieved by using a high averaging number and slow acquisition speed (long pixel dwell time) in Zen, and adjusting the recording time in SPCM to fit the expected scan time. This imposes an additional limit on the duration of FLIM acquisition. To overcome the potential loss of accuracy due to the lower number of recorded photons, we reduce the spatial resolution of the data (using a 256 × 256 image size). This protocol significantly streamlines FLIM measurements to save time and user manual involvement, and allows more frequent data collection and/or bigger sample size than those possible with Basic Protocol 2.The pitfalls include potential loss of in-focus information if cells migrate out of focus (as there is no possibility to re-focus between measurements), the danger of blocked recording of all subsequent measurements due to even a temporary detector overload in one of the positions, the need to choose cells with similar level of fluorescence intensity to be measured with the same laser settings, and the sequential naming of the recorded output files that does not discriminate between positions and time points.
Materials
- Same as for Basic Protocol 2
1.Follow steps 1-9 of Basic Protocol 2. In step 7, recording positions of the cells into the Position List is required and not optional.
2.Using Zen and SPCM settings and the procedure outlined in steps 10-21 of Basic Protocol 2 for the brightest and dimmest among selected cells, adjust the laser power to a level suitable for all measurements.
3.In Zen, set up the following acquisition parameters in the Acquisition Mode window:
1.Frame size: 256 × 256 (pixels) 2. Scan Mode: Frame 3.Direction to —-> (unidirectional) 4.Averaging: Number – 16, Mode – Frame, Method – “mean” 5.Bit depth to 8 bit
It might be necessary to click on “Show all” at the top of the tab to see some of these options.
4.Adjust Speed/Pixel Dwell to achieve Scan Time that is at least 5 s longer than the desired individual FLIM acquisition time. (e.g., for 80 s FLIM acquisition, Scan Time can be 1 min 29 s).
5.Select “Time Series” in the multidimensional acquisition area—this will open the Time Series panel. Within it, select the number of cycles (i.e., the number of times each cell to be measured) and the interval between repeated measurements of the same cell.
6.Set up laser power to the number optimized in step 2.
7.In the System Parameters window of SPCM, choose:
- Operation Mode: “FIFO image”
- StopT: enabled
- Cycles: type the total number of measurements (a product of the number of stage positions multiplied by the number of measurements for each position)
Note that the meaning of “cycle” in SPCM is different from that in Zen
- Autosave: “Each cycle”
- Spec data file: write a name for the measurement file and select the location where to save it (the folder icon next to the name).
With these settings, each measurement will be recorded with the same name followed by number, sequentially throughout the time-lapse.
6.In the field “Trigger”: select the arrow pointing up-right

This ensures that the start of each FLIM acquisition is synchronized with Zen time-lapse.
7.In the field “Collection”: type the desired duration of acquisition (which is shorter than the Scan Time set up in Zen by more than 5 s) 8.In the field “Repeat”: type the same number as in the “Collection” field 9.In Data Format , ADC Resolution: 256 10.In Page Control , Image pixels X: 256; Image pixels Y: 256
The reduced spatial resolution compensates for the decreased duration of acquisition, to achieve similar numbers of photons per pixel.
Keep the rest of the parameters in System Parameters window as set during installation.
8.In the DCC-100 Detector Control panel, click “Enable Output.”
9.In the main panel of SPCM, click “Start.”
10.In Zen, double-check that the latest version of the Positions List is loaded and the laser power is set up to the level established in step 2. Click “Start Experiment.”
11.If treatment is required: prepare treatment solution or vehicle at a 10-fold higher concentration than the desired final concentration in phenol red−free complete medium and add to the dish, as in steps 26-28 of Basic Protocol 2.
12.At the desired time, re-focus using standard lasers, and further adjust focus as in step 20 of Basic Protocol 2, if necessary. Update and save the Positions List.
13.Return to parameters set in steps 3, 4, 6, and 7.
14.In the Time Series window in Zen, choose new number of measurements (“number of cycles”) and the interval between repeated measurements of the same cell as in step 5.
15.In the System Parameters window of SPCM, write a new number of cycles and a new file name in Spec data file.
16.Repeat steps 8-10.
Support Protocol: MEASURING INSTRUMENT RESPONSE FUNCTION (IRF)
The photon data traces generated in Basic Protocol 2 and Alternate Protocol 2 include Instrument Response Function (IRF), an instrument-generated signal that needs to be removed before further analysis. The shape of IRF can be estimated in the SPCImage analysis software from the rising edge of the data trace, giving the so-called synthetic IRF. However, using the directly measured IRF can improve the accuracy of fluorescence lifetime measurements (Dikovskaya et al., 2019). For 2-photon FLIM systems, IRF can be determined from the photon trace of gold nanorods that produce ultrafast luminescence that outlines instrument signal in a broad fluorescence spectrum (Talbot et al., 2011). This protocol describes preparation and measurement of a gold nanorod sample, as well as extracting the IRF curve from obtained data trace within SPCImage.
Materials
-
10-nm gold nanorod suspension (Sigma-Aldrich, cat. no 716820)
-
Nail polish
-
PAP pen
-
Glass slide
-
Glass coverslip
-
Additional reagents and equipment for FLIM measurements (Basic Protocol 2)
Slide preparation
1.Resuspend gold nanorod suspension by vortexing, take a small (10-100 µl) aliquot, and dilute in water (1:1).
2.Make a small circle with a PAP pen in the middle of a glass slide. Make a mark near it with a black marker.
3.Drop 10-20 µl of diluted gold nanorods into the center of the circle.
4.Let air dry.
5.Cover with glass coverslip and seal with nail polish.
Measurements
6.Set up the microscope in the same way as for FLIM measurements in cells (see Basic Protocol 2), except the slide holder is used instead of a dish holder:
-
Pre-warm environmental chamber to 37°C.
-
If the instrument contains a beam splitter/detector assembly that houses fluorescence filters, make sure the filters are correct: GFP emission bandpass filter (e.g., 500-550 nm or similar), and a short-pass filter (e.g., 690 nm) that blocks far red and infrared light above the cut-off wavelength.
-
Start Zen software.
-
Switch on the Simple-Tau module; start SPCM software.
-
Place the slide with nanorods in the slide holder, with high-magnification oil-immersion lens, ideally the same as used for FLIM measurements.
-
Using the “Locate” tab in Zen with transmitted light, focus on the mark near the dried nanorods.
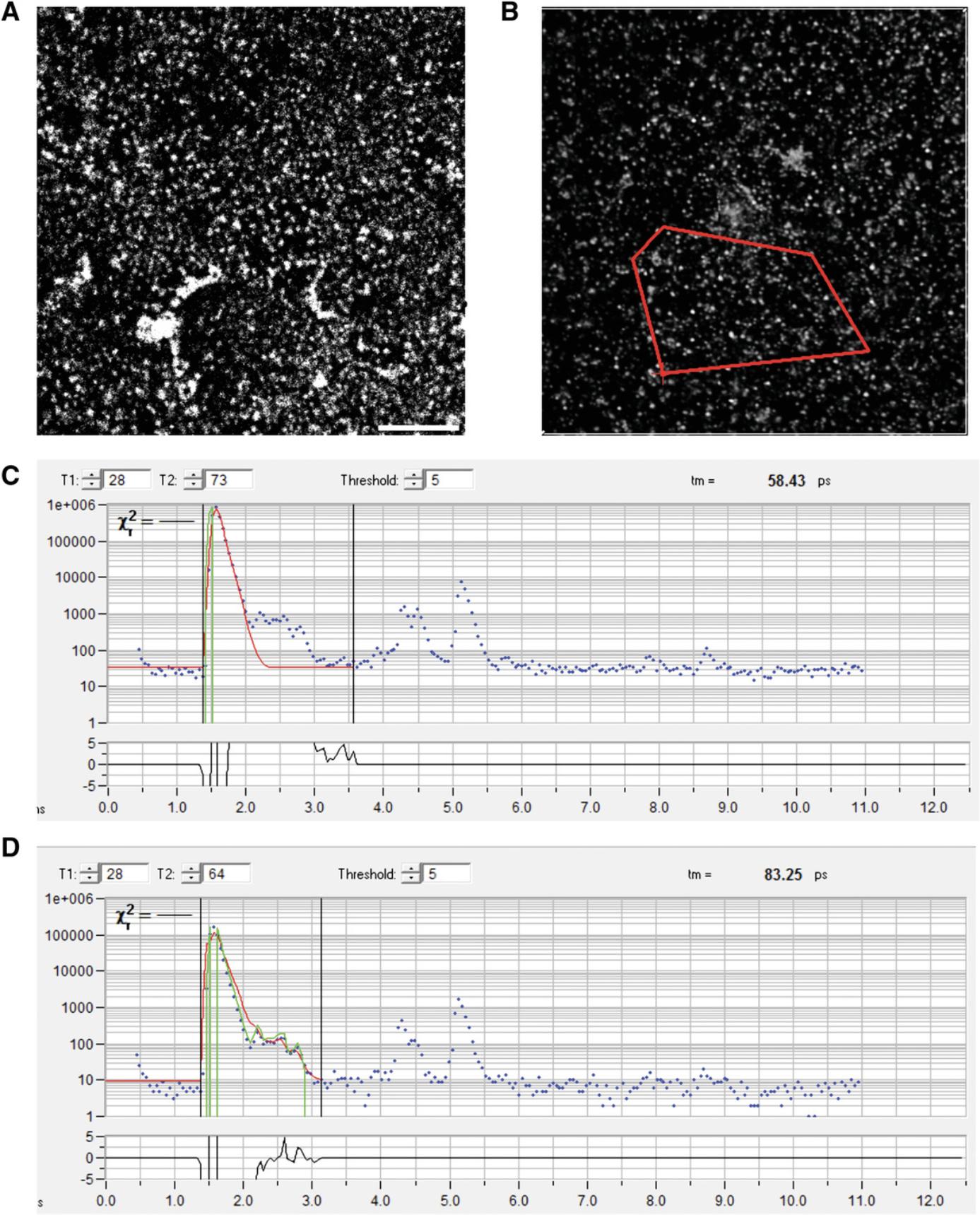
-
Using 2-photon laser, focus further on the nanorods, which will appear as fluorescent speckles (Fig.5A).
-
Set up Zen and SPCM software for FLIM, as in steps 9-14 and 17 of Basic Protocol2.
-
Set up 2-photon laser for 920 nm; reduce the power to minimal.
7.In the DCC-100 Detector Control pane, click “Enable Output.”
8.Click “continuous” in Zen. This will start the laser.
9.Optional : If necessary, refocus, guided by the low-level signal in Zen or CFD rate in SPCM (CFD rate is the highest when in focus).
10.If necessary, increase laser power to achieve CFD rates between 5 × 104 and 1 × 106 events per second.
11.In SPCM, click “Start.” The display window will become active to show the accumulation of photons.
12.When acquisition is finished (automatically in 2 min or manually by clicking “Stop” at an earlier time), manually stop the laser illumination in Zen. In the DCC-100 Detector Control panel, click “Disable Outputs.”
13.In SPCM, save the file.
Loading IRF into SPCImage settings
14.Open SPCImage and import file with IRF data (SPCImage, File -> Import).
15.Select an area and click on the Lock icon

(Fig. 5B).
16.Optional : Move the right black vertical line in the bottom panel closer to the beginning of the curve to remove smaller “waves” that follow the main part of the data trace (Fig. 5C).
17.Select IRF -> Copy from decay data, or click on “Curve to IRF” icon

.
18.Click on the camera icon

.
19.Save the file (File -> Save).
Basic Protocol 3: DATA ANALYSIS IN SPCImage
The fluorescence lifetime in each pixel of a FLIM data file acquired in Basic Protocol 2 or Alternate Protocol 2 is determined by fitting an exponential decay model to the photon data trace, after removing IRF by de-convolution (Becker, 2015; Lakowicz, 2006). This protocol explains how to do this in SPCImage. Each measurement is processed individually; however, in order to produce compatible results, the same analytical parameters should be used for the entire experiment (all apart from the Shift, see below). After initial optimization, the parameters can be stored with the settings and reused, to speed up the workflow. We suggest using a single-exponential model that has the lowest requirement for the size of the dataset (the number of photons in trace) for accurate evaluation. The protocol generates two export files (one with photon numbers, and another with fluorescence lifetimes in each pixel), as well as an optional illustration, from each measurement for further processing. Selection of areas of interest and recording the fluorescence lifetime statistics are not required at this stage, as it will be done outside SPCImage as described in Basic Protocols 4 and 5.More detailed description of SPCImage can be found in the BH TCSPC Handbook (Becker, 2017).
Materials
- SPCImage software. The protocol below is for versions 5 and 6.Other versions might vary.
1.Optional : If IRF was measured and loaded into SPCImage settings (Support Protocol), open the file in which the IRF has been saved into settings (SPCImage, File -> Open), and reload settings using the curled arrow icon

.
2.Open the data file in SPCImage, either by importing a recorded .sdt file (SPCImage, File -> Import), or by sending the active file from SPCM to SPCImage (SPCM, Main -> Send Data to SPCImage).
3.Optional : If using measured IRF that has been loaded into SPCImage settings (Support Protocol), load the stored conditions using the curled arrow icon

.
Alternatively, the software will calculate synthetic IRF based on the fluorescence decay data in the pixel marked by the blue cross within the image (Fig. 6A, 1). By default, this is the brightest pixel of the image. If this method for calculating IRF is used, check if the blue cross in the image represents the artifact-free area of the image. If necessary, use the cursor to re-position the blue cross, and use IRF -> Auto to re-calculate synthetic IRF from the newly chosen pixel.
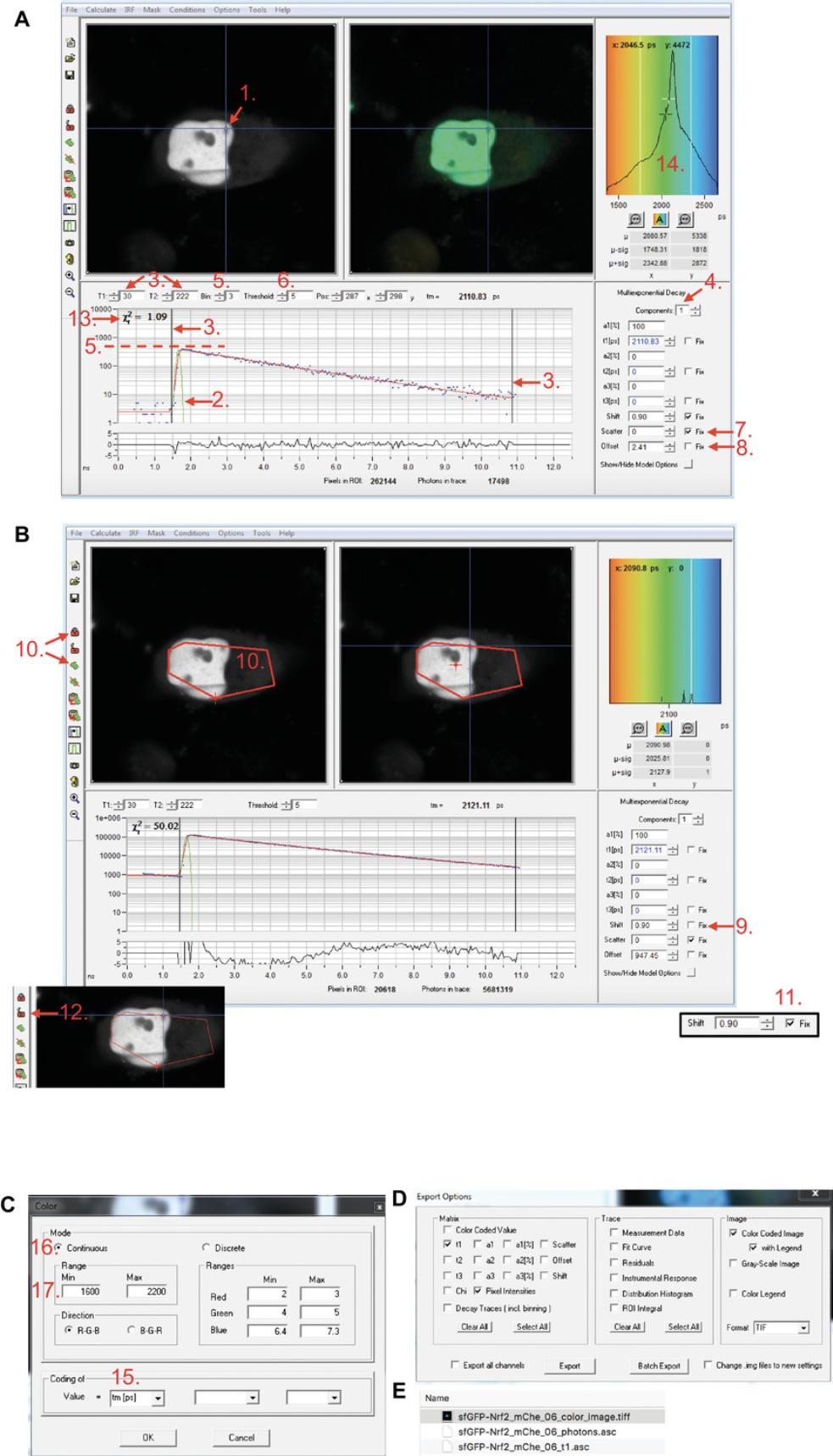
Uploaded (measured) or synthetic IRF will be displayed as a green line overlaying the data points in the data window (Fig. 6A, 2).
4.Set up the following parameters:
1.Under Option -> Model, choose either Multiexponential Decay or Incomplete Decay (with 12.5 ns Repetition Time for 80 MHz pulsed laser or 25 ns Repetition Time for 40 MHz pulsed laser); both models are suitable for sfGFP FLIM. However, the model should be consistent throughout the experiment for comparable results). Click “OK.” 2.In the line above the data curve, set T1 to position the first black line at the start of the rising edge of the data, and T2 to position the second black line at the end but still within the data range (Fig. 6A, 3), either by typing the numbers, or by dragging the lines. Keep these values the same throughout the experiment for comparable results. 3.In the right part of the data panel, choose the number of components in the exponential fitting (Fig. 6A, 4).
For the procedure described here, we typically use 1-component analysis.
4.In the line above the data curve, choose the Bin number that increases the number of events in the dataset to the required level: for a single-component exponential decay fitting, the peak of the data curve should be equal to or above 100 (photons) on the Y axis, and for a 2-component analysis, it should be equal or above 1000 (Fig. 6A, 5). This binning should be kept the same throughout the experiment for comparable results.
Typically, we select Bin 3 or 4.
- In the line above the data curve, type the threshold that defines the minimum photon number in a pixel below which the fluorescence lifetime will not be determined (Fig. 6A, 6).
Typically, for binning 3 or 4, we use a threshold of 100.
- If the synthetic IRF is used, check that the green line roughly outlines the rising edge of the data curve. If it is far away from the rising edge, re-calculate IRF: make sure that everything on the right side of the window is deselected, move the cursor to reposition the blue cross in the image window, and update IRF as in 3. Repeat until the IRF line is close to the data curve. 7.Fix Scatter to zero (Fig. 6A, 7). 8.If Incomplete decay model is used, fix Offset to zero; otherwise, leave unfixed (Fig. 6A, 8)
- Estimate “Shift” value: make sure that “Shift” is left unfixed (Fig. 6B, 9). In the image window, roughly outline the area of interest (or select a representative area) using the selection tool (

), and click the Lock icon (

) to pool the data together into one dataset (Fig. 6B, 10). This calculates the best-fitting model for selected area and populates the parameters at the right part of the panel with fitted values. Fix Shift to the value that is displayed (Fig. 6B, 11). Click the Unlock icon (

) (Fig. 6B, 12). 10. Optional : if 2-component analysis is used, the t2 values (fluorescence lifetime of unquenched donor) could be fixed to the fluorescence lifetime of sfGFP or sfGFP-Nrf2-expressing cells, determined in a separate experiment.
Fixing t2 value speeds up the fitting process and reduces the required number of photons, compared to the unfixed 2-component model.
- The goodness of fit χ2 value displayed in the left top corner of the data panel (Fig. 6A, 13) could be used to optimize the fitting parameters (the lower the χ2, the better the fit). The chosen model should be applied to all data within an experiment for comparable results.
Note that in this software application, the χ2 value increases with the size of the dataset and the total brightness of the binned pixel or analyzed area; therefore, the fitting model should be optimized on the same image area.
- Once the settings are optimized, they can be saved using the camera icon

and reused in the next imported file by clicking on the curled arrow icon

. If measured IRF is used, saved settings should include it, since it will overwrite the previous settings containing imported IRF.
5.Select Calculate -> Decay matrix.
6.If illustrations of fluorescence lifetime images are required : In the fluorescence lifetime frequency histogram at the right top corner of the software, right-click to open the Color control panel (Fig. 6A, 14). This will open a new panel (Fig. 6C). In “Value = ”, select “tm” (Fig. 6C, 15). In “Mode,” choose Continuous (Fig. 6C, 16). In “Range,” type the desired min and max values for color display (Fig. 6C, 17). This should be the same in the entire experiment, for comparable display results.
7.Export the data : Choose File -> Export. In the Export Option window that will open (Fig. 6D), select “t1” and “Pixel Intensities” under Matrix, and “Color Coded Image” and “with Legend” under Image. This will generate two data files (name_t1.asc and name_photons.asc) and the color image with the legend (Fig. 6E).
Optional : if 2-component analysis is used, the “a1,” “t2,” and “a2” need to be checked in addition to “t1” and “Pixel intensities” under Matrix. This will generate four files that will need to be further processed to generate the matrix with mean fluorescence lifetimes (see Basic Protocol 4).
8.Save the analyzed file. This will generate a new file with an .img extension that contains the settings and the calculated fluorescence lifetimes. This file serves as a record for the fluorescence lifetime quantification parameters.
Basic Protocol 4: DATA PROCESSING IN ImageJ/FIJI
This protocol describes the procedure for selecting areas of interest in the images exported from SPCImage using the freely available ImageJ/FIJI software. It uses SPCImage export files containing photon numbers (name _photons.asc) and generates a new file for each selection. For ease of further analysis, we recommend naming the new files according to the type of selection, and storing all files derived from the same measurement in the same folder (see Basic Protocol 5, step 6). In addition, we describe processing of the four SPCImage export files from 2-component analysis to generate a single file containing mean fluorescence lifetimes in pixel positions.
Materials
- ImageJ or FIJI software. We use versions 1.52p or 1.52q. Other versions have not been tested.
- Exported files from SPCImage containing matrices with photon number per pixel (name _photons.asc files) generated in Basic Protocol 3
- Optional : Exported files from FLIM measurement analyzed with a 2-component model in SPCImage, containing matrices with individual fluorescence lifetime components and their amplitudes in pixel positions (name _t1.asc, name _t2.asc, name _a1.asc, and name _a2.asc files) generated in Basic Protocol 3
1.In ImageJ/FIJI, import the “ name _photons.asc” file using File -> Import -> text image.
2.Outline the cell, using either the polygon (A) or the freehand (B) selection tool.

3.Set up all pixel values outside selected area to zero using Edit -> Clear Outside.
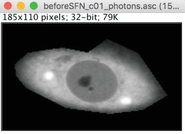
4.Save as new file using File -> Save As -> text image. When saving, modify the name to indicate the type of selection (e.g., original “ name _photons.asc” file will be normally saved as “ name _photons.txt” file. Modify to “ name _photons_ez.txt” for selection of entire cell area).
5.Optional : the file can be further modified to select subcellular regions of interest. For this, deselect existing selection (Edit -> Selection -> Select none, or click on the area outside the selection). Make a new selection (as in A). Repeat “Edit -> Clear Outside” to retain outlined area (see B), or “Edit -> Clear” to retain area of the cell that excludes selected region (C).
Save each file as in step 4, modifying the name to reflect selected area (e.g., “ name _photons_cz.txt” for cytoplasm or “ name _photons_nz.txt” for nucleus).
6.Optional : If a two-component multi-exponential model was used in SPCImage (Basic Protocol 3), the matrix containing mean fluorescence lifetimes at the pixel positions needs to be generated from exported t1, t2, a1, and a2 files for each measurement, using the following formula:
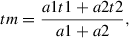
where tm is mean fluorescence lifetime in the pixel position, t1 and t2 are fluorescence lifetimes of the first and second components in the corresponding pixel, and a1 and a2 are the relative amplitudes of these components in the same pixel [adapted from the BH TCSPC handbook (Becker, 2017)]. A file with these tm values in pixel positions can be produced using an ImageJ/FIJI macro, tm_2cFLIM.ijm, that we developed for this purpose (available at https://github.com/DinaDikovskaya/FLIMacro).
Basic Protocol 5: EXPERIMENT ANALYSIS IN FLIMDAST
Visualization and quantification of fluorescence lifetime changes are performed within the R-based FLIM DAtaSet Tool (FLIMDAST). Since the design of a FLIM time course is usually complex and varies between experiments, we developed FLIMDAST as a user interface that collects information about the layout of FLIM experiment and links the names/locations of the processed data files to the elements within this layout. Crucially, it assigns each measurement specific reference data to which the measurement should be compared. Within the user interface, any pair of reference/non-reference measurements can be displayed and visually compared in the overlay scatterplot that plots fluorescence lifetime in each pixel against photon number detected in the same pixel (Fig. 1B). The overlay settings are user-defined and can be applied to the entire experimental dataset (Fig. 1C). Similarly, the user defines parameters for calculating intensity-independent changes in fluorescence lifetime that are then applied to the entire experiment (Fig. 1C). The outputs are a PDF file with the abovementioned scatterplots, and/or a CSV file with calculated intensity-independent changes in fluorescence lifetimes, as well as mean fluorescence lifetimes and mean photon numbers per pixel for all measurements within the time-course experiment.
Materials
- SPCImage export files containing fluorescence lifetime matrices, generated in Basic Protocol 3 (“ name _t1.asc” files) or files derived from processing the t1, t2, a1, and a2 export files (if the 2-component analysis was used)
- Files generated in ImageJ/FIJI containing photon numbers in pixels within areas of interest, produced in Basic Protocol 4, or “ name _photons.asc” files exported from SPCImage in Basic Protocol 3
- R software: FLIMDAST was developed in R version 3.3.3; versions other than 3.3.3 and 3.6.3 have not been tested
- RStudio: We use version 1.1.383; other versions have not been tested
1.Download the FLIMDAST folder containing the app.R file from the GitHub repository (https://github.com/DinaDikovskaya/FLIMDAST) according to the instructions in the accompanying README.md file, and place the folder into a working directory on your computer.
2.Open RStudio and start FLIMDAST: From inside RStudio, set up the correct working directory [in the top ribbon, select Session -> Set Working Directory -> Choose Directory, navigate to the directory that contains the FLIMDAST folder (but not to the FLIMDAST folder itself!), and click “Open”]. In the Console panel, type library(“shiny”) and click Enter. Type runApp(“FLIMDAST”) and click Enter. This will open a FLIMDAST user interface in a local host window.
3.In FLIMDAST, open “Experiment Layout” tab. Choose the number of experimental conditions (measurement groups) in your experiment. Give them names in the corresponding fields.
4.Select number (up to two) of additional regions of interest in each cell to be analyzed. Give them names in the corresponding fields.
5.Indicate if the time course is divided into separate parts (up to three parts are possible), and select the number of time points in each part.
6.Open the “Single Object Time Course” tab. Choose the experimental condition from the choices provided earlier on the “Experiment Layout” tab. For each measurement, select its Time point from the menu build based on the experimental layout, assign Reference status, and select the location of the requested files, using the corresponding “Choose file” buttons. The Fluorescence lifetime file is the “ name _t1.asc” SPCImage export file containing the fluorescence lifetime matrix (see Basic Protocol 3) or the “ name _tm.txt” file generated by the tm_2cFLIM.ijm macro from the t1, t2, a1, and a2 outputs of 2-component analysis (see Basic Protocol 4). Photon number files are the .txt files generated in ImageJ from the photon matrix file exported from SPCImage (see Basic Protocol 4) or the “ name _photons.asc” export file from SPCImage. If “ autofill files” is switched on, the file names for the photon data can be automatically generated from the name of the fluorescence lifetime file, based on the replacement patterns that can be typed in the fields opposite from the corresponding “Choose file” buttons. The locations/names of selected files will be indicated underneath the “Choose file” buttons.
7.Once all files are located, click Add. This will assemble all information about the measurement into one table entry.
8.Repeat steps 6-7 for all measurements of the same cell within the entire time course. Within such a time course, each time point can be chosen only once, and one of them should be a reference.
9.If added by mistake, the last table entry may be removed by clicking “Delete last entry.”
10.Once the data for the entire time course of one object/cell are collected in the table, click “Save to main table.” This will pass all information to the “Main Table” tab, reset the current table, and give the next cell a new ID number. The cell ID numbers are sequential within each experimental condition.
11.Repeat steps 6-10 for all cells in the experiment, for all experimental conditions.
12.All measurements will be assembled in the Experiment data location table in the “Main Table” tab. It can be saved, cleared, or uploaded from a previously saved file using “Download Main Table,” “Empty Main Table,” and “Upload table from file” buttons, respectively. The “Upload table from file” button opens a browser bar for selecting the file. The selected file will be uploaded into a separate table. To replace the current table with the uploaded table, click “Use as Main Table.” This will reset the numbers and names of the additional regions in the “Experiment Layout” tab to match their values in the uploaded table.
13.To set plotting and quantification parameters, highlight two table entries in the “Single Object Time Course” or the “Main Table” tab, including one reference and one non-reference measurement. Click “Use for settings.” This will pass selected datasets to the “Plot” and “Quantify” tabs, and activate the plot displays.
14.Open the “Plot” tab. Select the reference dataset for the first layer of scatterplot, and choose its color. Indicate if you want to overlay it with another dataset. Repeat for overlay dataset(s)—up to 3 overlays are possible. Adjust scales for fluorescence lifetime and photon number in the scatterplot. The displayed scatterplot can be downloaded at this point using the “Download plot” button. If you would like to use this scatterplot layout for the entire experiment, click “Use these settings.” Repeat if more than one layout is desired. The last added design can be removed by clicking “Remove last settings.”
15.To apply the plot settings to the entire experiment, click “Plot all data.” This button becomes visible in the “Plot” and “Main Table” tabs when the Experiment data location table in the “Main Table” tab is not empty and plot settings are selected in the “Plot” tab. This will generate and download a multi-page PDF file with plots for the entire experiment.
16.In the “Quantify” tab, select regions for calculating the intensity-independent change in fluorescence lifetime relative to the reference dataset.
17.In the “Quantify” tab, using the “data” and “model” switches, choose the display items for the scatterplot that help to visualize the quantification algorithm. These switches do not affect quantification. If any “scatter” is on, the value of intensity-independent difference between selected datasets will be displayed under the title of the plot. The plot can be downloaded using the “download plot” button.
18.Optional : In the “Quantify” tab, select lower and/or upper limits for the intensity region included in quantification. Without these limits, the fluorescence lifetime change will be quantified within the overlapping range of intensities of the reference and non-reference datasets trimmed by removing the brightest 0.5% and the dimmest 0.5% of the pixels.
19.Click “Use these settings” to select the chosen quantification parameters for the entire experiment. Repeat if more than one set of settings is desired. The last added settings can be removed by clicking “Remove last settings.”
20.To apply the quantification settings to the entire experiment, click “Quantify all data.” This button becomes visible in the “Quantify” and “Main Table” tabs when the Experiment data location table in the “Main Table” tab is not empty and quantification settings are selected in the “Quantify” tab. This will generate a Result Table with the changes in fluorescence lifetime (columns named “shift”) for each non-reference measurement in the Main Table. In addition, it contains mean values for fluorescence lifetime (mean_t1) and photon number per pixel (mean_photons) for each analyzed region of the cell. Download the Result Table by clicking on the “Download Results” button.
21.To close FLIMDAST, close the local host window, or click the red “STOP” button at the top of the Console window in RStudio.
REAGENTS AND SOLUTIONS
HSB, 2×
- 50 mM HEPES, pH 7.4
- 280 mM NaCl
- 1.5 mM Na2HPO4
- For HeLa and HEK293 cells : Adjust pH to 7.0 and filter sterilize. Store at room temperature for a few months, or frozen for longer times.
- For other cell types : Prepare 100-ml aliquots, adjust pH to 7.0, 7.05, 7.1, 7.15, and 7.2, and filter sterilize. Test transfection efficiency using each preparation of 2× HBS under otherwise identical conditions, as described in Alternate Protocol 1 for transfecting sfGFP or mCherry plasmids, and use the solution that gives the best transfection efficiency.
COMMENTARY
Background Information
Among the existing methods for monitoring oxidative stress responses, microscopy-based live cell technologies offer several advantages, including the ability to report changes in real time, an intact (intra)cellular context free of artifacts introduced by tissue/cell disruption and artificial buffer components, and the possibility to relate readout to cell/tissue morphology, long-term cellular behavior, or other parameters, combined with the subcellular resolution common to all cell imaging techniques. However, the number of molecular events in the oxidative stress response pathways that can be monitored in live cells is limited in comparison with biochemical assays. Most live reporters provide a readout of an activated Nrf2-driven stress response program at its execution stage. Several reporters that place GFP (Zhu & Fahl, 2000) or luciferase (Boerboom et al., 2006; Dohlen et al., 2008; Simmons, Fan, Yeoman, Wakefield, & Ramabhadran, 2011; Wang, Hayes, & Wolf, 2006) under transcriptional control of multiple Nrf2-responsive elements [termed antioxidant response elements (ARE)/electrophile response elements (EpRE)] have been generated; however, only rarely have these been used for live imaging (Dohlen et al., 2008; Zhu & Fahl, 2000). One notable exception is the OKD48 transgenic mouse with a luciferase reporter that combines ARE-dependent transcription and Keap1-mediated protein regulation (Oikawa, Akai, Tokuda, & Iwawaki, 2012), which is more widely used as a whole mouse imaging−based reporter of Nrf2-dependent oxidative stress response (Forootan et al., 2017; Suzuki et al., 2019). Fluorescence intensity of GFP-labeled Nrf2 and the GFP-labeled Nrf2 target gene sulfiredoxin 1 (Bischoff et al., 2019) also provides a readout of the execution stage of the oxidative stress response, although imaging of the fluorescently labeled Nrf2 is more often used for measuring nuclear accumulation of Nrf2 (Lau et al., 2010; Xue et al., 2015), an essential step directly preceding the execution of the oxidative stress response and a common marker of Nrf2 activation.
Research into the mechanisms of Nrf2 activation in health and disease requires live imaging tools that can assess initiation of this pathway at its early stages. Fluorescence imaging of Nrf2 labeled with photoactivatable fluorophore Dedra2 has been used to measure Nrf2 stability in live cells (Burroughs et al., 2018), the physiological target of Nrf2 regulatory pathways. An even earlier step in one such pathway is the interaction between Nrf2 and Keap1 that recruits Nrf2 to the Cul3-RBX1 E3 ubiquitin ligase complex, which targets Nrf2 for degradation. Keap1 is the cellular sensor that ultimately controls the Nrf2-mediated oxidative stress response, as Keap1 modifications by oxidants or electrophiles stop Nrf2 ubiquitination by Cul3, thereby leading to Nrf2 stabilization (Dinkova-Kostova et al., 2017). A split luciferase complementation assay was developed for high-throughput imaging-based screening of the Keap1-Nrf2 interaction (Ramkumar, Sekar, Foygel, Elango, & Paulmurugan, 2013), based on the premise that the Keap1-Nrf2 complex needs to dissociate for Nrf2 activation, an initial view (Wakabayashi et al., 2004) that has since been disputed (Eggler, Liu, Pezzuto, van Breemen, & Mesecar, 2005; Zhang, Lo, Cross, Templeton, & Hannink, 2004). An alternative FLIM-FRET-based assay to measure Keap1-Nrf2 interaction was developed by our group (Baird, Lleres, Swift, & Dinkova-Kostova, 2013). Among other methods, FRET is uniquely sensitive to minor changes in the distance between FRET donor and FRET acceptor, since FRET efficiency is inversely proportional to the sixth power of the distance between contributing fluorophores (Lakowicz, 2006; Vogel, van der Meer, & Blank, 2014). This sensitivity allows the FRET-FLIM assay to detect not only complete complex disassembly but also more subtle changes in the conformation of the Keap1-Nrf2 complex. Using this assay, the effect of several Nrf2 inducers on Keap1-Nrf2 complex has been established (Baird et al., 2014; Bertrand et al., 2015; Quinti et al., 2017). Perhaps unexpectedly, this assay has revealed that many Nrf2 inducers that inhibit the ability of Keap1 to destabilize Nrf2 increased FRET efficiency between labeled Keap1 and Nrf2, indicating that these molecules (or their respective N- and C-terminal fluorescently tagged fusion proteins) are positioned more closely when the Keap1-Nrf2 complex is inactive. On the other hand, Nrf2 inducers that target the Keap1-Nrf2 binding interface can create an inactive Keap1-Nrf2 complex in which the distance between Keap1 and Nrf2 is increased ((Bertrand et al., 2015, and our unpublished data). Thus, the FLIM-FRET assay serves not only as a readout for Keap1-Nrf2 complex remodeling in the process of Nrf2 activation, but also provides unique mechanistic insights into the apparently variable nature of such remodeling.
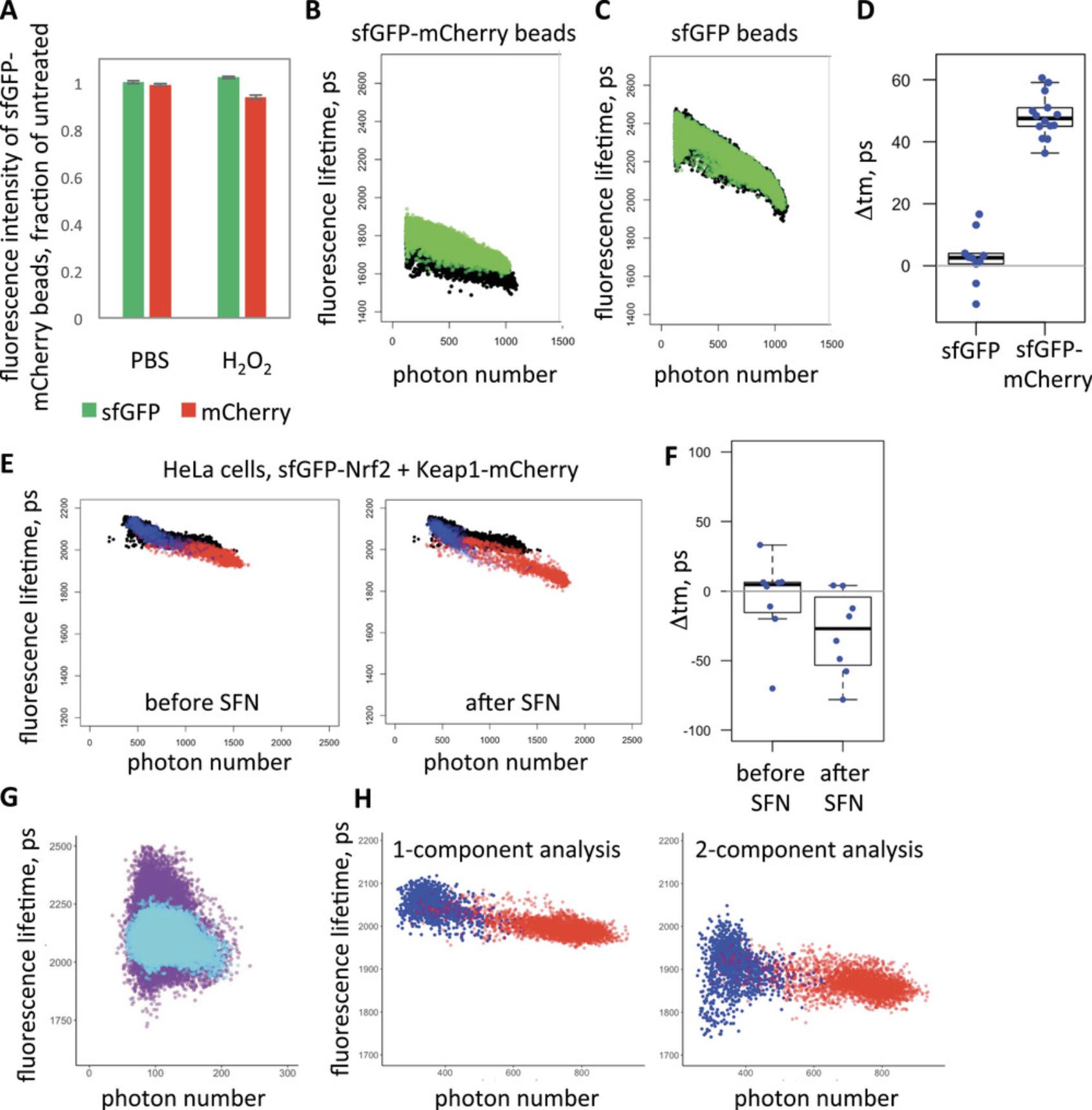
Recently, we have further improved this assay by optimizing the linker between Keap1 and the FRET acceptor mCherry, and by replacing the FRET donor EGFP with a super-folder GFP (sfGFP) whose maturation speed matches the short half-life of Nrf2 (Dikovskaya et al., 2019). We also developed more streamlined data processing that allows increased sample size. Analysis of factors contributing to cell-to-cell variability within these samples revealed that, contrary to dogma, there was a consistent intensity-dependent bias in our measurements (Dikovskaya et al., 2019), despite all technical precautions. Further investigation identified the so-called pile-up effect as a likely cause for this phenomenon (manuscript in preparation).
The critical importance of removing intensity-dependent bias from the Nrf2-Keap1 FLIM-FRET assay stems from two factors: (1) the conformational change within the Nrf2-Keap1 complex produces only a small shift in fluorescence lifetime, which is similar in magnitude to intensity-dependent aberrations, and could be easily obscured by the latter; and (2) most inducers stabilize Nrf2, thus increasing the intensity of the FRET donor; if the contribution of the intensity-dependent bias is not accounted for, the results could easily be misleading.
Critical Parameters and Troubleshooting
1.Expression levels of the constructs. It is crucially important to obtain at least some cells expressing an appropriate level of sfGFP-Nrf2, one that is sufficient for FLIM measurements yet not excessive to a degree that would cause artifacts. FLIM measurements need lower fluorescence intensity than “standard” fluorescence imaging, and often the most suitable cells are barely visible through the eyepiece (though the sensitivity of the optical settings will vary between systems). High expression levels are not only damaging for cells, but are also incompatible with TCSPC-based FLIM-FRET measurements due to a requirement to keep the acquisition rate below 1 × 106 photons per second, and an increased risk of homo-FRET, an energy transfer between identical fluorophores that can lower fluorescence lifetime at high fluorophore density. The correct ratio between Keap1 and labeled Nrf2 is another critical factor: if it is too high, Nrf2 could be completely degraded; if too low, the majority of overexpressed Nrf2 will not be in complex with Keap1, making the assay results inconclusive. Inspecting the localization of sfGFP-Nrf2 can help estimate whether this ratio is sufficient for Keap1 to recruit Nrf2 to cytoplasm (see Fig. 2 for examples), which is indicative of sfGFP-Nrf2-Keap1 complex formation. Achieving such fine-tuned levels of expression requires optimization. In general, healthy growing cells that have not been allowed to reach full confluence for several passages before transfection are more easily transfected. The lower cell density at transfection increases transfection efficiency, and vice versa. If the expression is too high, reducing DNA amount (with a parallel reduction in lipofectamine 2000 reagent), or switching to calcium phosphate transfection will bring the expression level down. Since only a few transfected cells per sample are required, we suggest keeping expression levels low.
2.Avoiding cytotoxicity. Unless cell death is the process under investigation, cytotoxicity should be kept to a minimum before and during imaging. If the cause of cell death is transfection, increase cell density at transfection, reduce amount of DNA and/or Lipofectamine, make sure that no antibiotics are present during transfection, and replace the medium 4-6 hr after transfection. Trypsinization of cells too soon after transfection can, in our experience, stimulate cell death; we generally leave cells for at least 20-24 hr after transfection before re-seeding. If excessive cell death is detected during imaging (see Fig. 2O,P for examples), it might be caused by incompatibility of a particular cell type with the imaging medium, or by lack of CO2. The choice of commercially available phenol-free CO2-independent media is limited, and cell survival in such media needs to be tested in advance. Another likely reason for cell death during imaging is phototoxicity caused by prolonged and/or too intense illumination. In this case, reduce laser power during FLIM acquisition. Also, instead of inspecting the sample through the eyepiece under continuous illumination, use tile-scan images of the sample. Finally, cytotoxicity could be caused by treatment. A parallel experiment done under conditions identical to those used in FLIM measurements (such as medium, confluency, and prior transfections) has to be conducted to establish if this is the case, and to identify the non-toxic concentration of inducer/vehicle.
3.Precision and consistency of FLIM measurements. The accuracy and precision of fluorescence lifetime determined by fitting an exponential decay model to the photon micro-time trace strongly depends on the number of photons in that trace. In the pixel-by-pixel analysis described here, such a trace is composed of all photons that were detected in a particular pixel of the image during FLIM acquisition. An inadequately low number of photons results in a much broader distribution of fluorescence lifetime values, and could be recognized by an increase in the “thickness” of the distribution of fluorescence lifetime versus photon number in scatterplots generated in Basic Protocol 5 (Fig. 7G). This can obscure any existing clusters of fluorescence lifetimes and, in some cases, may affect calculations of fluorescence lifetime change between FLIM measurements. The number of photons in the trace can, in principle, be increased by choosing a cell with higher fluorescence intensity, and by increasing laser power and duration of acquisition, all of which have serious technical and biological restrictions (see above). The photon trace content could also be increased by reducing spatial resolution, which makes each pixel bigger. A similar effect can be achieved by binning the acquired FLIM data during data analysis in SPCImage, which effectively pools the photon traces of several pixels into one. Another approach is to limit the demands on the number of photons in the trace by choosing the simplest possible fitting model (such as a one-exponential decay model, which can offer a good approximation despite not fully matching the data; Fig. 7H). Any chosen strategy negatively affects some other aspects of the measurement, and thus, the final choice is a practical compromise, which can be achieved in many different ways. Importantly, for comparison between measurements, the same settings have to be applied to all data throughout the entire experiment. For the imaging part, we keep the duration of acquisition, image size, and resolution constant, while limiting the CFD acquisition rate within 5 × 104 to 1 × 106 (ideally, 1−5 × 105) photons/s by selecting cells with suitable level of fluorescence and, for Basic Protocol 2, by adjusting the laser power. For the analysis part, the level of binning and the choice of model are kept constant for all measurements, while the Shift parameter is adjusted individually in each measurement.
Understanding Results
The aim of this workflow is to monitor changes in Nrf2-Keap1 complex in individual live cells by repetitive measurements of the relative distance between fluorescently labeled Nrf2 and Keap1 proteins in cells, using fluorescence lifetimes of labeled Nrf2.Depending on the type of intervention, such changes move Keap1 and Nrf2 either closer or further apart. Either of these conformational changes can result in functional inactivation of the complex and consequential induction of an Nrf2-mediated stress response. If the aim of the experiment is to establish kinetics of Nrf2 induction, then the timing of the change in fluorescence lifetime, rather than its direction, is important. If the experiment aims to understand the mechanism of action of an Nrf2 inducer, then both the timing and the direction of change are important outcomes. A reduction in fluorescence lifetime indicates that Nrf2 and Keap1 move closer to each other during complex remodeling, and vice versa. The magnitude of the change is another factor of potential interest. In principle, fluorescence lifetime is affected both by the distance between the labeled molecules and by the proportion of interacting molecules. Thus, an increase in fluorescence lifetime could mean that all sfGFP-Nrf2 and Keap1-mCherry molecules move further apart, or that there is an increased fraction of sfGFP-Nrf2 protein that is not bound by Keap1-mCherry, and further investigation is required to distinguish between these two possibilities.
The unique advantage of this workflow is the ability to detect a subtle change in fluorescence lifetime that normally would be obscured by the noise in the measurements. On scatterplots, it appears as a minor shift of the entire fluorescence lifetime distribution, or a subset of it, either downwards (for higher FRET) or upwards (for lower FRET). The sensitivity of this method can be seen by the change caused by a minor loss of FRET acceptor in beads with immobilized sfGFP-mCherry (Fig. 7A-D). The application of this method to measure changes in the Keap1-Nrf2 complex caused by the classical inducer sulforaphane (SFN) is exemplified in Fig. 7E and F. The downward shift in fluorescence lifetime of cells after sulforaphane treatment indicates that the Keap1-Nrf2 complex becomes tighter in this condition. We quantify this by measuring the vertical distance between curves fitted to both distributions, within an overlapping range of intensities. Such comparison between two measurements produces a single value. The statistical significance of the change needs to be assessed in the context of the entire experimental dataset (Fig. 7D,F).
Inspection of the scatterplots gives further indication of the nature of the assessed changes. Color-coding of selected cellular regions makes it possible to identify the region responsible for the change (Fig. 7E,H).
Time Considerations
Cell preparation takes 3-4 days, starting from a healthy, logarithmically growing cell culture. The entire “microscope time” depends on the experimental design, usually from a few hours to a full day. Setting up the microscope might take an hour. Locating suitable cells for FLIM can take considerable time, often more than an hour. When considering the time interval for the time course, both duration of each acquisition and the sample size need to be taken into account. Individual FLIM measurements take ∼5 min each, unless using the time-lapse procedure described in Alternate Protocol 2, which can save time. Data processing and analysis can take several more days.
Acknowledgments
We thank Paul Appleton and Iain Porter (University of Dundee Imaging Facility) for expert technical help, and Vladislav Shcheslavskiy (Becker-Hickl GmbH) for help with time-lapse FLIM setup, Marek Gierlinski (University of Dundee School of Life Sciences) and Graeme Ball (University of Dundee Imaging Facility) for advice on the data analysis algorithm and software publishing, and the Biotechnology and Biological Sciences Research Council (BB/L01923X/1), Tenovus Scotland (T17/14), and Cancer Research UK (C20953/A18644) for financial support.
Literature Cited
- Baird, L., Lleres, D., Swift, S., & Dinkova-Kostova, A. T. (2013). Regulatory flexibility in the Nrf2-mediated stress response is conferred by conformational cycling of the Keap1-Nrf2 protein complex. Proceedings of the National Academy of Sciences of the United States of America , 110(38), 15259–15264. doi: 10.1073/pnas.1305687110.
- Baird, L., Swift, S., Lleres, D., & Dinkova-Kostova, A. T. (2014). Monitoring Keap1-Nrf2 interactions in single live cells. Biotechnology Advances , 32(6), 1133–1144. doi: 10.1016/j.biotechadv.2014.03.004.
- Becker, W. (2015). Advanced time-correlated single photon counting applications (Vol. 111). New York: Springer International Publishing.
- Becker, W. (2017). The bh TCSPC handbook ( 7 ed.). Berlin, Germany: Becker&Hickl GmbH.
- Bertrand, H. C., Schaap, M., Baird, L., Georgakopoulos, N. D., Fowkes, A., Thiollier, C., … Wells, G. (2015). Design, synthesis, and evaluation of triazole derivatives that induce Nrf2 dependent gene products and inhibit the Keap1-Nrf2 protein-protein interaction. Journal of Medicinal Chemistry , 58(18), 7186–7194. doi: 10.1021/acs.jmedchem.5b00602.
- Bischoff, L. J. M., Kuijper, I. A., Schimming, J. P., Wolters, L., Braak, B. T., Langenberg, J. P., …van de Water, B. (2019). A systematic analysis of Nrf2 pathway activation dynamics during repeated xenobiotic exposure. Archives of Toxicology , 93(2), 435–451. doi: 10.1007/s00204-018-2353-2.
- Boerboom, A. M., Vermeulen, M., van der Woude, H., Bremer, B. I., Lee-Hilz, Y. Y., Kampman, E., … Aarts, J. M. (2006). Newly constructed stable reporter cell lines for mechanistic studies on electrophile-responsive element-mediated gene expression reveal a role for flavonoid planarity. Biochemical Pharmacology , 72(2), 217–226. doi: 10.1016/j.bcp.2006.04.002.
- Burroughs, A. F., Eluhu, S., Whalen, D., Goodwin, J. S., Sakwe, A. M., & Arinze, I. J. (2018). PML-nuclear bodies regulate the stability of the fusion protein Dendra2-Nrf2 in the nucleus. Cellular Physiology and Biochemistry , 47(2), 800–816. doi: 10.1159/000490033.
- Dikovskaya, D., Appleton, P. L., Bento-Pereira, C., & Dinkova-Kostova, A. T. (2019). Measuring the interaction of transcription factor Nrf2 with its negative regulator keap1 in single live cells by an improved FRET/FLIM analysis. Chemical Research in Toxicology , 32(3), 500–512. doi: 10.1021/acs.chemrestox.8b00354.
- Dinkova-Kostova, A. T., Kostov, R. V., & Canning, P. (2017). Keap1, the cysteine-based mammalian intracellular sensor for electrophiles and oxidants. Archives of Biochemistry and Biophysics , 617, 84–93. doi: 10.1016/j.abb.2016.08.005.
- Dohlen, G., Odland, H. H., Carlsen, H., Blomhoff, R., Thaulow, E., & Saugstad, O. D. (2008). Antioxidant activity in the newborn brain: A luciferase mouse model. Neonatology , 93(2), 125–131. doi: 10.1159/000107777.
- Eggler, A. L., Liu, G., Pezzuto, J. M., van Breemen, R. B., & Mesecar, A. D. (2005). Modifying specific cysteines of the electrophile-sensing human Keap1 protein is insufficient to disrupt binding to the Nrf2 domain Neh2. Proceedings of the National Academy of Sciences of the United States of America , 102(29), 10070–10075. doi: 10.1073/pnas.0502402102.
- Forootan, S. S., Mutter, F. E., Kipar, A., Iwawaki, T., Francis, B., Goldring, C. E., … Copple, I. M. (2017). Real-time in vivo imaging reveals localised Nrf2 stress responses associated with direct and metabolism-dependent drug toxicity. Scientific Reports , 7(1), 16084. doi: 10.1038/s41598-017-16491-2.
- Lakowicz, J. (2006). Principles of fluorescence spectroscopy (Vol. 1). New York: Springer International Publishing.
- Lau, A., Wang, X. J., Zhao, F., Villeneuve, N. F., Wu, T., Jiang, T., … Zhang, D. D. (2010). A noncanonical mechanism of Nrf2 activation by autophagy deficiency: Direct interaction between Keap1 and p62. Molecular and Cellular Biology , 30(13), 3275–3285. doi: 10.1128/MCB.00248-10.
- Oikawa, D., Akai, R., Tokuda, M., & Iwawaki, T. (2012). A transgenic mouse model for monitoring oxidative stress. Scientific Reports , 2, 229. doi: 10.1038/srep00229.
- Phelan, K., & May, K. M. (2015). Basic techniques in mammalian cell tissue culture. Current Protocols in Cell Biology , 66, 1.1.1–1.1.22. doi: 10.1002/0471143030.cb0101s66.
- Quinti, L., Dayalan Naidu, S., Trager, U., Chen, X., Kegel-Gleason, K., Lleres, D., … Kazantsev, A. G. (2017). KEAP1-modifying small molecule reveals muted NRF2 signaling responses in neural stem cells from Huntington's disease patients. Proceedings of the National Academy of Sciences of the United States of America , 114(23), E4676–E4685. doi: 10.1073/pnas.1614943114.
- Ramkumar, K. M., Sekar, T. V., Foygel, K., Elango, B., & Paulmurugan, R. (2013). Reporter protein complementation imaging assay to screen and study Nrf2 activators in cells and living animals. Analytical Chemistry , 85(15), 7542–7549. doi: 10.1021/ac401569j.
- Simmons, S. O., Fan, C. Y., Yeoman, K., Wakefield, J., & Ramabhadran, R. (2011). NRF2 oxidative stress induced by heavy metals is cell type dependent. Current Chemical Genomics , 5, 1–12. doi: 10.2174/1875397301105010001.
- Suhling, K., Hirvonen, L. M., Levitt, J. A., Chung, P.-H., Tregidgo, C., le Marois, A., … Krstajic, N. (2015). Fluorescence lifetime imaging (FLIM): Basic concepts and some recent developments. Medical Photonics , 27, 3–40. doi: hTtps://doi.org/10.1016/j.medpho.2014.12.001.
- Suzuki, T., Muramatsu, A., Saito, R., Iso, T., Shibata, T., Kuwata, K., … Yamamoto, M. (2019). Molecular mechanism of cellular oxidative stress sensing by Keap1. Cell Reports , 28(3), 746–758 e744. doi: 10.1016/j.celrep.2019.06.047.
- Talbot, C. B., Patalay, R., Munro, I., Warren, S., Ratto, F., Matteini, P., … Dunsby, C. (2011). Application of ultrafast gold luminescence to measuring the instrument response function for multispectral multiphoton fluorescence lifetime imaging. Optics Express , 19(15), 13848–13861. doi: 10.1364/OE.19.013848.
- Vogel, S. S., van der Meer, B. W., & Blank, P. S. (2014). Estimating the distance separating fluorescent protein FRET pairs. Methods , 66(2), 131–138. doi: 10.1016/j.ymeth.2013.06.021.
- Wakabayashi, N., Dinkova-Kostova, A. T., Holtzclaw, W. D., Kang, M. I., Kobayashi, A., Yamamoto, M., … Talalay, P. (2004). Protection against electrophile and oxidant stress by induction of the phase 2 response: Fate of cysteines of the Keap1 sensor modified by inducers. Proceedings of the National Academy of Sciences of the United States of America , 101(7), 2040–2045. doi: 10.1073/pnas.0307301101.
- Wang, X. J., Hayes, J. D., & Wolf, C. R. (2006). Generation of a stable antioxidant response element-driven reporter gene cell line and its use to show redox-dependent activation of nrf2 by cancer chemotherapeutic agents. Cancer Research , 66(22), 10983–10994. doi: 10.1158/0008-5472.CAN-06-2298.
- Xue, M., Momiji, H., Rabbani, N., Barker, G., Bretschneider, T., Shmygol, A., … Thornalley, P. J. (2015). Frequency modulated translocational oscillations of Nrf2 mediate the antioxidant response element cytoprotective transcriptional response. Antioxidant Redox Signaling , 23(7), 613–629. doi: 10.1089/ars.2014.5962.
- Zhang, D. D., Lo, S. C., Cross, J. V., Templeton, D. J., & Hannink, M. (2004). Keap1 is a redox-regulated substrate adaptor protein for a Cul3-dependent ubiquitin ligase complex. Molecular and Cellular Biology , 24(24), 10941–10953. doi: 10.1128/MCB.24.24.10941-10953.2004.
- Zhu, M., & Fahl, W. E. (2000). Development of a green fluorescent protein microplate assay for the screening of chemopreventive agents. Analytical Biochemistry , 287(2), 210–217. doi: 10.1006/abio.2000.4875
Internet Resources
The repository containing the ImageJ/FIJI macro for processing SPCImage export files.
The repository containing the FLIM DAtaSet Tool (FLIMDAST).
Citing Literature
Number of times cited according to CrossRef: 2
- Tony Heurtaux, David S. Bouvier, Alexandre Benani, Sergio Helgueta Romero, Katrin B. M. Frauenknecht, Michel Mittelbronn, Lasse Sinkkonen, Normal and Pathological NRF2 Signalling in the Central Nervous System, Antioxidants, 10.3390/antiox11081426, 11 , 8, (1426), (2022).
- Sharadha Dayalan Naidu, Takafumi Suzuki, Dina Dikovskaya, Elena V. Knatko, Maureen Higgins, Miu Sato, Miroslav Novak, José A. Villegas, Terry W. Moore, Masayuki Yamamoto, Albena T. Dinkova-Kostova, The isoquinoline PRL-295 increases the thermostability of Keap1 and disrupts its interaction with Nrf2, iScience, 10.1016/j.isci.2021.103703, 25 , 1, (103703), (2022).

