Making Bioinformatics Training Events and Material More Discoverable Using TeSS, the ELIXIR Training Portal
Carole Goble, Carole Goble, Finn Bacall, Finn Bacall, Aitor Apaolaza, Aitor Apaolaza, Munazah Andrabi, Munazah Andrabi, Chris Child, Chris Child, Olivier Sand, Olivier Sand, Alexander Botzki, Alexander Botzki
bioinformatics training
Bioschemas annotation
TeSS
training catalog
training event
training material
Abstract
Many trainers and organizations are passionate about sharing their training material. Sharing training material has several benefits, such as providing a record of recognition as an author, offering inspiration to other trainers, enabling researchers to discover training resources for their personal learning path, and improving the training resource landscape using data-driven gap analysis from the bioinformatics community. In this article, we present a series of protocols for using the ELIXIR online training registry Training eSupport System (TeSS). TeSS provides a one-stop shop for trainers and trainees to discover online information and content, including training materials, events, and interactive tutorials. For trainees, we provide protocols for registering and logging in and for searching and filtering content. For trainers and organizations, we also show how to manually or automatically register training events and materials. Following these protocols will contribute to promoting training events and add to a growing catalog of materials. This will concomitantly increase the FAIRness of training materials and events. Training registries like TeSS use a scraping mechanism to aggregate training resources from many providers when they have been annotated using Bioschemas specifications. Finally, we describe how to enrich training resources to allow for more efficient sharing of the structured metadata, such as prerequisites, target audience, and learning outcomes using Bioschemas specification. As increasing training events and material are aggregated in TeSS, searching the registry for specific events and materials becomes crucial. © 2023 The Authors. Current Protocols published by Wiley Periodicals LLC.
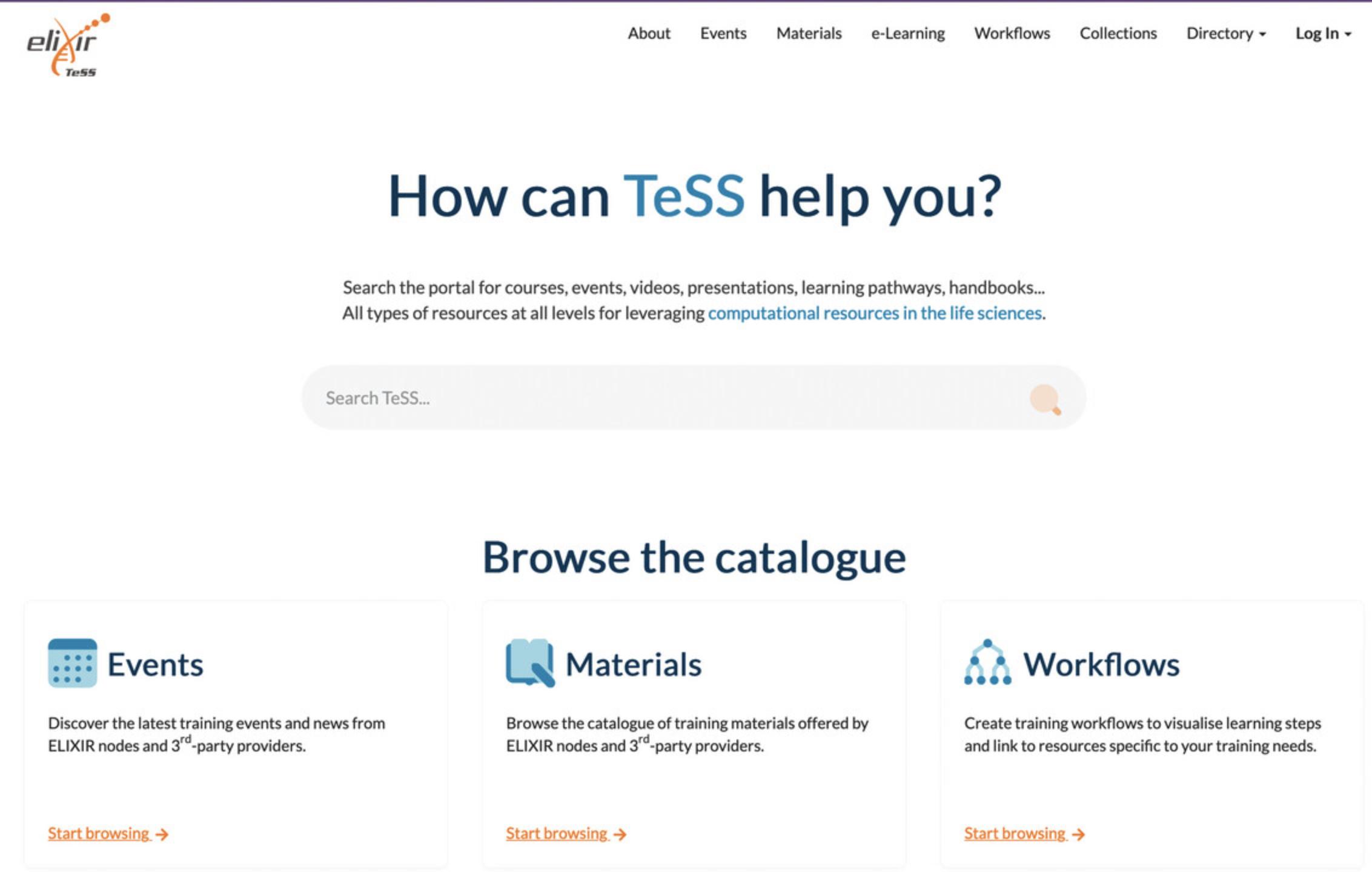
Basic Protocol 1 : Searching for training events and materials in TeSS
Support Protocol : Integrating TeSS widgets on your website
Basic Protocol 2 : Logging in to TeSS using an institutional account
Alternate Protocol : Creating and logging in to a TeSS account
Basic Protocol 3 : Manual registration of training events in TeSS
Basic Protocol 4 : Manual registration of training materials in TeSS
Basic Protocol 5 : Registration of a content provider in TeSS
Basic Protocol 6 : Automated harvesting of training events and materials in TeSS
INTRODUCTION
The Training eSupport System (TeSS) is an ELIXIR online training registry (Beard et al., 2020) that provides a one-stop shop for trainers and mostly (but not only) graduate-level trainees to discover online information and content, including training materials, events, and interactive tutorials. TeSS provides opportunities to promote training events and news and to contribute to a growing catalog of materials. For trainers, the portal offers an environment for sharing materials and event information; for trainees, it offers a convenient gateway to identify relevant training events and resources. Thanks to our search-engine optimization and widgets, registering an event on TeSS will allow it to appear in more searches and be seen across all websites that display site resources.
The ELIXIR training platform promotes sharing of training materials and exchange of training best practices. Many trainers are passionate about providing open-access training material. Such sharing has several benefits, including providing a record of recognition as an author, offering inspiration to other trainers, enabling researchers to discover training resources for their personal learning path, and improving the training resource landscape using data-driven gap analysis from the bioinformatics community (Garcia et al., 2020).
Now that you are convinced you should register your events and training materials in TeSS, let's see how it is done. Registration can be performed manually or automatically. In either case, it is important to describe and annotate training resources using relevant keywords from controlled vocabularies, taxonomies, thesauruses, or ontologies. This reduces ambiguity and facilitates discovery and retrieval of information by improving the efficacy of metadata filtering. An example ontology is EDAM (Ison et al., 2013), which includes terms for data types, data identifiers, data formats, operations, and topics. Other ontologies can be found in resources like FAIRsharing (Sansone et al., 2019), the Ontology Lookup Service (Jupp et al., 2015), or BioPortal (Whetzel et al., 2011).
The preferred and hence primary mechanism for collecting training metadata in TeSS is to collect data from content providers automatically. This involves the use of a “scraper” that extracts information from a variety of trusted websites. In this case, describing digital objects with structured metadata is fundamental (not to mention that it will also make them FAIR). Regardless of the type of object, adding appropriate standardized metadata will help make it both machine- and human-readable.
What is the most recommended mechanism to increase the FAIRness of training events announcements and training materials? By using metadata schemas for structured data from the collaborative project Schema.org, adding structured metadata to web pages has been greatly facilitated (Guha, Brickley, & MacBeth, 2015). The community has developed annotation schemas for a vast variety of data types, including creative works, non-text objects, organizations, and products, all described by extensive lists of properties such as author, audience, and context. Search engines like Google crawl the internet to index those annotated websites and provide snippets of information via linked data objects. To make life science resources more discoverable, the Bioschemas community initiative (Gray, Goble, & Jiménez, 2017) builds upon the foundation of the Schema.org standard. For automatic registration of training resources, the recently released and first official version of the specifications for Course (a training course), CourseInstance (an event of said training course), and TrainingMaterial (Castro et al., 2022) are used in the automatic collection of structured metadata. Those three standards are community-endorsed, which facilitates a broader and more homogeneous adoption of the standard through life sciences content providers. The Bioschemas specification offers guidance to content providers on how to enrich their training resources to allow for more efficient sharing of structured metadata like prerequisites, target audience, and learning outcomes. Training registries like TeSS use a scraping mechanism to aggregate training resources from many providers when they have been annotated using Bioschemas specifications.
In this article, we describe how to search for specific training events and material in TeSS (Basic Protocol 1 and Support Protocol), gain access to TeSS (Basic Protocol 2 and Alternate Protocol), manually and automatically register training events and materials (Basic Protocols 3, 4, and 6), and register a content provider (Basic Protocol 5).
Basic Protocol 1: SEARCHING FOR TRAINING EVENTS AND MATERIALS IN TeSS
Training resources (both events and materials) may be searched in TeSS in several ways. A general search can be performed based on keywords, which will return separate lists of events and materials. Alternatively, events or materials can be searched for independently of each other. The second approach allows more precise filtering on several parameters (e.g., event type, country, and target audience) alone or in combination. In this protocol, we provide examples of searches for specific events or materials.
Necessary Resources
- Desktop or a laptop computer or a mobile device with fast Internet connection
- Up-to-date web browser such as Google Chrome, Mozilla Firefox, Apple Safari, or Microsoft Edge
Search for specific events
1a. Select “Events” in the top menu.
2a. Enter “Single-cell” as the keyword in the search text box and click on the magnifier icon.
3a. In the side menu, scroll down to the “Country” facet. Select “United Kingdom”.
4a. In the side menu, scroll to the “Target audience” facet. Select “Graduate students”.
Search for specific materials
1b. Select “Materials” in the top menu.
2b. Enter a keyword in the search text box. We will enter “Data retrieval”.
3b. In the side menu, select a difficulty level. We will select “Beginner”.
4b. Scroll to the “Resource type” facet and select “e-learning”.
Support Protocol: INTEGRATING TeSS WIDGETS ON YOUR WEBSITE
Widgets are chunks of JavaScript code that can be copied into the website source code to display TeSS content. Several different styles and functionalities are available from our configurable widget suite. Widgets can be used to enhance your site and offer your community lists of relevant events or training resources. Examples of code can be found at https://elixirtess.github.io/TeSS_widgets/.
Necessary Resources
- Desktop or a laptop computer or a mobile device with fast Internet connection
- Up-to-date web browser such as Google Chrome, Mozilla Firefox, Apple Safari, or Microsoft Edge
1.For a tabular layout of events with filtering options, choose the specific widget type Events table with a sidebar.
2.Manually annotate the HTML website (e.g., index.html) with the code snippet for the TeSS widget. More specifically, enter the code snippet as shown below as a sibling of a div element.
- function initTeSSWidgets() {
- TessWidget.Events(document.getElementById('tess-widget-events-table'),
- 'FacetedTable',
- {
- opts: {
- columns: [{name: 'Date', field: 'start'},
- opts: {
- TessWidget.Events(document.getElementById('tess-widget-events-table'),
{name: 'Name', field: 'title'},
{name: 'Location', field: 'location'}],
* allowedFacets: ['scientific-topics', 'country', 'city', 'target-audience'],
* facetOptionLimit: 5
* },
* params: {
* q: 'Python',
* country: ['Belgium', 'United Kingdom']
* }
* });
- }
3.Save the HTML page and serve the HTML page for visual inspection.
4.Visually validate that the rendered HTML page includes a table with the events extracted from TeSS.
5.Remove the term “Python” from the general search.
Basic Protocol 2: LOGGING IN TO TeSS USING AN INSTITUTIONAL ACCOUNT
In order to be able to register resources in TeSS, you need to log in to the registry. The following steps are specific for the login procedure using a Life Science RI account. Alternatively, you can use another existing account like Google, Apple, or ORCID. In this case, you will be redirected to the respective login portal, but we cannot provide a detailed procedure for all the federated authentication mechanisms involved.
Necessary Resources
- Desktop or a laptop computer or a mobile device with fast Internet connection
- Up-to-date web browser such as Google Chrome, Mozilla Firefox, Apple Safari, or Microsoft Edge
- Access to an email account
1.Open your browser and go to https://tess.elixir-europe.org/.
2.Click the “Log In” dropdown button on the top-right corner of the page.
3.Choose “Log in with LS Login” from the dropdown menu.

4.Start typing the name of your institution (e.g., “CNRS”) into the text box, choose the appropriate option when it appears, and proceed with your usual institutional login procedure.
Alternate Protocol: CREATING AND LOGGING IN TO A TeSS ACCOUNT
In order to register resources in TeSS as a unaffiliated provider, you can create a local account that will allow you to log in to the registry. The following steps show how this is done.
Necessary Resources
- Desktop or a laptop computer or a mobile device with fast Internet connection
- Up-to-date web browser such as Google Chrome, Mozilla Firefox, Apple Safari, or Microsoft Edge
- Access to an email account
1.Open your browser and go to https://tess.elixir-europe.org/.
2.Click the “Log In” dropdown button on the top-right corner of the page.
3.Click the “Register” option.
4.Fill out the username, email, and password fields, complete the captcha if present, and click the data processing consent checkbox after reading the privacy policy.
5.Wait for a confirmation email to arrive, then click the “Confirm my account” link.
6.Click the “Log In” dropdown button on the top-right corner of the page and then choose the “Login” option.
7.Fill out your username (or email address) and password, then click the “Log in” button.
Basic Protocol 3: MANUAL REGISTRATION OF TRAINING EVENTS IN TeSS
An event in TeSS is a link to a single training event sourced by a provider along with a description and related meta information (e.g., date, location, audience, ontological categorization, keywords). Training events can be added manually or harvested automatically from a provider's website. Here is the procedure to register training events manually.
Necessary Resources
- Desktop or a laptop computer or a mobile device with fast Internet connection
- Up-to-date web browser such as Google Chrome, Mozilla Firefox, Apple Safari, or Microsoft Edge
1.Log in to TeSS at https://tess.elixir-europe.org.
2.Click on the “Events” menu.
3.Click on “Register event” to be directed to the form.
4.In the “Type” field, select “Face-to-face” for a face-to-face event.
5.In the “Title” field, enter the title of the event: “Bulk RNASeq: from counts to differential expression”.
6.In the optional “Subtitle” field, enter any additional information for the event, such as “Spring edition” for a regularly repeated course.
7.In the “URL” field, enter the URL where the announcement and registration for the event can be found: “https://training.vib.be/all-trainings/bulk-rnaseq-counts-differential-expression-2”.
8.In the “Description” field, provide general information and relevant context about the event in the form of short text:
“The course consists of introductory online material on producing count matrices and two face-to-face sessions on differential expression analysis in R and all the questions that arise when trying the analysis on your own data.”
9.Specify the start date and door time of the event in the “Start” field via the pop-up “Date and Time Picker” (in our case, 29 September, 2022).
10.Specify the end date in the “End” field via the pop-up Date and Time Picker (4 October, 2022).
11.In the “Timezone” field, choose the time zone of the location where the event will occur: “(GMT:+01:00) Amsterdam” for our event in Leuven, Belgium.
12.In the “Duration” field, indicate how long the event will last in hours, days, or whatever applies best. In our example: “2 days”.
13.In the “Prerequisites” field, enter the necessary prerequisites for the training event:
“Background knowledge of NGS data formats and the first steps in the analysis workflow (fastqc -> bam files). If you are a newbie in the field, you have to follow the NGS introduction training first. Experience in R programming is highly recommended. If you never worked with R, you should attend the R introduction training first.”
14.In the “Learning objectives” field, list the learning objectives of the event, such as:
- Learn how RNA-seq reads are converted into counts
- Understand QC steps that can be performed on RNA-seq reads
- Generate interactive reports to summarize QC information with MultiQC
- Use the Galaxy Rule-based Uploader to import FASTQs from URLs
- Create a Galaxy Workflow that converts RNA-seq reads into counts
We recommend applying the Bloom hierarchy of cognitive skills to formulate the learning objectives. For more detailed information about our pedagogical model, please browse the course material of the ELIXIR Train the Trainer course at https://github.com/TrainTheTrainer/ELIXIR-EXCELERATE-TtT.
This field supports markdown syntax.
15.In the “Address” and related fields, start to type the address. Once you select an address, the Google maps view will be shown and the fields Venue, City, Region, Country, and Postcode will be filled in automatically. For our example:
| Address: | Herestraat 49 |
| Venue: | Campus Gasthuisberg |
| City: | Leuven |
| Region: | Flemish region |
| Country: | Belgium |
| Postcode: | 3000 |
16.In the “Eligibility” field, enter “First come first served”.
17.In the “Organiser” field, enter the organizer of the event: “ELIXIR-Belgium”.
18.In the “Contact” field, enter the name of the training coordinator.
19.In the “Host institutions” field, start typing “VIB” to bring up a dropdown menu to choose from.
20.In the “Keywords” field, enter “transcriptomics”.
21.In the “Target audience” field, enter “Life Science Researchers”.
22.In the “Scientific topics” field, enter “RNA-Seq, Sequencing”.
23.In the “Operations” field, enter “Differential gene expression profiling”.
24.In the “Capacity” field, enter the maximum number of attendants (e.g., 20).
25.In the “Event type” field, choose “Workshops and courses”.
26.In the “Tech requirements” field, enter “R, Rstudio, various R packages”.
27.For the type of credit or recognition of attendance that will be delivered, enter “Certificate of attendance recognized by Doctoral School”.
28.In the “External resources” field, search for “DESeq2” in the tools section. Once the tool has been found in bio.tools, click on the “+” sign to add it to the entry.
29.In the “Cost basis” field, select “Cost to non-members”.
30.In the “Content Provider” field, enter “VIB Training”.
31.In the “Materials search” box, search for “Transcriptomics data analysis made easy” and select suggested training materials to associate with the course.
32.Select the “Node” the organizer belongs to from the dropdown list: “ELIXIR Belgium”.
33.Enter the Sponsors/Funders of your training event: “VIB”.
34.Click the “Add event” button to finalize the registration.
Basic Protocol 4: MANUAL REGISTRATION OF TRAINING MATERIALS IN TeSS
In the context of TeSS, a training material is a link to a single online training material sourced by a content provider (such as text on a webpage, presentation, video, etc.) along with a description and related meta information (e.g., ontological categorization, keywords). Materials can be added manually or harvested automatically from a provider's website. Here is the procedure to register training materials manually.
Necessary Resources
- Desktop or a laptop computer or a mobile device with fast Internet connection
- Up-to-date web browser such as Google Chrome, Mozilla Firefox, Apple Safari, or Microsoft Edge
1.Log in to TeSS at https://tess.elixir-europe.org.
2.Click on the “Material” menu.
3.Click on “Register training material” to be directed to the form.
4.In the “Title” field, enter “How to use AlphaFold for prediction of single proteins or protein complexes using the HPC”.
5.In the “URL” field, enter “https://elearning.bits.vib.be/courses/alphafold/”.
6.In the “Description” field, enter:
"Architectural details, code and trained AlphaFold models were released by DeepMind in 2021.Given the high computational cost of deep learning algorithms, specialized hardware and software are required. Online solutions are available but come with considerable disadvantages. Therefore, the Flemish Supercomputer Center (VSC) provides high performance computing facilities, on which AlphaFold is installed and fully operational. This course gives a solid introduction on how AlphaFold can be easily and swiftly accessed using the HPC.
This tutorial material was created by Jasper Zuallaert (VIB-UGent), with the help of Alexander Botzki (VIB) and Kenneth Hoste (UGent). For questions and remarks, feel free to contact jasper.zuallaert@vib-ugent.be or training@vib.be."
7.In the “Keywords” field, enter “Structure prediction”, “AlphaFold Database”, and “Protein complex prediction”.
8.In the “License” field, select “Creative Commons Attribution Non commercial Share Alike 4.0 International”.
9.In the “Status” field, select “Active”.
10.In the “Contact” field, enter “training@vib.be”.
11.In the “DOI” field, enter the URL of the DOI if you have created one for your training material.
12.In the “Version” field, enter “1.0”.
13.In the “Content Provider” field, select the content provider from the dropdown list: “VIB Training”.
14.In the “Authors” field, enter the name(s) of the author(s) of the material: “Jasper Zuallaert” and “Alexander Botzki”.
15.In the “Contributors” field, enter the name(s) of contributor(s) to the material: “Kenneth Hoste”.
16.In the “Events” field, select “Structural Prediction of Proteins using AlphaFold on the HPC” from the dropdown list.
17.In the “Target audience” field, select “Life scientists with programming skills” from the dropdown list.
18.In the “Prerequisites” field, add:
“You are encouraged to use your own laptop. For those who do not have a laptop, the YASARA software can be run in a remote Linux environment (access to cloud via web browser). Knowledge of command line and basic Python skills are recommended.”
19.For “Competency” level, select “Intermediate”.
20.For “Learning Objectives”, enter:
“Understand the technical methodology of AlphaFold2, Understand the technical setup at the Flemish SuperComputer, Predict three-dimensional protein models with AlphaFold2 using the HPC at the VSC UGent.”
21.Enter the “Date created” (1 April, 2022).
22.Enter “no value” in “Date modified”.
23.Enter the “Date published” (28 April, 2022).
24.In the “Resource types” field, enter “Tutorial” and “Slides” from the dropdown menu.
25.In the “Scientific topics” field, select “Structure prediction, Protein Structure Analysis, Machine Learning”.
26.In the “External resources” field, click on “Suggested tools to associate with this resource”. In the “Search” field, enter “AlphaFold”. Wait a couple of seconds until the result from the request to the ELIXIR Tools and Services Registry is shown. Select the entry “AlphaFold 2” by clicking on the “+” sign.
27.In the “Nodes” field, use the dropdown list to select “Belgium” as the node to which the authors and contributors belong.
28.In the “Operations” field, select “Multiple Sequence Alignment, Structure Visualisation”.
29.Click on “Register training material” to finalize the registration.
Basic Protocol 5: REGISTRATION OF A CONTENT PROVIDER IN TeSS
TeSS features content providers, which are entities such as academic institutions, non-profit organizations, or portals that provide training materials of relevance to the life sciences and ELIXIR. In order to have a training event automatically harvested in TeSS (see Basic Protocol 6), a content provider must first be registered. This also adds visibility for content providers. Here is the procedure to register a new content provider in TeSS.
Necessary Resources
- Desktop or a laptop computer or a mobile device with fast Internet connection
- Up-to-date web browser such as Google Chrome, Mozilla Firefox, Apple Safari, or Microsoft Edge
1.Log in to TeSS at https://tess.elixir-europe.org.
2.Under “Directory”, click on the “Providers” menu.
3.Click on “Register content provider” to be directed to the form.
4.In the “Title” field, enter “NanoCommons”.
5.In the “URL” field, enter the URL of the content provider: “https://www.nanocommons.eu/”.
6.In the “Contact” field, enter “Jean Dupont” as the contact person.
7.In the “Description” field, provide general information and relevant context about the content provider in the form of short text:
“NanoCommons will deliver a sustainable and openly accessible nanoinformatics framework (knowledgebase and integrated computational tools, supported by expert advice, data interpretation and training), for assessment of the risks of NMs, their products and their formulations. NanoCommons combines Joint Research Activities to implement the nanoinformatics Knowledge Commons, Networking Activities to facilitate engagement with the research community, industry and regulators, and provision of funded Access to the nanoinformatics tools via funded calls for Transnational Access.”
8.To provide a logo for the Content Provider, in the “Image” field under “or enter the URL”, enter the URL of the image location: https://www.nanocommons.eu/wp-content/uploads/2018/04/NanoCommons-Logo-Sphere-Trans-White-circle-512px.png
9.In the “Type” field, enter “Project” as type of content provider.
10.In the “Approved Editors” field, enter “Alexander” and select “alexander.botzki@vib.be (Alexander Botzki)”.
11.In the “Keywords” field, enter “nanotechnology”, “commons”, “product safety”, and “toxicology”.
12.In the “Nodes” field, select an associated ELIXIR node, if applicable.
13.Click on “Register content provider” to confirm the creation of the content provider.
Basic Protocol 6: AUTOMATED HARVESTING OF TRAINING EVENTS AND MATERIALS IN TeSS
TeSS features several options for automatically harvesting resources from external sources. This can be helpful if you maintain a large collection of events and materials that changes frequently. To register resources in TeSS automatically, data must be able to be reliably extracted from target sources. To this end, it is helpful if the data are structured according to a globally used standardized format. The following are examples of the kinds of structured data that are compatible with TeSS.
If your website currently includes no structured data and you want your resources added to TeSS, we recommend using Bioschemas to structure your site. Schema.org is a project run by a consortium of search engines. Schema.org has created an extensive library of schemas that webmasters can use to explicitly mark up website content in order to improve search engine visibility and interoperability. Bioschemas is an initiative to supplement the work of Schema.org to help improve the findability of online resources in the life sciences. TeSS supports the following Bioschemas profiles: (1) CourseInstance and Course for events that are courses, (2) Event for other events, and (3) TrainingMaterial for training materials. Figure 3 provides a schematic overview of the protocol steps.
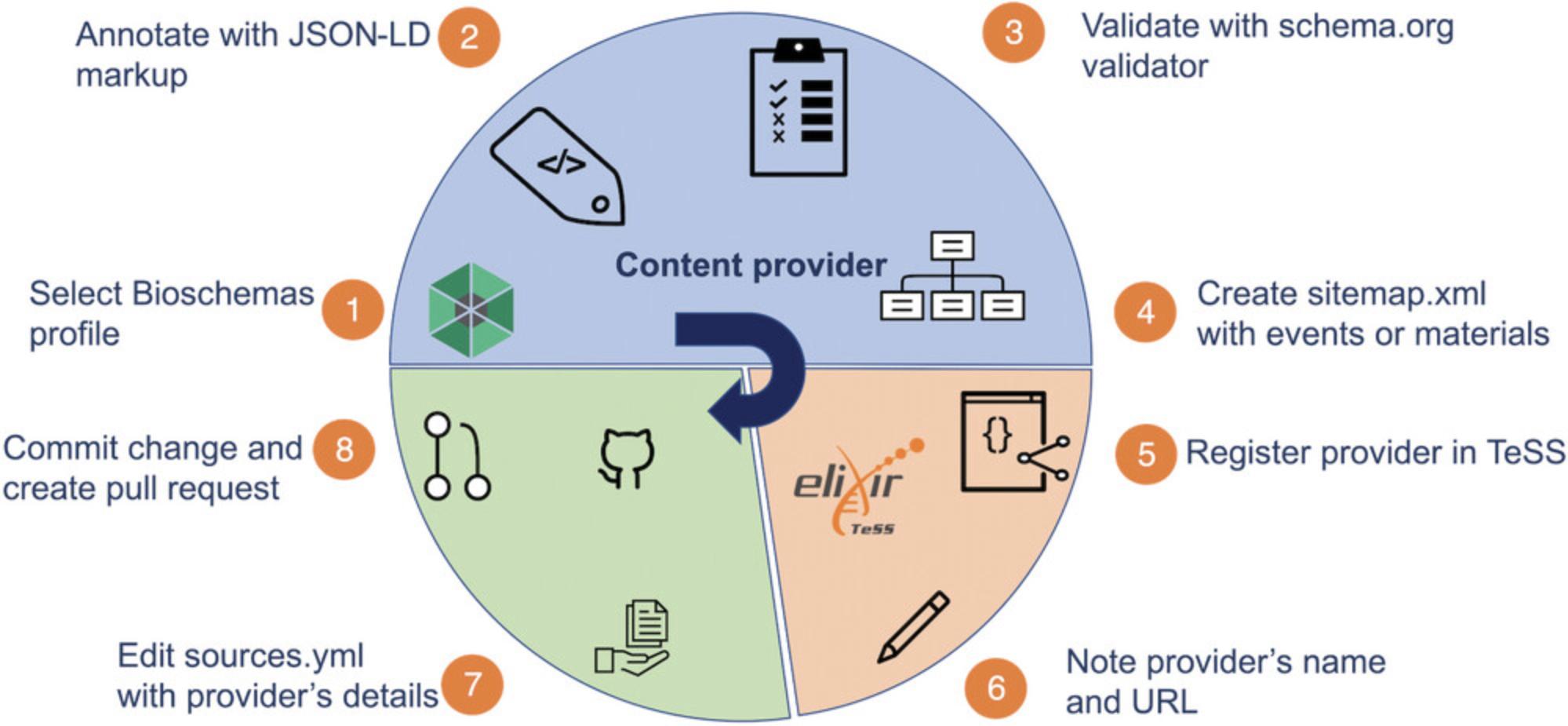
Necessary Resources
- Desktop or a laptop computer or a mobile device with fast Internet connection
- Up-to-date web browser such as Google Chrome, Mozilla Firefox, Apple Safari, or Microsoft Edge
1.Select the profile Learning resource https://bioschemas.org/profiles/TrainingMaterial/1.0-RELEASE for the automated harvesting of a specific training material from the NanoCommons initiative (https://nanocommons.github.io/user-handbook/) located at https://nanocommons.github.io/tutorials/enteringData/index.html.
2.Manually annotate the HTML website containing the training material (here, index.html; https://nanocommons.github.io/tutorials/enteringData/index.html) using JSON-LD markup. More specifically, enter a JSON object as shown below in a script element with the attribute type="application/ld+json". This script element needs to be placed as a child of the head element of the HTML page. This results in the following script HTML element.
- <script type="application/ld+json">
- {
- "@context": "https://schema.org/",
- "@type": "LearningResource",
- "http://purl.org/dc/terms/conformsTo": { "@type": "CreativeWork", "@id": "https://bioschemas.org/profiles/TrainingMaterial/1.0-RELEASE" },
- "name": "Adding nanomaterial data",
- "version": "0.9.3",
- "description": "This tutorial describes how nanomaterial data can be added to an eNanoMapper server using an RDF format.",
- "license": "https://creativecommons.org/licenses/by/4.0/",
- "keywords": "ontologies, enanomapper, RDF",
- "url": "https://nanocommons.github.io/tutorials/enteringData/",
- "provider": {
- "@type": "Organization",
- "name": "NanoCommons",
- "url": "https://www.nanocommons.eu/"
- },
- "audience": {
- "@type": "EducationalAudience",
- "educationalRole": "Graduates"
- },
- "inLanguage": {
- "@type": "Language",
- "name": "English",
- "alternateName": "en"
- },
- "author": [
- {
- "@context": "https://schema.org",
- "@type": "Person",
- "name": "Egon Willighagen",
- "identifier": "https://orcid.org/0000-0001-7542-0286",
- "orcid": "https://orcid.org/0000-0001-7542-0286"
- }
- ]
- }
- {
3.Validate the individual page with the Schema.org validator at https://validator.schema.org by pasting the URL https://nanocommons.github.io/tutorials/enteringData/index.html into the Fetch URL tab (Fig. 4).
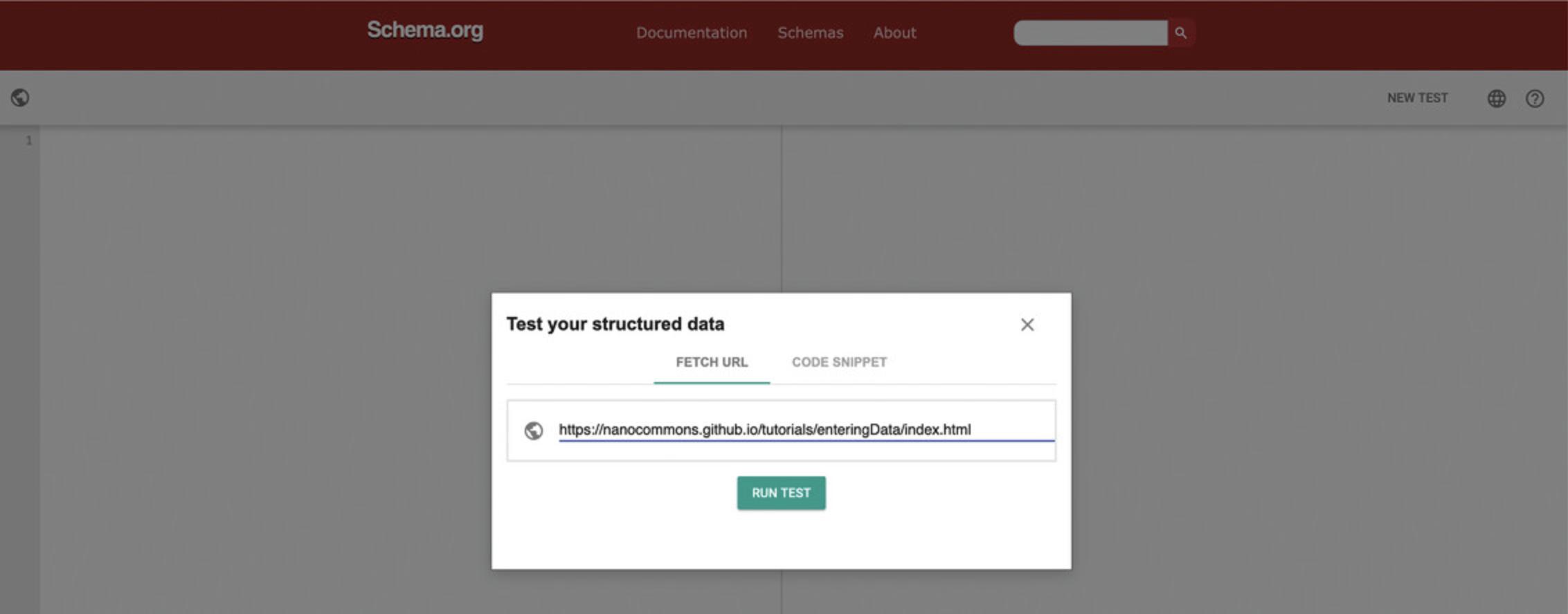
4.Create a sitemap listing the material page URL and save it as sitemap.xml. The sitemap.xml file needs to be publicly browsable on the internet. In our example, the manually created sitemap.xml is published at https://nanocommons.github.io/sitemap.xml.
5.Enter “NanoCommons” as the “Content Provider”.
6.Make note of the Content Provider's exact Title and URL. In our example, this is NanoCommons and https://www.nanocommons.eu/.
7.Go to https://github.com/ElixirTeSS/bioschemas_sources/edit/main/sources.yml to edit the sources.yml file and add your Content Provider details along with the URL to your sitemap:
- title: NanoCommons
8.To commit your change, click the button to open a pull request, which will then be reviewed. After review, the new training material will appear in TeSS.
COMMENTARY
Background Information
During the ELIXIR-EXCELERATE project, the TeSS platform was established as the key application for bioinformatics training events and material aggregation. During further adoption of the TeSS platform as such on the international level (e.g., in Australia and the Netherlands), it became obvious that the implementation of individual automatic content scrapers for more than 30 content providers lead to an enormous diversity of the code base and finally to a maintenance challenge. Therefore, during the last two to three years, a strategic reorientation of the automated harvesting procedure has been proposed and implemented in TeSS using the Bioschemas profiles for Course, CourseInstance, and TrainingMaterial. This approach will put content providers in a more prominent position, since they are in turn responsible for more structured delivery of more comprehensive metadata of their training events and materials to TeSS as the aggregation platform. In the long run, this will increase the quality level of aggregated resources and their usefulness to the TeSS user base.
Content providers looking to promote the use of particular tools and databases via their course offerings or specific training materials can find information in other ELIXIR registries, such as bio.tools (https://bio.tools) for tools and web services and FAIRsharing.org (https://fairsharing.org) for databases, standards, and policies. Lately, such registries have started to track training relating to the resources they list. Therefore, it is mutually beneficial that TeSS provides the relevant links (allowing users to search for resource-specific training) and the resource pages in bio.tools and FAIRsharing that contain reciprocal links for users to discover relevant training in TeSS.
Related to the provisioning of persistent identifiers, TeSS provides the Digital Object Identifier (DOI) as a unique means to identify training materials. Given that minting services by, for example, university libraries are not yet broadly available for teaching and learning material in contrast to research datasets and collections, associated workflows, software, and models, we recommend using services like OSF or Zenodo to get own unique, persistent URLs and DOIs.
Given the engagement of the ELIXIR Train the Trainer instructor's community to create a vibrant environment for reciprocal support and exchange of experiences via the ELIXIR Train the Trainer Programme, TeSS content providers are encouraged to follow best-practice guidance on course and training material development (Via et al., 2020). Therefore, we recommend applying the Bloom hierarchy of cognitive skills to formulate the learning objectives for courses and training materials. For more detailed information about our pedagogical model, please browse the course material of the ELIXIR Train the Trainer course (https://github.com/TrainTheTrainer/ELIXIR-EXCELERATE-TtT).
Critical Parameters
Curation of manual content for automatically harvested events or training materials
In Basic Protocols 3 and 4, it is important to note that much of the content in TeSS is retrieved and kept up to date via automated scrapers (see Basic Protocol 6) that pull information from public webpages and application programming interfaces (APIs) in regular intervals. To prevent TeSS from overwriting a field you have just changed, click the “locker” icon to lock the field. Fields marked as locked will not be overwritten by an automatic procedure.
For training events and materials, mandatory fields are marked with an * symbol. Unmarked fields are optional.
Automated harvesting of training events
In step 2 of Basic Protocol 6, we recommend following the guidelines from the Bioschemas community on strategies for marking up your internet sites by using this tutorial: https://bioschemas.org/tutorials/howto/howto_add_markup. If your website is hosted on GitHub, you can follow https://bioschemas.org/tutorials/howto/howto_add_github instead.
In step 3, if you get an error during the validation procedure by the Schema.org validator, consult the properties definitions in the respective profile on Schema.org and Bioschemas.org.
In step 7, you must have a GitHub account to edit the sources.yml file. It is important to make sure the title and URL exactly match the Content Provider's title and URL on TeSS. You will make changes in the GitHub project ElixirTeSS/bioschemas_sources, for which you do not have write access. Submitting a change will write it to a new branch in your fork
If you have implemented Bioschemas markup on your website and would like your content to appear in TeSS, see our “Bioschemas sources” repository for detailed information on how to proceed: https://github.com/ElixirTeSS/bioschemas_sources#readme.
Troubleshooting
See Table 1 for troubleshooting if you experience issues with missing concepts in the EDAM ontology, potentially new properties in the form on the TeSS website, a missing hosting institution, or synchronizations issues with the automated procedure.
| Problem | Possible cause | Solution |
|---|---|---|
| Missing concept in the EDAM ontology | EDAM does not cover all scientific topics | Select a closely related term or submit a request to add new concepts, if appropriate |
| There are new properties in the TeSS interface for events or material | Sometimes new properties are added to the TeSS interface | Browse the documentation on the TeSS website |
| Your host institution is missing when you go to register an event or material | The dropdown of host institutions is a fixed list | Enter a new host organisation in the field |
| After using the automated harvesting protocol, your training material does not appear in TeSS the following day | The title and/or URL did not exactly match the Content Provider's title and URL on TeSS | Double check that the title and URL match exactly the Content Provider's title and URL on TeSS |
| You have provided a validated Bioschemas markup, but the harvesting procedure in TeSS did not recognize all the properties correctly | Submit an issue on the ELIXIRTeSS/Bioschemas_sources GitHub repository |
Understanding Results
As a content provider in TeSS, it is important to realize that adding training events or materials manually will take roughly 10-15 min per item if all properties are filled in. Given that this can be a considerable time investment, we strongly encourage content providers to implement the automated harvesting procedure based on Bioschemas annotation. While this can also be very time consuming, depending on the nature of the content provider's technical platform, the upfront investment of the technical setup will pay back in the long run. From a consumers’ perspective of the TeSS content, a high annotation level is very much appreciated. The higher the annotation level, the more useful the search experience will be for the TeSS audience. In particular, this greatly facilitates the filtering process after an initial search using facets, e.g., for country, difficulty level, resource type, or audience.
Acknowledgments
The fundamental work for building TeSS as a platform was part of the ELIXIR-EXCELERATE project 2014-2018 and we are grateful for the efforts provided from across the ELIXIR Nodes and ELIXIR Platforms. We are also grateful for support and funding of this work by the ELIXIR Scientific Programme 2019-2023 (https://elixir-europe.org/about-us/what-we-do/elixir-programme). Furthermore, we would like to acknowledge Christof De Bo (ELIXIR-BE, VIB Bioinformatics Core) for his support creating the illustration in Figure 3.
Author Contributions
Finn Bacall : Conceptualization, investigation, methodology, software, validation, writing review and editing; Aitor Apaolaza : Investigation, methodology, software; Munazah Andrabi : Investigation, writing review and editing; Chris Child : Investigation, methodology, software; Carole Goble : Methodology, project administration, supervision, writing review and editing; Olivier Sand : Conceptualization, project administration, supervision, validation, writing original draft, review and editing; Alexander Botzki : Conceptualization, project administration, supervision, visualization, writing original draft, review and editing.
Conflict of Interest
The authors declare no conflict of interest.
Open Research
Data Availability Statement
The data that support the findings of this study are openly available in the ELIXIR TeSS GitHub repository at https://github.com/ElixirTeSS/bioschemas_sources.
Literature Cited
- Beard, N., Bacall, F., Nenadic, A., Thurston, M., Goble, C. A., Sansone, S.-A., & Attwood, T. K. (2020). TeSS: A platform for discovering life-science training opportunities. Bioinformatics , 36(10), 3290–3291. doi: 10.1093/bioinformatics/btaa047
- Garcia, L., Batut, B., Burke, M. L., Kuzak, M., Psomopoulos, F., Arcila, R., … Palagi, P. M. (2020). Ten simple rules for making training materials FAIR. PLoS Computational Biology , 16(5), e1007854. doi: 10.1371/journal.pcbi.1007854
- Castro, L., Palagi, P. M., Beard, N., Bioschemas Training Profiles Group Members, ELIXIR FAIR Training Focus Group, The GOBLET Foundation, … Brazas, M. (2022). Bioschemas Training Profiles: A set of specifications for standardizing training information to facilitate the discovery of training programs and resources. bioRxiv , 2022.11.24.516513. doi: 10.1101/2022.11.24.516513
- Gray, A. J., Goble, C., & Jiménez, R. C. (2017). Bioschemas: From potato salad to protein annotation. International Semantic Web Conference; Berlin. Accessed 2019 Mar 11. Retrieved from https://pdfs.semanticscholar.org/74ec/a9c89622bff731b21b03acb4f2400a0f00fa.pdf
- Guha, R. V., Brickley, D., & MacBeth, S. (2015). Schema.org: Evolution of structured data on the web: Big data makes common schemas even more necessary. Queue , 13(9), 10–37. doi: 10.1145/2857274.2857276
- Ison, J., Kalas, M., Jonassen, I., Bolser, D., Uludag, M., McWilliam, H., … Rice, P. (2013). EDAM: An ontology of bioinformatics operations, types of data and identifiers, topics and formats. Bioinformatics , 29(10), 1325–1332. doi: 10.1093/bioinformatics/btt113
- Jupp, S., Burdett, T., Malone, J., Leroy, C., Pearce, M., McMurry, J., & Parkinson, H. (2015). A new ontology lookup service at EMBL-EBI. In J. Malone, R. Stevens, K. Forsberg, & A. Splendiani (Eds.), Proceedings of the 8th SWAT4LS International Conference, Cambridge, UK, December 7-10, 2015.
- Sansone, S.-A., McQuilton, P., Rocca-Serra, P., Gonzalez-Beltran, A., Izzo, M., … the FAIRsharing Community. (2019). FAIRsharing as a community approach to standards, repositories and policies. Nature Biotechnology , 37(4), 358–367. doi: 10.1038/s41587-019-0080-8
- Via, A., Palagi, P. M., Lindvall, J. M., Tractenberg, R. E., Attwood, T. K., & The GOBLET Foundation. (2020). Course design: Considerations for trainers – A professional guide. F1000Res , 9, 1377. doi: 10.7490/f1000research.1118395.1
- Whetzel, P. L., Noy, N. F., Shah, N. H., Alexander, P. R., Nyulas, C., Tudorache, T., & Musen, M. A. (2011). BioPortal: Enhanced functionality via new web services from the National Center for Biomedical Ontology to access and use ontologies in software applications. Nucleic Acids Research , 39, (suppl), W541–W545. doi: 10.1093/nar/gkr469
Internet Resources
Main information site for EDAM, the ontology of bioscientific data analysis and data management. EDAM is a comprehensive ontology of well-established, familiar concepts that are prevalent within bioscientific data analysis and data management (including computational biology, bioinformatics, and bioimage informatics).
Schema.org is a collaborative community activity with a mission to create, maintain, and promote schemas for structured data on the internet, on web pages, in email messages, and beyond. On this website, you can find https://bioschemas.org/, the homepage of the Bioschemas community. Bioschemas aims to improve the findability on the web of life sciences resources such as datasets, software, and training materials. Information about the three Bioschemas profiles implemented in this protocol can be found at https://bioschemas.org/profiles. Various tutorials can be found at https://bioschemas.org/tutorials.
Site for easy validation of websites that have been marked up with structured data as, e.g., a JSON-LD object.
This is the README file of the GitHub repository bioschemas_sources for the TeSS portal hosted in the ELIXIR TeSS GitHub organization.
GitHub repository hosted by ELIXIR-EXCELERATE-TtT in the TrainTheTrainer GitHub organization.
GitHub repository for TeSS widgets where several implementation scenarios are documented.

