Intravital Microscopy to Visualize Murine Small Intestinal Intraepithelial Lymphocyte Migration
Matthew Fischer, Matthew Fischer, Karen L. Edelblum, Karen L. Edelblum
Abstract
Intraepithelial lymphocytes (IELs) are critical sentinels involved in host defense and maintenance of the intestinal mucosal barrier. IELs expressing the γδ T-cell receptor provide continuous surveillance of the villous epithelium by migrating along the basement membrane and into the lateral intercellular space between adjacent enterocytes. Intravital imaging has furthered our understanding of the molecular mechanisms by which IELs navigate the epithelial compartment and interact with neighboring enterocytes at steady state and in response to infectious or inflammatory stimuli. Further, evaluating IEL migratory behavior can provide additional insight into the nature and extent of cellular interactions within the intestinal mucosa. Three protocols describe methodology to visualize small intestinal IEL motility in real time using fluorescent reporter–transgenic mice and/or fluorophore-conjugated primary antibodies and spinning-disk confocal microscopy. Using Imaris image analysis software, a fourth protocol provides a framework to analyze IEL migration and quantify lymphocyte/epithelial interactions. Together, these protocols for intravital imaging and subsequent analyses provide the basis for elucidating the spatiotemporal dynamics of mucosal immune cells and interactions with neighboring enterocytes under physiological or pathophysiological conditions. © 2022 The Authors. Current Protocols published by Wiley Periodicals LLC.
Basic Protocol 1 : Mouse preparation and laparotomy
Support Protocol : Antibody labeling of cell surface markers
Basic Protocol 2 : Image acquisition by spinning-disk confocal microscopy
Basic Protocol 3 : 4D analysis of images
INTRODUCTION
Intraepithelial lymphocytes (IELs) are located within the intestinal epithelium, which allows these sentinel immune cells to rapidly respond to luminal microorganisms (Cheroutre, Lambolez, & Mucida, 2011). IELs can be characterized by their expression of the αβ or γδ T-cell receptor (TCR) and markers of tissue residence, including CD103, CD69, or CD8α (Olivares-Villagómez & Van Kaer, 2018). γδ IELs comprise ∼60% of the murine IEL compartment and are important in limiting intestinal injury, inflammation, and infection (Hu, Jia, & Edelblum, 2018; Nielsen, Witherden, & Havran, 2017). Although the γδTCR interacts with butyrophilin-like proteins (Di Marco Barros et al., 2016; Jandke et al., 2020), a ligand for the murine γδTCR has yet to be identified. The absence of a specific γδTCR ligand has made elucidating the functional role of these cells within the intestinal mucosa somewhat challenging.
These limitations led us to develop a live imaging approach to visualize the dynamics of γδ IEL interactions with neighboring enterocytes using TcrdH2BeGFP (TcrdEGFP) (Prinz et al., 2006) or TcrdGDL (Sandrock et al., 2018) mice, which express GFP downstream of the Tcrd promoter in either the nucleus or the cytoplasm, respectively. Using intravital microscopy, we were the first to report that γδ IELs survey the villous epithelium by continuously migrating along the basement membrane and into the lateral intercellular space (LIS) between adjacent epithelial cells (Edelblum et al., 2012). Intravital imaging has been essential in broadening our understanding of how γδ IELs regulate the epithelial barrier (Fischer, Golovchenko, & Edelblum, 2020; Hu et al., 2022). Not only is intravital microscopy useful in identifying novel functions of mucosal immune cells, but these studies are now employed as a standard readout of IEL function (Sumida & Cyster, 2018; Sumida et al., 2017; Wang, Sumida, & Cyster, 2014).
The protocols described herein provide step-by-step instructions regarding how to externalize the small intestinal mucosa (Basic Protocol 1) and visualize IELs using a fluorescent reporter and/or a fluorophore-conjugated primary antibody (Support Protocol and Basic Protocol 2). We also provide a protocol for quantifying the dynamics of IEL motility (Basic Protocol 3). Taken together, these protocols encompass the procedures required to evaluate the spatiotemporal kinetics of IELs at steady state or in response to pathophysiological stimuli, such as intestinal inflammation or microbial pathogens.
STRATEGIC PLANNING
Before beginning, it is important to ensure that the enclosed incubator on the microscope has had sufficient time to reach 37°C. Once the surgery has begun (Basic Protocol 1), there will be no extended breaks until image acquisition (Basic Protocol 2) has been completed (Fig. 1). However, image analysis (Basic Protocol 3) can be performed at any time following video acquisition.
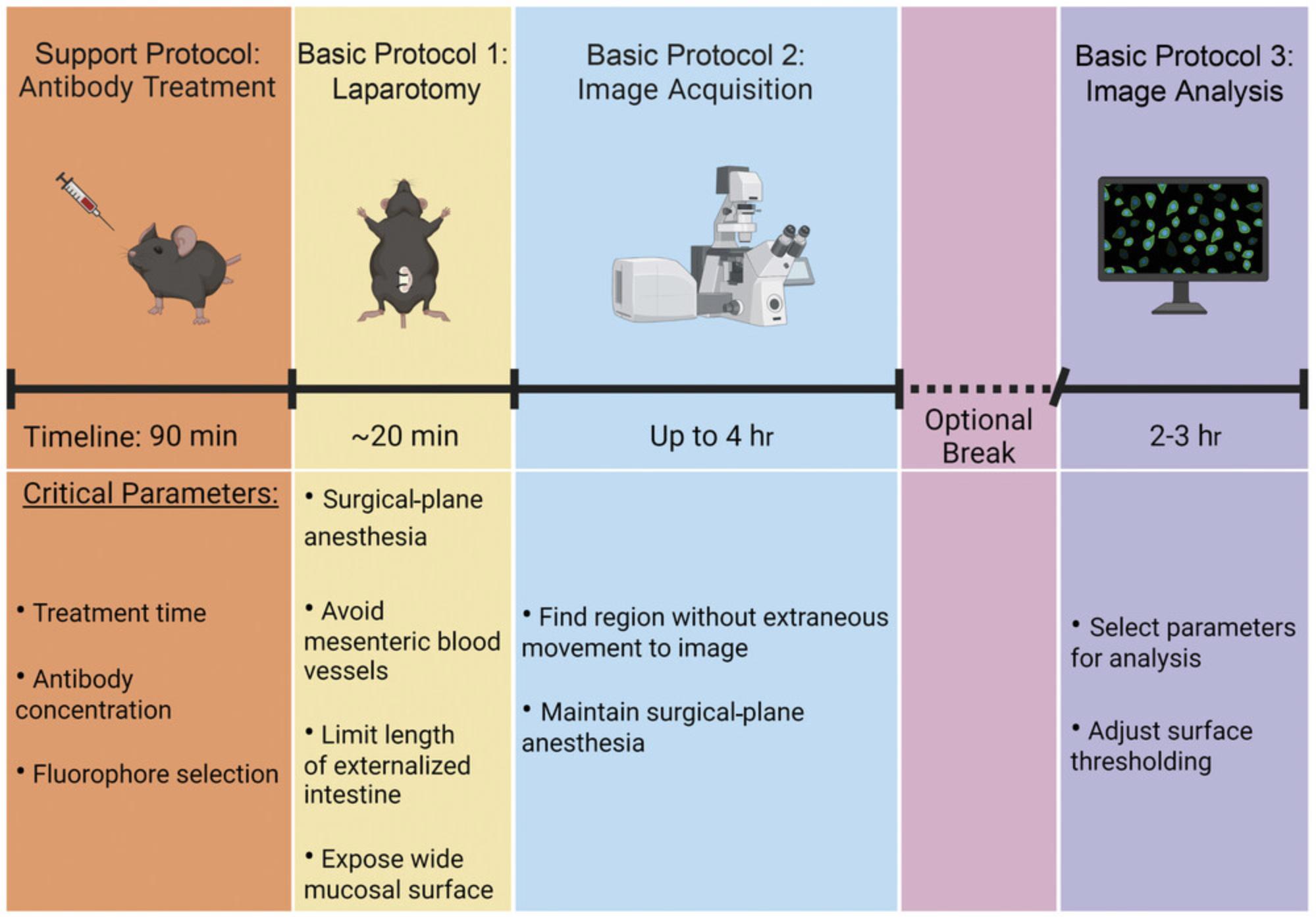
CAUTION : Ensure that all procedures have been approved by your local animal care and biosafety review committees. If using common equipment in a core facility, check with facility staff to determine if there are additional protocols that must be followed if performing intravital microscopy, especially under BSL2 conditions.
Basic Protocol 1: MOUSE PREPARATION AND LAPAROTOMY
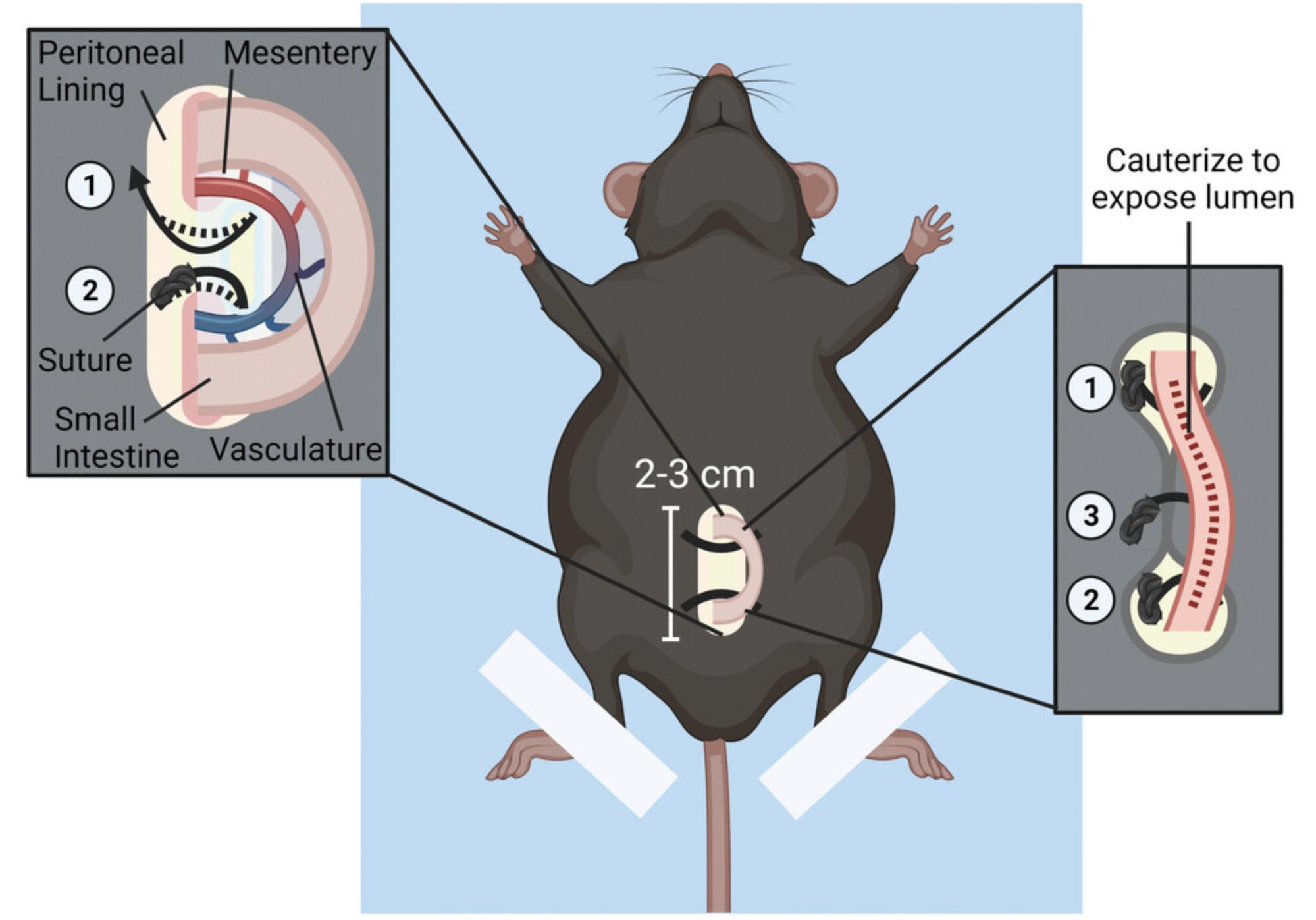
Intravital imaging allows for visualization of immune cell migration dynamics in the intestinal mucosa. This protocol describes how to identify and expose a section of murine small intestine for intravital imaging while maintaining an intact neurovascular supply (Fig. 2). Although the surgery and imaging protocols are separated here, Basic Protocol 2 should be started immediately after the surgery is complete. Mice must be maintained in a surgical plane of anesthesia throughout both protocols.
Materials
-
8- to 12-week-old Tcrd-H2BeGFP C57BL/6 mice (The Jackson Laboratory, strain no. 016941)
-
Ketamine-xylazine solution (see recipe)
-
Lubricant (MWI Veterinary Puralube Ointment, MWI Veterinary, cat. no. 027505)
-
27G hypodermic needles (27G PrecisionGlide Hypodermic Needles, Becton Dickinson, cat. no. 305109)
-
Sterile 1-ml syringes (1 ml BD Slip Tip Sterile Syringes, Becton Dickinson, cat. no. 309659)
-
Sterile 0.5-ml tuberculin syringes with attached 27G needle (Becton Dickinson, cat. no. 305620)
-
Hoechst 33342–saline solution (see recipe)
-
Heating pad [12 × 24–in. (30 × 60–cm); Sunbeam, cat. no. 732-500-000R]
-
Absorbent underpads [6 × 6–in. (15.25 × 15.25–cm); Fisher Scientific, cat. no. 14-206-62]
-
Laboratory tape (12.7-mm General Purpose Laboratory Tape, VWR, cat. no. 89097-920)
-
Rectal probe (Kent Scientific, cat. no. RET-3) with thermocouple thermometer (Type J/K/T Thermocouple Thermometer, Kent Scientific, cat. no. WD-20250-91)
-
Operating scissors (4.5-in. length, 41-mm blade length; ROBOZ, cat. no. RS-6802)
-
Angled forceps (two pairs; Nugent Forceps, Angular; 1-mm tip width, 4.25-in. length; ROBOZ, cat. no. RS-5228)
-
Curved, taper-point needles (Eye Needles, Size #3, 1/2 Circle, Taper Point, 12 mm Chord Length, ROBOZ, cat. no. RS-7983-3)
-
5-cm sutures (black braided silk, non-absorbable, non-sterile surgical sutures, LOOK™, cat. no. SP116)
-
Electrocautery (Change-A-Tip Deluxe High Temperature Cautery kit, Bovie, cat. no. DEL1)
-
Kimwipes [11.2 × 21.3–cm (4.4 × 8.4–in.); Kimberly Clark, cat. no. 21905-026]
-
Vannas scissors (McPherson-Vannas Micro Dissecting Spring Scissors, 3" Overall Length, ROBOZ, cat. no. RS-5600)
-
Covered container (for mouse transport)
1.Anesthetize a 8- to 12-week-old Tcrd-H2BeGFP C57BL/6 mouse by i.p. injection of ketamine-xylazine solution based on weight of the animal (120 mg/kg ketamine and 10 mg/kg xylazine) with a 27G hypodermic needle attached to a sterile 1-ml syringe according to IACUC-approved procedure.
2.Once the mouse is anesthetized, slowly inject Hoechst 33342–saline solution nuclear dye (37.5 mg/kg) through retro-orbital venous sinus using a sterile 0.5-ml tuberculin syringe with an attached 27G needle. Use lubricant to prevent drying of mouse's eyes.
3.Set heating pad warmth setting to medium and tape a 6 × 6–in. (15.25 × 15.25–cm) absorbent underpad to surface of the heating pad using laboratory tape. Then, use laboratory tape to secure hind legs of the mouse to the absorbent underpad (Fig. 2, center). Monitor body temperature using a rectal probe with a thermocouple thermometer.
4.Use operating scissors to make a 2-cm vertical incision along lower midline of the mouse abdomen (Fig. 2, center).
5.Identify a suitable region of jejunum (2 to 3 cm) for imaging. Use angled forceps to carefully pull out cecum and small intestine until a region of jejunum that is devoid of Peyer's patches and luminal contents is exposed.
6.Leave this area of jejunum tissue externalized while gently returning cecum and any remaining exposed small intestine into the peritoneal cavity.
7.Create a hole in membrane supporting the mesenteric vasculature by placing a pair of opened angled forceps on each side of the membrane and gently rubbing the forceps tips together.
8.To close the incision, use forceps to guide a curved, taper-point needle attached to a 5-cm suture through one side of the peritoneal lining, through the hole in the mesenteric membrane under the externalized intestine made in step 7, and back up through the other side of the peritoneal lining (Fig. 2, left box).
9.Close skin beneath the intestinal loop by repeating the same process as described in step 8.
10.Lightly cauterize a line of perforation along length of the externalized jejunum (∼1.5 cm) with an electrocautery (Fig. 2, right box). Using a moistened Kimwipe, immediately apply a few drops of water to cauterized surface to limit heat-induced tissue damage. Wick away residual water with a dry Kimwipe.
11.Use Vannas scissors to first cut a small horizontal slit at distal edge of the cauterized line. Insert scissors to cut along the length of the cauterized tissue to expose the luminal mucosal surface for imaging.
12.Cover abdomen with a damp Kimwipe to prevent dehydration of the tissue. Transport mouse to the microscope in Basic Protocol 2 in a covered container.
Support Protocol: ANTIBODY LABELING OF CELL SURFACE MARKERS
Identifying a cell of interest using a reporter mouse strain is the most efficient strategy for labeling and tracking cell migration during intravital imaging (Basic Protocol 2); we use TcrdEGFP and TcrdGDL mice to track γδ IELs (Hu et al., 2018; Hu et al., 2022). However, various immune cells, including CD8α+ IELs, can be labeled with fluorophore-conjugated primary antibodies prior to imaging to identify specific cell populations of interest (Sumida & Cyster, 2018; Sumida et al., 2017; Wang et al., 2014).
The antibody labeling protocol described below has been optimized for labeling CD8α+ IELs; however, fluorophore, concentration, and labeling time for an individual antibody are important considerations. Antibodies labeled with far-red fluorophores such as APC or Alexa Fluor 647 are preferred due to their brightness and resistance to photobleaching. Depending on the laser configuration of the microscope and the number of channels to be imaged, it is possible to use one laser channel to image two distinct features within a region of interest. For example, because CD8α+ T cells are spatially distinct from the lumen, Alexa Fluor 633 hydrazide and APC-CD8α can be imaged simultaneously without affecting downstream analysis. This has advantages because the simultaneous acquisition of two features will decrease the length of time it takes to acquire each Z-stack, thus increasing the temporal resolution. However, if the fluorophore-conjugated antibody is dimmer than the similarly labeled hydrazide or if the labeling efficiency is low, the intensity of the brighter feature (lumen) would be oversaturated in post-processing. This would make it challenging to resolve the lumen and the surface of the cell of interest when generating surfaces during analysis. Further, if using fluorophore-conjugated antibodies for intravital imaging, the relative expression of the antigen should be considered. The amount of antibody should be in excess to ensure that the cell of interest will be labeled appropriately. Labeling of tissue-resident immune cells, such as IELs, requires more time for the antibody to penetrate into the tissue compared to labeling of infiltrating peripheral immune cells.
Materials
-
8- to 12-week-old Tcrd-H2BeGFP C57BL/6 mice (The Jackson Laboratory, strain no. 016941)
-
Ketamine-xylazine solution (see recipe)
-
10 µg anti-CD8α antibody (clone 53-6.7) APC (eBioscience, cat. no. 17-0081-81) diluted in 100 µl endotoxin-free Dulbecco's phosphate-buffered saline (DPBS; 1×, without Ca2+ and Mg2+; MilliporeSigma, cat. no. TMS012A)
-
27G hypodermic needles (27G PrecisionGlide Hypodermic Needles,Becton Dickinson, cat. no. 305109)
-
Sterile 1-ml syringes (1 ml BD Slip Tip Sterile Syringes, Becton Dickinson, cat. no. 309659)
-
Sterile 0.5-ml tuberculin syringes with attached 27G needle (0.5 ml BD Tuberculin Syringes with attached 27G needle, Becton Dickinson, cat. no. 305620)
1.Anesthetize a 8- to 12-week-old Tcrd-H2BeGFP C57BL/6 mouse by i.p. injection of ketamine-xylazine solution based on weight of the animal (120 mg/kg ketamine and 10 mg/kg xylazine) with a 27G hypodermic needle attached to a sterile 1-ml syringe according to IACUC-approved procedure.
2.Once surgical-plane anesthesia has been reached (see Basic Protocol 1, annotation to step 1), slowly inject 10 μg rat anti-CD8α antibody (clone 53-6.7) APC primary antibody diluted in 100 µl endotoxin-free DPBS through retro-orbital venous sinus using a sterile 0.5-ml tuberculin syringe with an attached 27G needle.
3.Proceed to step 3 of Basic Protocol 1.
Basic Protocol 2: IMAGE ACQUISITION BY SPINNING-DISK CONFOCAL MICROSCOPY
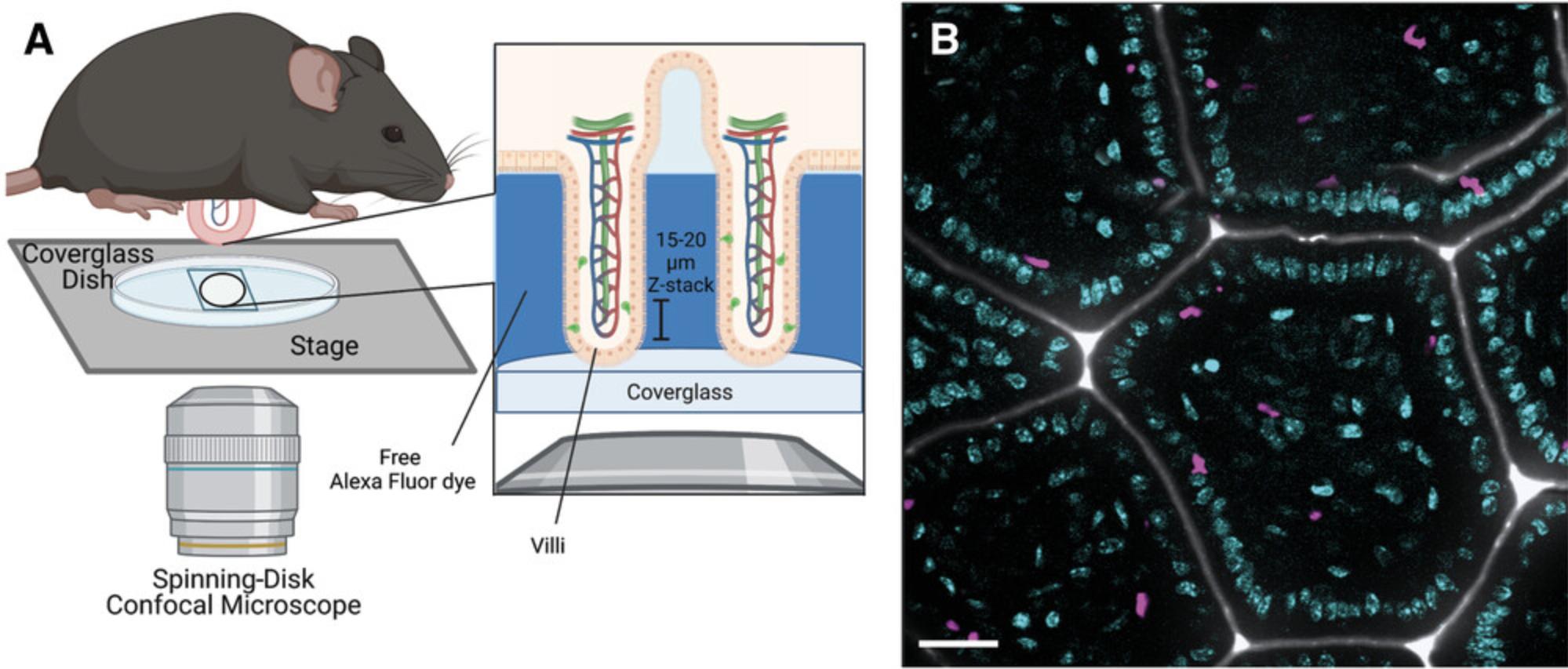
Successful visualization of mucosal immune cells using intravital microscopy requires finding a stable field to image, minimizing motion artifacts (such as peristalsis), and optimizing the temporal resolution of image acquisition. Although the weight of the mouse should push the exposed villi against the coverglass, excessive motion due to respiration, peristalsis, and blood flow may make it difficult to find a stable field to image. This protocol will describe how to properly position the exposed intestinal mucosal surface and identify an appropriate field for image acquisition and also provides tips for troubleshooting artifactual motion (Fig. 3). Although we describe the settings used on an inverted spinning-disk confocal microscope, many of the same parameters can be used to successfully visualize IEL migration using two-photon laser-scanning confocal microscopy.
Materials
-
Alexa Fluor 633 (AF633) hydrazide (Invitrogen, cat. no. A30634)
-
Glucose-free Ca2+ Mg2+ HBSS solution (see recipe)
-
Anesthetized post-surgical mice (see Basic Protocol 1)
-
Ketamine-xylazine solution (see recipe)
-
Type G immersion liquid (Leica Microsystems, cat. no. 11513910)
-
35-mm petri dish with coverglass bottom (14 mm Microwell No. 0 coverglass, MatTek, cat. no. P35G-0-14-C)
-
Microscope incubation enclosure (cage incubator, Okolab), 37°C
-
Spinning-disk confocal microscope [inverted DMi8 microscope (Leica Microsystems) equipped with Yokogawa CSU-W1 spinning disk (Andor, Oxford Instruments)]
-
Imaging software (Revolution Fusion acquisition software, Andor, Oxford Instruments)
-
20× objective (20× 0.4 HC PL FLUOTAR, Leica Microsystems)
-
63× glycerol immersion objective (63× 1.3 N.A. HC PLAN APO glycerol immersion objective, Leica Microsystems)
-
Electron multiplication (EM) CCD camera (EMCCD; iXon Life 888 EMCCD camera, Andor, Oxford Instruments)
-
Additional reagents and equipment for mouse euthanasia
1.Add 1 μl AF633 hydrazide to 2 ml glucose-free Ca2+ Mg2+ HBSS solution.
2.Use a P200 pipet to pipet 150 μl of the 1 μM AF633-HBSS solution from step 1 into the center of a 35-mm petri dish with a coverglass bottom.
3.Tightly hold the anesthetized post-surgical mouse by pinching skin along either flank, with the exposed lumen facing down, and center mucosal surface flat against the coverglass.
4.Open microscope incubation enclosure (pre-warmed to 37°C) and place mouse laying in the dish into the 35-mm holder on the stage of the spinning-disk confocal microscope.
5.Launch imaging software and set excitation intensity between 10 and 15 mW and the exposure time to 120 to 150 ms for each laser (405, 488, and 640 nm). Select a frame average of “2” and turn on EM gain function to reduce background noise.
6.Use 20× objective and 405-nm laser to visualize the nuclei and identify a field of villi with little to no visible movement by eye. Once identified, use XY scan to record the coordinates for the field of interest. Switch to 63× glycerol immersion objective (with type G immersion liquid) and return to original field of view using the recorded coordinates.
7.Set first plane for the Z-stack using the 405-nm channel by focusing on the orthogonal plane just beneath the epithelium at the villus tip. Once GFP+ γδ IELs and/or a distinct epithelial monolayer are visible along the outer edge of each villus, adjust focus going down the length of the villus until it is difficult to accurately resolve individual epithelial nuclei. Acquire a 15- to 20-μm Z-stack with 1.5-μm steps (Fig. 3A, right box).
8.Confirm that signal in other channels is visible in the selected field and begin image acquisition with EMCCD camera. Record for up to 30 to 60 min in 2 to 3 fields for each mouse. Monitor mouse every 5 min throughout image acquisition, looking for twitching or whisker movement.
9.Once image acquisition is complete, euthanize mouse by cervical dislocation followed by bilateral pneumothorax.
Basic Protocol 3: 4D ANALYSIS OF IMAGES
Once images have been acquired (Basic Protocol 2), the resulting videos can be analyzed using Imaris image analysis software. There are multiple software programs available for tracking cell migration, including those that are commercially available (ImarisTrack) and open source software (Fiji/ImageJ, CellTrack). A recent review (Masuzzo, Van Troys, Ampe, & Martens, 2016) provides a general overview of the computational capabilities of these programs to assist in decision-making regarding which platform is best suited to one's experimental needs. Although this review focuses on tracking cell migration in vitro , these same principles apply to the intravital imaging described here.
This protocol uses ImarisTrack because it has a comprehensive suite that facilitates cell tracking in a complex 3D environment. The auto-regressive motion algorithm for tracking cell migration is specifically designed to predict the best path for each cell (IEL) to provide unbiased data analysis. ImarisTrack renders surfaces around objects within the time-lapse image, which assists in the downstream analyses described below. Finally, the software can accommodate plugins for other open-source software, such as Matlab, to provide access to additional analysis tools. Imaris calculates a wide range of parameters; however, only a select few are described in detail here (Table 1). This protocol will describe the parameters for tracking IEL motility and metrics of IEL/epithelial interactions, which allows for an unbiased comparison of the migratory behavior of IELs across technical replicates and experimental conditions.
| IEL | Lumen | |
|---|---|---|
| Surface creation | ||
| Source channel | 488 or 640 nm | 560 or 640 nm |
| Thresholding type | Absolute intensity | Background subtraction |
| Surface detail (μm) | 0.5 | 0.5 |
| Diameter of largest sphere (μm) | - | 2-10 |
| Seed point diameter (μm) | 7 | - |
| Filter seed points | Quality | Quality |
| Filter surfaces | Number of voxels | Number of voxels |
| Surface tracking | ||
| Algorithm | Autoregressive motion | - |
| Max distance (μm) | 8 | - |
| Max gap (μm) | 3 | - |
| Filter track | Track duration | - |
Materials
- Imaris image analysis software [Bitplane, Oxford Instruments Group; on computer with Windows OS (minimum Windows OS requirements: 8 GB RAM, 3 GHz CPU dual core with 64 bit support, NVIDA Quadro P400 2 GB Graphics]
1.Import imaging files into Imaris image analysis software.
2.Identify a 30- to 60-min section of video for cell tracking.
3.Generate an IEL surface with “Add new surfaces” tool in the left selection pane.
4.Select “Track surface (over time)” under “Algorithm Settings” menu.
5.Use “Source Channel” to select channel corresponding to IELs to create the surface. Set “Surface detail” to 0.5 μm and “Thresholding” to “Absolute Intensity.”
6.Set “Absolute Intensity” to select all IELs while excluding autofluorescence or background noise. Enable “Split touching objects (region growing)” and set seed diameter points to ∼7 μm.
7.Once the surface calculations are completed, use “Quality” filter to select for IELs and remove any background noise.
8.Skip “Edit Surface” tab and continue to “Tracking Selection” menu.
9.Use “Autoregressive Motion” algorithm for IEL tracking. Select 8.0 μm as “Max Distance” parameter and 3 for “Max gap” size. Complete surface creation by selecting “Finish.”
10.Open “Edit Tracks” tab and deselect “Focus tracks” so all tracks are displayed. Follow IEL surface along the mapped track to ensure accurate tracking throughout the video. If incorrect, disconnect selected object at the frame with the observed error and connect selected objects to reconnect the track.
11.After correcting all IEL surface tracks, create a second surface to denote lumen by selecting “Track Surfaces (over time).”
12.Select channel of the luminal dye and select a 0.5 ± 0.25–μm surface detail. Adjust surface thresholding to “Background Subtraction” and set “Diameter of the largest object” to 2 to 10 μm. After checking the accuracy of the surface rendering, skip the rest of the surface creation menus with the “Finish” button (see Table 1 for quick reference).
13.Use “Edit track” tool to remove any incorrectly mapped surfaces that were missed by the filter.
14.Select “Object-Object Statistics” under “Edit” tab for both the IEL and the luminal surfaces.
15.Open “Preferences” menu and use “Statistics” tab to select the appropriate statistics for the analysis. Turn on “Object-Object Statistics” under “Edit” tab for each surface/spot.
REAGENTS AND SOLUTIONS
Glucose-free Ca2+ Mg2+ HBSS solution
- Add the following to 750 ml ddH2O while stirring:
- 8.06 g NaCl (138 mM)
- 0.048 g Na2HPO4 (0.3 mM)
- 0.10 g MgSO4•7H2O (0.4 mM)
- 0.10 g MgCl2•6H2O (0.5 mM)
- 0.40 g KCl (5.0 mM)
- 0.06 g KH2PO4 (0.3 mM)
- 3.57 g HEPES (15 mM)
- Add 0.14 g CaCl2 (1.3 mM) to 10 ml ddH2O
- Add CaCl2 solution dropwise to stirring solution to avoid calcium phosphate precipitate
- Adjust pH to 7.4 using 1 M NaOH or HCl
- ddH2O to 1 L
- Sterilize using 0.22-μm filter
- Store ≤1 year at room temperature
Hoechst 33342–saline solution
- 50 ml ddH2O
- 0.45 g NaCl (MW 58.44)
- Store ≤1 year at room temperature
- Before use, add 50 μl 10 mg/ml Hoechst 33342 (1.4 mg/ml, Invitrogen, cat. no. H3570) and 315 μl 0.9% (w/v) saline solution
- Prepare dye within 30 min of injection
The final injection volume should be calculated based on the weight of the mouse.
Ketamine-xylazine solution
- 7.5 ml ddH2O
- 1 ml 10× PBS (Fisher Scientific, cat. no. BP399-20)
- 1 ml 100 mg/ml ketamine (10 mg/ml, NDC: 11695-0703-1)
- 0.5 ml 20 mg/ml xylazine (1 mg/ml, NDC: 59399-110-20)
- Store ≤3 months at room temperature
COMMENTARY
Background Information
Although the analysis of cellular relationships in fixed tissue samples only reveals cell/cell interactions at one snapshot in time, intravital microscopy affords the opportunity to investigate the kinetics of these interactions in real time (Chieppa, Rescigno, Huang, & Germain, 2006; Chojnacki et al., 2019; Sullivan, Bui, Muller, Butin-Israeli, & Sumagin, 2018). Observing molecular and/or cellular biology in vivo has changed how we view many aspects of intestinal biology, including the reorganization of epithelial tight junction proteins (Marchiando et al., 2011; Marchiando et al., 2010; Watson et al., 2005), immune/epithelial interactions (McDole et al., 2012; Rescigno et al., 2001; Sumida et al., 2017; Wang et al., 2014), and the regulation of lymphocyte migration within the gut (Hoytema van Konijnenburg et al., 2017; Hu et al., 2018; Sujino et al., 2016; Sumida & Cyster, 2018). Whereas IELs were first observed interacting with apoptotic intestinal epithelial cells using electron microscopy in the late 1990s (Shibahara et al., 1995), we have recently shown that γδ IELs make extended contact with apoptotic enterocytes to facilitate their extrusion into the intestinal lumen (Hu et al., 2022). Thus, visualization of complex cellular dynamics within a tissue is advantageous in identifying novel functions or cellular responses in health and disease.
In the context of leukocytes, migration is frequently assessed in 2D using transwells and chemotaxis assays, yet these overly simplified model systems do not account for many of the structural features and cellular interactions that occur within the tissue microenvironment (Doyle, Petrie, Kutys, & Yamada, 2013). The protocols described here led to the first demonstration of γδ IEL surveillance behavior (Edelblum et al., 2012). These cells were previously thought to be sessile because their migration speed is reduced compared to T cells found in lymph nodes (Chennupati et al., 2010); however, these results are unsurprising based on the restrictive architecture of the epithelial compartment.
Different imaging modalities can be used for intravital microscopy depending on the location of the cells of interest and the temporal resolution required. Two-photon laser-scanning microscopy is commonly used for intravital imaging and relies on photomultiplier tubes (PMTs) to detect individual pixels with a 40% to 50% quantum efficiency, thus allowing deeper penetration into the tissue with low levels of photobleaching (Kolesnikov, Farache, & Shakhar, 2015). In contrast, spinning-disk confocal microscopy takes advantage of an EMCCD camera to detect 1 million pixels simultaneously to increase the quantum efficiency to 95%. Although spinning-disk confocal microscopy cannot penetrate deep into tissues, the temporal resolution of image acquisition of superficial tissues is greatly enhanced. Moreover, the reduction in the likelihood of photobleaching using spinning-disk confocal microscopy coupled with the faster rate of acquisition is useful when assessing the migratory dynamics of multiple cell populations. Integration of multispectral intravital imaging with spinning-disk confocal microscopy would further expand the potential to simultaneously visualize different fluorescence signals (Janssen, Najiminaini, Zhang, Omidi, & Carson, 2021).
Adding a fourth channel to the three described in the basic setup in Basic Protocol 2 can increase the acquisition time from 20 s/frame to 30 s/frame. To achieve finer temporal resolution during image acquisition, spatially distinct markers can be imaged using the same laser. For example, imaging a fluorophore-labeled antibody that binds to an immune cell and luminal dye in the same channel is an excellent option because the immune cell is unlikely to come into close contact with the apical surface of the epithelium. Even as IELs migrate into the LIS, spatial resolution between the IEL and the lumen will be clear. However, the fluorophore used in the shared channel should be chosen with care. The Alexa Fluor hydrazide in the lumen is typically very bright, so a weakly labeled antigen or a dim fluorophore may require a higher laser voltage, which, in turn, would result in the luminal signal becoming oversaturated. Therefore, it is important to balance the relative intensity of the two markers, if imaging both in one channel, with the need for more rapid image acquisition when deciding how to best proceed with an individual experiment.
Rapid capture of high-resolution images is ideal for visualizing the dynamics of mucosal immune cells and interactions between leukocytes, as well as with the intestinal epithelium. The protocols included here were designed for the analysis of γδ IEL migratory behavior using GFP γδ T-cell reporter mice; however, these methods can be easily adapted to investigate the interactions between any fluorescently labeled cells and/or their interaction with the villous epithelium. Alternatively, the inclusion of fluorescently labeled microorganisms in the luminal space or analysis following acute oral infection would allow for investigation of the spatiotemporal kinetics of cells within the intestinal mucosa in response to enteric pathogens (Edelblum et al., 2015).
Critical Parameters
A certain degree of variability is to be expected when imaging biological replicates; however, there are critical steps throughout the protocols that will reduce this variability and increase the likelihood of acquiring videos of suitable quality for analysis. During the laparotomy (Basic Protocol 1), it is important to try to keep the vasculature of the mouse intact and to avoid larger blood vessels when creating a hole in the mesentery. This will reduce the likelihood of nicking a vessel or restricting blood flow when the sutures are tied off; otherwise, inhibiting blood flow can result in hypoxia and tissue necrosis.
The greatest challenge in intravital imaging of the intestine (Basic Protocol 2) is finding a suitable area to image with minimal extraneous movement due to peristalsis. Ideally, having a sizable region of mucosa positioned against the coverslip increases the likelihood of identifying a region of stable villi to image. However, if too much intestine is externalized, this will increase artifactual motion due to peristaltic contractions. Therefore, externalizing only 2 to 3 cm of intestine during the laparotomy and exposing the entire length of the externalized gut is key. If it is difficult to locate a region of villi with minimal extraneous movement, it is helpful to remove the mouse and coverglass dish from the stage, clean the dish, replace the Alexa Fluor hydrazide solution, and reposition the mouse. It may be useful to lightly press down on the mouse's back to help push the exposed mucosa flat against the coverglass, as tight packing of the villi against the glass helps to limit the impact of peristalsis. It is possible that not every mouse will yield a video of sufficient quality for further downstream analysis.
If the IELs stop moving or develop a rounded morphology, this may be indicative of a drop in the body temperature of the mouse. Depending on the setup of the environmental chamber on the inverted microscope, cooler air may enter near the objective base. Therefore, increasing the incubation chamber temperature a degree or two will help to maintain the correct temperature around the exposed tissue. In any case, using a rectal thermometer to monitor body temperature is helpful to prevent these issues.
Throughout the surgery and imaging protocols, it is imperative to continuously monitor the sedation level of the mouse. To re-administer anesthesia without disrupting image acquisition, use forceps to gently scruff the mouse's neck and inject ketamine-xylazine solution subcutaneously. Isoflurane can also be administered to maintain surgical-plane anesthesia.
Troubleshooting
Additional attention paid to the critical parameters listed in the previous section will help avoid technical issues during surgery or imaging. Table 2 can be used to help diagnose many of the common problems that users encounter throughout each of the protocols.
| Problem | Possible cause | Solution |
|---|---|---|
| Basic Protocol Basic Protocol 1: Mouse preparation and laparotomy | ||
| Exposed mucosal surface does not stay open | Cauterized area of the exposed mucosa is too small | Extend the opening of the exposed mucosa or increase the length of externalized intestine |
| Feces in the lumen | Region selected for imaging contained feces | Gently remove feces using water and forceps |
| Support Protocol: Antibody labeling of cell surface markers | ||
| No signal in the antibody channel | Selected fluorophore is not bright enough for visualization | Choose a green or far-red fluorophore; adjust the laser voltage or exposure |
| Signal in vasculature but not on target cells | Antibody has not sufficiently penetrated into the tissue |
Increase the length of time for antibody labeling |
| Weak antibody labeling | Antibody target protein expression is low | Increase the antibody concentration; try a different antibody clone |
| Basic Protocol Basic Protocol 2: Image acquisition by spinning-disk confocal microscopy | ||
| “Bouncing” motion during imaging | Artifact of respiration, peristalsis, or heartbeat | Reposition the mouse on the coverglass |
| High background noise | Blood in the hydrazide-HBSS solution | Replace the hydrazide-HBSS solution |
| Rounded, sessile IELs at steady state | Low body temperature | Monitor mouse body temperature; adjust the incubation chamber temperature |
| Basic Protocol Basic Protocol 3: 4D analysis of images | ||
| Slow, persistent drift in a single direction throughout the video | Image drift after image acquisition | Create a spot of IEC nuclei > select 4-6 nuclei visible throughout the entire video at multiple points across the field of view > select “Correct drift” under “Edit tracks” > select “Correct image and selected object” and “Translational drift” |
| Surface not rendered for a cell although signal is visible by eye | Rendering settings cannot detect the fluorescence intensity for the object in question | Under the “Draw” tab, select the “Specific channel” and “Magic wand” options > select (hold Shift + left click) the position at which the new surface should be added |
| Multiple IELs combined into one surface | IELs are migrating close to each other, and the software is recognizing them as one IEL | Orient the surface to be cut vertically > under the “Edit” tab, select (hold Shift + left click) the point to be cut and then “Cut surface” |
| Single IEL split between multiple surfaces | IEL length is longer than the “max object diameter” setting | Disconnect the surfaces from their respective tracks > select both surfaces > select “Unify” in the “Edit” tab under selection |
| Surface rendering is larger than the visible volume | Thresholding setting is too low | Adjust the thresholding (absolute intensity) value in the creation wizard |
| Luminal surface encroaches into tissue | Autofluorescence during image acquisition | Surfaces can be cut in each affected frame by orienting the surface to be cut vertically under the “Edit” tab > select (hold Shift + left click) the point to be cut and then “Cut surface” |
Statistical Analysis
Many of the metrics used to analyze IEL motility are generated for each individual cell; thus, a large sample size can affect the statistical analysis and result in statistically significant differences that may not be biologically relevant. Therefore, the statistical methods and representation of these data should be chosen carefully (Lord, Velle, Mullins, & Fritz-Laylin, 2020). It may be more pertinent to take a nested average of overall track speed mean per mouse instead of the mean of each individual cell. An unpaired Student's t test or one-way ANOVA with Tukey's multiple comparison or Dunnett multiple comparison post-hoc test is used when comparing between 2 or 3+ experimental conditions, respectively.
Understanding Results
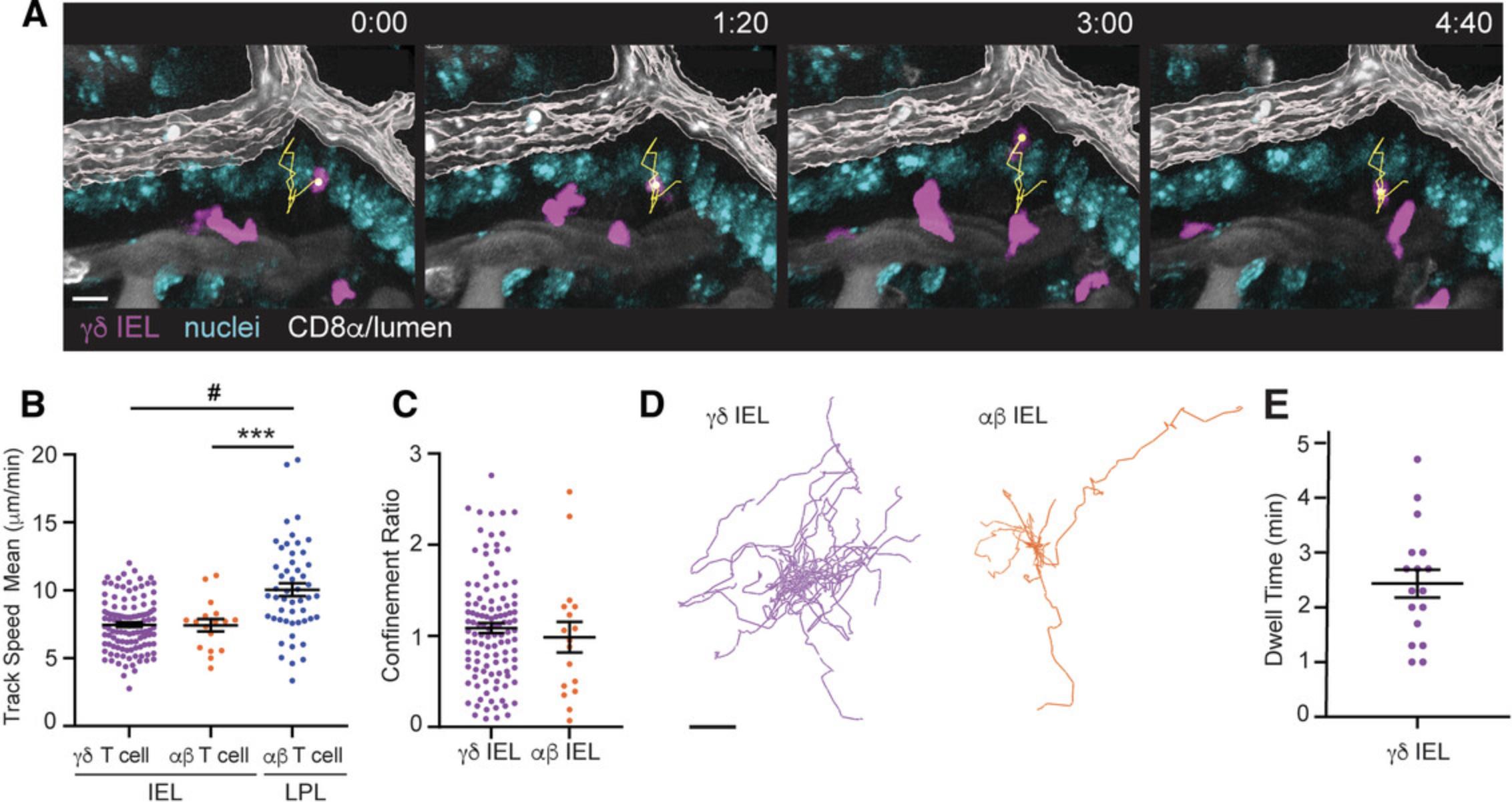
In the sample data, we imaged a TcrdEGFP mouse and administered APC-conjugated anti-CD8α antibody retro-orbitally (Video 1, and Fig. 4A). Because the anti-CD8α antibody labels both lamina propria lymphocytes (LPLs) and IELs, we applied a filter to delineate IELs based on their location relative to the lumen. We measured the distance between the lumen and the basolateral surface of an epithelial cell and found that it is ∼20 μm. Therefore, any T cells located >20 μm from the rendered luminal surface were excluded from analysis. This filter has the added advantage of excluding lymphocytes migrating just beneath the epithelium at the villous tip; these IEL/epithelial interactions would not be visible because these enterocytes are above the first plane of the Z-stack. It should be noted that the distance filter used to define IELs may vary between videos based on the precision of rendering the luminal surface and whether individual villi are directly perpendicular to the coverglass during image acquisition.
Open Research
Data Availability Statement
Data available on request from the authors.
Literature Cited
- Beltman, J. B., Maree, A. F., & de Boer, R. J. (2009). Analysing immune cell migration. Nature Reviews Immunology , 9(11), 789–798. doi: 10.1038/nri2638
- Chennupati, V., Worbs, T., Liu, X., Malinarich, F. H., Schmitz, S., Haas, J. D., … Prinz, I. (2010). Intra- and intercompartmental movement of gammadelta T cells: Intestinal intraepithelial and peripheral gammadelta T cells represent exclusive nonoverlapping populations with distinct migration characteristics. Journal of Immunology , 185(9), 5160–5168. doi: 10.4049/jimmunol.1001652
- Cheroutre, H., Lambolez, F., & Mucida, D. (2011). The light and dark sides of intestinal intraepithelial lymphocytes. Nature Reviews Immunology , 11(7), 445–456. doi: 10.1038/nri3007
- Chieppa, M., Rescigno, M., Huang, A. Y., & Germain, R. N. (2006). Dynamic imaging of dendritic cell extension into the small bowel lumen in response to epithelial cell TLR engagement. Journal of Experimental Medicine , 203(13), 2841–2852. doi: 10.1084/jem.20061884
- Chojnacki, A., Wojcik, K., Petri, B., Aulakh, G., Jacobsen, E. A., Lesuer, W. E., … Patel, K. D. (2019). Intravital imaging allows real-time characterization of tissue resident eosinophils. Communications Biology , 2(1), 181. doi: 10.1038/s42003-019-0425-3
- Di Marco Barros, R., Roberts, N. A., Dart, R. J., Vantourout, P., Jandke, A., Nussbaumer, O., … Hayday, A. (2016). Epithelia use butyrophilin-like molecules to shape organ-specific γδ T cell compartments. Cell , 167(1), 203–218.e217. doi: 10.1016/j.cell.2016.08.030
- Doyle, A. D., Petrie, R. J., Kutys, M. L., & Yamada, K. M. (2013). Dimensions in cell migration. Current Opinion in Cell Biology , 25(5), 642–649. doi: 10.1016/j.ceb.2013.06.004
- Edelblum, K. L., Shen, L., Weber, C. R., Marchiando, A. M., Clay, B. S., Wang, Y., … Turner, J. R. (2012). Dynamic migration of γδ intraepithelial lymphocytes requires occludin. Proceedings of the National Academy of Sciences of the United States of America , 109(18), 7097–7102. doi: 10.1073/pnas.1112519109
- Edelblum, K. L., Singh, G., Odenwald, M. A., Lingaraju, A., El Bissati, K., McLeod, R., … Turner, J. R. (2015). γδ Intraepithelial lymphocyte migration limits transepithelial pathogen invasion and systemic disease in mice. Gastroenterology , 148(7), 1417–1426. doi: 10.1053/j.gastro.2015.02.053
- Fischer, M. A., Golovchenko, N. B., & Edelblum, K. L. (2020). γδ T cell migration: Separating trafficking from surveillance behaviors at barrier surfaces. Immunological Reviews , 298(1), 165–180. doi: 10.1111/imr.12915
- Hoytema van Konijnenburg, D. P., Reis, B. S., Pedicord, V. A., Farache, J., Victora, G. D., & Mucida, D. (2017). Intestinal epithelial and intraepithelial T cell crosstalk mediates a dynamic response to infection. Cell , 171(4), 783–794.e713. doi: 10.1016/j.cell.2017.08.046
- Hu, M. D., Ethridge, A. D., Lipstein, R., Kumar, S., Wang, Y., Jabri, B., … Edelblum, K. L. (2018). Epithelial IL-15 is a critical regulator of γδ intraepithelial lymphocyte motility within the intestinal mucosa. Journal of Immunology , 201(2), 747–756. doi: 10.4049/jimmunol.1701603
- Hu, M. D., Golovchenko, N. B., Burns, G. L., Nair, P. M., Kelly, T. J. t., Agos, J., … Edelblum, K. L. (2022). γδ Intraepithelial lymphocytes facilitate pathological epithelial cell shedding via CD103-mediated granzyme release. Gastroenterology , 162(3), 877–889.e877. doi: 10.1053/j.gastro.2021.11.028
- Hu, M. D., Jia, L., & Edelblum, K. L. (2018). Policing the intestinal epithelial barrier: Innate immune functions of intraepithelial lymphocytes. Current Pathobiology Reports , 6(1), 35–46. doi: 10.1007/s40139-018-0157-y
- Jandke, A., Melandri, D., Monin, L., Ushakov, D. S., Laing, A. G., Vantourout, P., … Hayday, A. (2020). Butyrophilin-like proteins display combinatorial diversity in selecting and maintaining signature intraepithelial γδ T cell compartments. Nature Communications , 11(1), 3769. doi: 10.1038/s41467-020-17557-y
- Janssen, B. G. H., Najiminaini, M., Zhang, Y. M., Omidi, P., & Carson, J. J. L. (2021). Multispectral intravital microscopy for simultaneous bright-field and fluorescence imaging of the microvasculature. Applied Microscopy , 51(1), 12. doi: 10.1186/s42649-021-00059-6
- Kolesnikov, M., Farache, J., & Shakhar, G. (2015). Intravital two-photon imaging of the gastrointestinal tract. Journal of Immunological Methods , 421, 73–80. doi: 10.1016/j.jim.2015.03.008
- Lord, S. J., Velle, K. B., Mullins, R. D., & Fritz-Laylin, L. K. (2020). SuperPlots: Communicating reproducibility and variability in cell biology. Journal of Cell Biology , 219(6), e202001064. doi: 10.1083/jcb.202001064
- Marchiando, A. M., Shen, L., Graham, W. V., Edelblum, K. L., Duckworth, C. A., Guan, Y., … Watson, A. J. (2011). The epithelial barrier is maintained by in vivo tight junction expansion during pathologic intestinal epithelial shedding. Gastroenterology , 140(4), 1208–1218.e1-2. doi: 10.1053/j.gastro.2011.01.004
- Marchiando, A. M., Shen, L., Graham, W. V., Weber, C. R., Schwarz, B. T., Austin, J. R. 2nd, … Turner, J. R. (2010). Caveolin-1-dependent occludin endocytosis is required for TNF-induced tight junction regulation in vivo. Journal of Cell Biology , 189(1), 111–126. doi: 10.1083/jcb.200902153
- Masuzzo, P., Van Troys, M., Ampe, C., & Martens, L. (2016). Taking aim at moving targets in computational cell migration. Trends in Cell Biology , 26(2), 88–110. doi: 10.1016/j.tcb.2015.09.003
- McDole, J. R., Wheeler, L. W., McDonald, K. G., Wang, B., Konjufca, V., Knoop, K. A., … Miller, M. J. (2012). Goblet cells deliver luminal antigen to CD103+ dendritic cells in the small intestine. Nature , 483(7389), 345–349. doi: 10.1038/nature10863
- Nielsen, M. M., Witherden, D. A., & Havran, W. L. (2017). γδ T cells in homeostasis and host defence of epithelial barrier tissues. Nature Reviews Immunology , 17(12), 733–745. doi: 10.1038/nri.2017.101
- Olivares-Villagómez, D., & Van Kaer, L. (2018). Intestinal intraepithelial lymphocytes: Sentinels of the mucosal barrier. Trends in Immunology , 39(4), 264–275. doi: 10.1016/j.it.2017.11.003
- Prinz, I., Sansoni, A., Kissenpfennig, A., Ardouin, L., Malissen, M., & Malissen, B. (2006). Visualization of the earliest steps of gammadelta T cell development in the adult thymus. Nature Immunology , 7(9), 995–1003. doi: 10.1038/ni1371
- Rescigno, M., Urbano, M., Valzasina, B., Francolini, M., Rotta, G., Bonasio, R., … Ricciardi-Castagnoli, P. (2001). Dendritic cells express tight junction proteins and penetrate gut epithelial monolayers to sample bacteria. Nature Immunology , 2(4), 361–367. doi: 10.1038/86373
- Sandrock, I., Reinhardt, A., Ravens, S., Binz, C., Wilharm, A., Martins, J., … Prinz, I. (2018). Genetic models reveal origin, persistence and non-redundant functions of IL-17-producing gammadelta T cells. Journal of Experimental Medicine , 215(12), 3006–3018. doi: 10.1084/jem.20181439
- Shibahara, T., Sato, N., Waguri, S., Iwanaga, T., Nakahara, A., Fukutomi, H., & Uchiyama, Y. (1995). The fate of effete epithelial cells at the villus tips of the human small intestine. Archives of Histology and Cytology , 58(2), 205–219. doi: 10.1679/aohc.58.205
- Sujino, T., London, M., Hoytema van Konijnenburg, D. P., Rendon, T., Buch, T., Silva, H. M., … Mucida, D. (2016). Tissue adaptation of regulatory and intraepithelial CD4(+) T cells controls gut inflammation. Science , 352(6293), 1581–1586. doi: 10.1126/science.aaf3892
- Sullivan, D. P., Bui, T., Muller, W. A., Butin-Israeli, V., & Sumagin, R. (2018). In vivo imaging reveals unique neutrophil transendothelial migration patterns in inflamed intestines. Mucosal Immunology , 11(6), 1571–1581. doi: 10.1038/s41385-018-0069-5
- Sumida, H., & Cyster, J. G. (2018). G-protein coupled receptor 18 contributes to establishment of the CD8 effector T cell compartment. Frontiers in Immunology , 9, 660. doi: 10.3389/fimmu.2018.00660
- Sumida, H., Lu, E., Chen, H., Yang, Q., Mackie, K., & Cyster, J. G. (2017). GPR55 regulates intraepithelial lymphocyte migration dynamics and susceptibility to intestinal damage. Science Immunology , 2(18), eaao1135. doi: 10.1126/sciimmunol.aao1135
- Wang, X., Sumida, H., & Cyster, J. G. (2014). GPR18 is required for a normal CD8αα intestinal intraepithelial lymphocyte compartment. Journal of Experimental Medicine , 211(12), 2351–2359. doi: 10.1084/jem.20140646
- Watson, A. J., Chu, S., Sieck, L., Gerasimenko, O., Bullen, T., Campbell, F., … Montrose, M. H. (2005). Epithelial barrier function in vivo is sustained despite gaps in epithelial layers. Gastroenterology , 129(3), 902–912. doi: 10.1053/j.gastro.2005.06.015
Key References
- Jia, L., & Edelblum, K. L. (2019). Intravital imaging of intraepithelial lymphocytes in murine small intestine. Journal of Visualized Experiments: JoVE , (148), e59853. doi: 10.3791/59853
Contains pictures and video recordings of the laparotomy procedure that are useful for observing critical steps in the procedure.
Internet Resources
Imaris learning center support website, containing articles and tutorials for enhancing Imaris image analysis.

