Interpreting Image-based Profiles using Similarity Clustering and Single-Cell Visualization
Anne E. Carpenter, Anne E. Carpenter, Beth A. Cimini, Beth A. Cimini, Shantanu Singh, Shantanu Singh, Fernanda Garcia-Fossa, Fernanda Garcia-Fossa, Mario Costa Cruz, Mario Costa Cruz, Marzieh Haghighi, Marzieh Haghighi, Marcelo Bispo de Jesus, Marcelo Bispo de Jesus
high-dimensional data
image-based profiling
Morpheus
morphological analysis
profiling
single-cell visualization
Abstract
Image-based profiling quantitatively assesses the effects of perturbations on cells by capturing a breadth of changes via microscopy. Here, we provide two complementary protocols to help explore and interpret data from image-based profiling experiments. In the first protocol, we examine the similarity among perturbed cell samples using data from compounds that cluster by their mechanisms of action. The protocol includes steps to examine feature-driving differences between samples and to visualize correlations between features and treatments to create interpretable heatmaps using the open-source web tool Morpheus. In the second protocol, we show how to interactively explore images together with the numerical data, and we provide scripts to create visualizations of representative single cells and image sites to understand how changes in features are reflected in the images. Together, these two tutorials help researchers interpret image-based data to speed up research. © 2023 The Authors. Current Protocols published by Wiley Periodicals LLC.
Basic Protocol 1 : Exploratory analysis of profile similarities and driving features
Basic Protocol 2 : Image and single-cell visualization following profile interpretation
INTRODUCTION
Automated microscopy allows biologists to acquire thousands of images from cells perturbed with drugs, small interfering RNA (siRNA), CRISPR-Cas9, and more. In a typical quantitative microscopy experiment, biologists select fluorescent biomarkers (such as antibodies or dyes for specific proteins or cell compartments) and measure only the features they hypothesize will be perturbed in the experiment. By contrast, in image-based profiling, the aim is to let the cells speak for themselves. Diverse stains are used (as in the Cell Painting assay, which stains eight cell components; Bray et al., 2016; Cimini et al., 2022) and then image analysis software segments the cells and measures all possible morphological features from single cells. The collection of features for a cell is called a profile (sometimes described as a morphological profile or image-based profile), and typically a thousand or more features are measured per cell. It is then possible to analyze whether features are modified in a treated sample of cells compared to controls. Afterward, samples can be grouped into clusters based on their image-based profiles (Fig. 1). However, the biological meaning behind clusters is difficult to interpret because there are thousands of features in the profile. This leads to a common bottleneck: given a sample or cluster of samples, how do you interpret what a given profile means biologically?
Here, we present two protocols: exploratory analysis using Morpheus software (Basic Protocol 1) and image and single-cell visualization following profile interpretation (Basic Protocol 2). In Basic Protocol 1, we show how to explore the overall large-scale associations of the data (after feature extraction and cleaning) using the free web-based software Morpheus. Using Morpheus, the data can be grouped in different ways, revealing how features and samples are correlated. Exploring the data is essential to gain insights into the biological interpretation of the profiles. In Basic Protocol 2, the goal is to help biologists create intuitions about differences between treatments by examining example cells. This notebook contains Python scripts to help crop representative or random single cells from each treatment and group the cropped images based on correlations of interest. In addition, representative images of each sample can be retrieved to understand how the cells are distributed across representative fields of view (e.g., those captured from different sites [locations] within a sample well), which can give insights into treatment toxicity and/or growth-stimulating effects. In Understanding Results, we provide insights on how visualizing example cells from the samples and linking them to the correlations between samples will provide extensive information that can be used to formulate new hypotheses and interpretations from the data. While these approaches are powerful, we note that they require high-dimensional image measurements and, as such, require the user to first use CellProfiler or a similar tool to identify objects and generate large numbers of measurements; they also unfortunately do not always lead to easily interpretable conclusions (see Understanding Results for further discussion).
The protocols described here yield a similarity matrix, hierarchical clustering for the samples, and representative example cells from their data. These outputs can easily be used for reports and publications. For the input data for both protocols, we use a dataset of images processed by CellProfiler to identify cells and extract features (Stirling et al., 2021) and by pycytominer to normalize and aggregate single-cell profiles into population-averaged profiles (Way, Chandrasekaran, et al., 2022). Extensive documentation is available online for feature extraction with CellProfiler (https://github.com/CellProfiler/tutorials) and for data aggregation, normalization, and feature selection with pycytominer (https://github.com/cytomining/pipeline-examples). In addition, we provide an example dataset in our GitHub repository, including comma-separated value (CSV) spreadsheets to be processed on Morpheus (https://github.com/ciminilab/2023_Garcia-Fossa_Cruz_CurrentProtocols). In our example dataset, each compound is annotated with its mechanism of action (MOA). However, these protocols can be used without having the MOA for every compound in the dataset, and instead by comparing treated cells with negative and/or positive controls, or comparing multiple perturbed samples with each other.
Basic Protocol 1: EXPLORATORY ANALYSIS OF PROFILE SIMILARITIES AND DRIVING FEATURES
The main goal of this tutorial is to examine the correlations between samples to check for their replicability, to explore correlations among them, to discern how features drive differences between samples or groups, and to interpret the biology behind the data.
After cell treatment, imaging, and feature extraction, some profiles are dramatic in only one or a few features and the feature names have obvious meanings (nucleus area or integrated intensity of the mitochondria channel in the cytoplasm, which corresponds to the total amount of staining in that channel); in these cases, looking at feature names will help to discern their connection to biological meaning. Other individual features have meanings that are more difficult to translate into plain language. Furthermore, the challenge is even greater to interpret a collection of feature names that all contribute strongly to a more complex morphological phenotype. For example, a collection of features from a channel stained for actin and wheat germ agglutinin together with DNA granularity was particularly important to predict 70 specific cell health phenotypes from Cell Painting data (Way et al., 2021). Even phenotypes that are visually obvious and distinctive by eye, such as cells stalled in a particular stage of the cell cycle, are often difficult to predict just by examining a list of distinctive features; the problem is even more acute for samples without a visual discernible phenotype yet quite distinguishable using image metrics.
To help us in the exploration and interpretation process, we often use Morpheus (available at https://software.broadinstitute.org/morpheus/), a free web-based open-source software that allows matrix visualization, analysis, clustering, filtering, and displaying of charts. The tool can be readily used without extensive computational or statistical experience. It allows for quick visualization of an entire dataset in different ways, so you can identify patterns in their data that could lead to new biological insights, or even use it as a data quality control step by examining replicability. Morpheus was originally designed at the Broad Institute for exploration of mRNA profiling data, but accepts a variety of matrix files from multiple formats (CSV, GCT, GMT, text file) to be imported. Although raw CellProfiler outputs tables can be input into Morpheus, here, we provide notebooks to preprocess the outputs from CellProfiler so the data can undergo aggregation and normalization (both of which can also be performed in Morpheus) followed by multiple feature reduction steps (some of which are not available in Morpheus).
More information can be found in the Morpheus documentation (https://software.broadinstitute.org/morpheus/documentation.html), as well as a two-part series of video tutorials on the Center for Open Bioimage Analysis (COBA) YouTube channel: “The beginner's guide to morphological profiling (Morphological profiling, part 1)” and “Practical exploration of morphological profiling data (Morphological profiling, part 2)”.
During this tutorial, we start by examining how similar each sample is to the other samples using per-well similarity matrices, sorting the data in a way that allows for interpretation. We provide a sample dataset in which drugs with known mechanisms of action (MOAs) have been added at various dose points prior to Cell Painting. To observe how MOAs are grouped, and if technical artifacts such as batch or plate-layout effects are playing a role in the distribution of the groups, we use hierarchical clustering. In the end, you will be able to identify whether drugs with similar MOAs have similar morphological profiles and the positive and negative connections between various MOA profiles. You will also learn how to determine what features drive the differences between the groups. We emphasize that this is just one of the data-exploration approaches that can be used to interpret image-based profiles, and produces comparative results rather than hard distinctions between similar and not.
Materials
- Laptop or desktop computer with at least 2 GB RAM and a suitable web browser such as Google Chrome
- Internet access to use Morpheus (https://software.broadinstitute.org/morpheus/)
- Data and Jupyter Notebooks (Kluyver et al., 2016), available at https://github.com/ciminilab/2023_Garcia-Fossa_Cruz_CurrentProtocols. The data are in a GCT format, a tab-separated value table containing the extracted features aggregated by well in a Cell Painting assay. In this assay, 1571 compounds were tested across six doses in A549 cells (Way, Natoli, et al., 2022).
- We randomly selected a plate map from this experiment (C-7161-01-LM6-011 plate map) and downloaded the CSV files for five of its replicate plates (SQ00015195, SQ00015218, SQ00015219, SQ00015220, SQ00015221) from the cpg0004-lincs dataset (Way, Natoli, et al., 2022) available from the Cell Painting Gallery on the Registry of Open Data on AWS (cellpainting-gallery). We then added annotations to the data (labels for each MOA, compound, and concentration) and normalized the features to the negative control (DMSO) in a Jupyter Notebook (Kluyver et al., 2016) using the pandas library (Reback et al., 2020) and pycytominer (Way, Chandrasekaran, et al., 2022). Next, we performed feature selection to exclude features with low variance (frequency cut = 0.05), high correlation to another feature in the profile (threshold = 0.9), features that have >5% NA (not available) values, blocklisted features, and outliers (features with minimum or maximum absolute values greater than threshold = 500). These parameters serve as useful starting values but may be adjusted as needed; for more details, see the data preparation notebook and pycytominer documentation (https://pycytominer.readthedocs.io/en/latest/). These steps are available in the basic_protocol_1/notebooks/data_processing folder using the Data_preparation.ipynb notebook in our GitHub repository (https://github.com/ciminilab/2023_Garcia-Fossa_Cruz_CurrentProtocols/blob/main/basic_protocol_1/notebooks/data_processing/Data_preparation.ipynb). We opened the CSV file obtained using Data_preparation.ipynb in Morpheus and clicked on Tools > Transpose, allowing the CSV table to be better visualized in Morpheus. To apply the protocol to your own data, we recommend using CellProfiler to extract features and pycytominer for data preparation.
- We calculated the average precision based on https://github.com/niranjchandrasekaran/profiling-workflow-demo/blob/master/analysis/0.calculate-ap.ipynb to enable us to remove weakly correlated pairs (defined as < 0 mean average precision between replicates) before analysis; no such profiles were found for this dataset. To reproduce our results, follow the instructions for creating an environment at https://github.com/niranjchandrasekaran/profiling-workflow-demo, and use our notebook WeakProfiles_Replicability.ipynb available at https://github.com/ciminilab/2023_Garcia-Fossa_Cruz_CurrentProtocols/tree/main/basic_protocol_1/notebooks to calculate the replicability between samples in our Morpheus_Example_FeatureSelected.csv dataset. For more information about removing weak profiles, see Critical Parameters.
1.To obtain the dataset for this protocol, clone the GitHub repository into your computer or download the repository at https://minhaskamal.github.io/DownGit/#/home?url=https://github.com/ciminilab/2023_Garcia-Fossa_Cruz_CurrentProtocols.
2.Access the website https://software.broadinstitute.org/morpheus/, click on “Select File” on the main screen, and select the Morpheus_Example_FeatureSelected.gct file you downloaded from GitHub. On the current tab, you will see a heatmap. Notice all the columns displayed for compound, concentration, etc.
3.Click on Options (gear symbol) > Annotations > Column annotations and click All to select all columns. Right-click on the column labels (Compound, Concentration, MOA, Wells, and Plate) and enable “Show color” for all the columns to color-code the columns.
4.Click on Options > Color Scheme and de-select the Relative color scheme. Change the minimum to –1000 and the maximum to 1000. Try also with –100 and 100. While the Relative color scheme converts values to colors based on each feature's minimum and maximum values (making every row range from blue to red based on their own min and max), overriding and changing the color scheme to these new values allows you to see raw feature values distributed within this new feature range. In this way, extreme feature values became visible.
5.Close the Option window, click the zoom tool, and select Fit To Window.
6.Use the mouse pointer to scroll throughout the row names in the right corner of the screen, highlighting the feature names. Any values colored in red or blue are unusual features that have high or low values compared to the rest of the features.
7.Open Options > Color Scheme and select Relative color scheme to use the minimum and maximum values in each row to convert values to colors.
8.Select Tools > Similarity Matrix > Pearson correlation on the rows. This will calculate the correlation between the wells for all pairs of features in the dataset and generate a similarity matrix for them. Click on Options > Display and select Link rows and columns.
9.Create a Hierarchical Clustering by selecting Tools > Hierarchical Clustering. In Metric, select “Matrix vales (for a pre-computed similarity matrix)”. Change Cluster to Rows and Columns and click OK. This will group the features depending on how similar their profiles are using the correlation metric you have chosen.
10.Go back to the first tab “Morpheus_Example_FeatureSelected” and select Tools > Similarity Matrix > Pearson correlation on the columns. This will calculate the correlation between features for all pairs of samples in the dataset and generate a similarity matrix for them.
11.Click on Options > Display and select Link rows and columns. This helps navigate the large matrix, showing the respective correlations.
12.While holding the shift key, click on the MOA, Compound, and Concentration columns (in this order) to sort them by value. This will display the samples in order, based on those categories of metadata (rather than based on the profile similarity itself). Focus on MOAs and the different compounds in each MOA. Can you see if compounds belonging to the same MOA have a similar morphological profile (Fig. 2)?
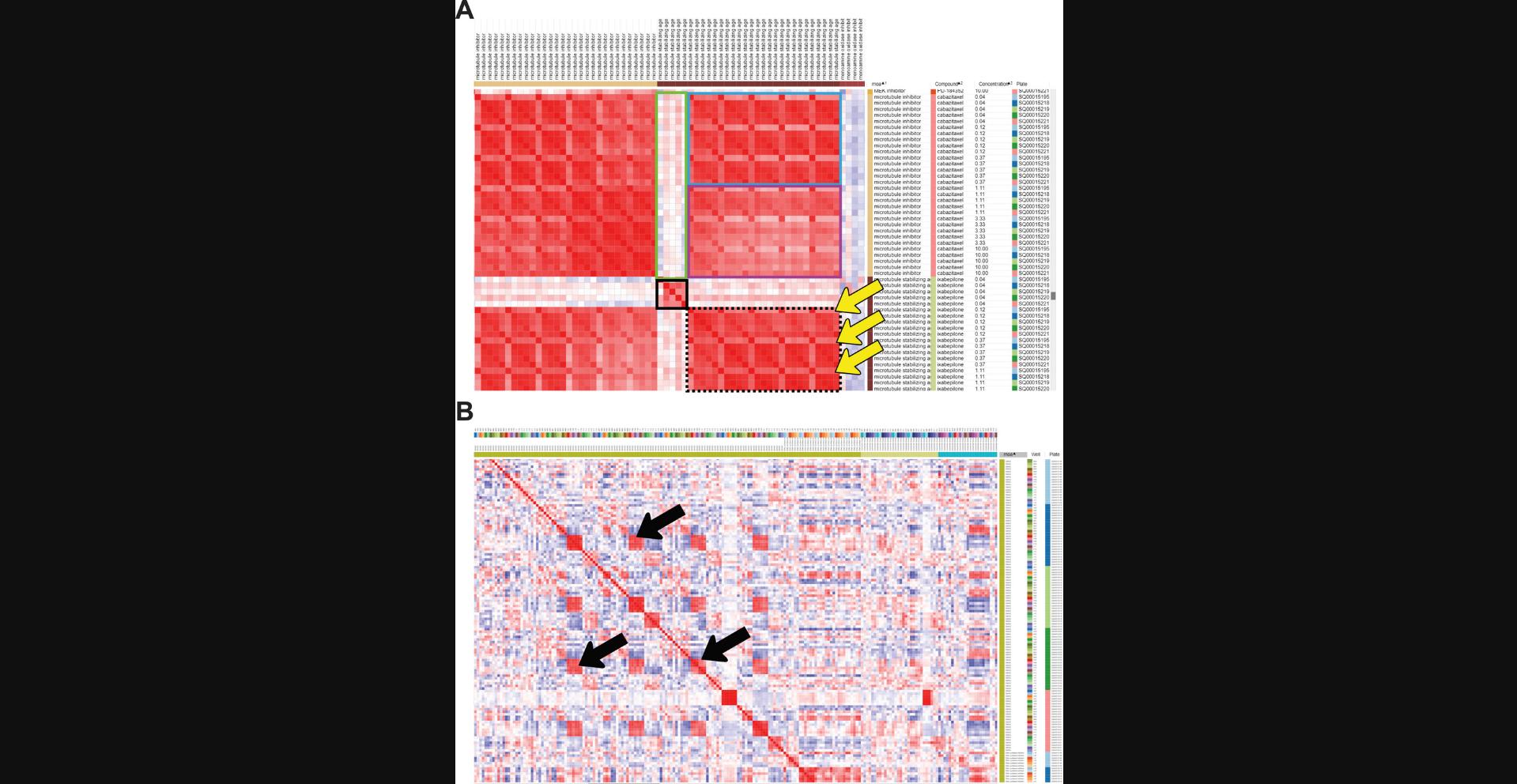
13.Using the same configuration as in the previous step (columns sorted by MOA > Compound > Concentration), continue to explore the similarity matrix and observe whether there are different MOAs with similar morphological profiles.
14.Sort the collapsed similarity matrix by MOA and by plate by holding the shift key. Zoom out (pressing the minus – key) to see a broader view of the matrix.
15.Roll over to the DMSO MOA (the negative control in this dataset).
16.Create a hierarchical clustering by selecting Tools > Hierarchical Clustering. In Metric, select “Matrix values (from a pre-computed similarity matrix)”. Change Cluster to Rows and Columns and click OK. This will group the samples depending on how similar their profiles are (using the correlation metric you have chosen). You can identify different groups and try to make sense of the groups.
17.Zoom out (using the – key) to see a broader view of the clustering. Scroll through and find large squares of red color in the matrix to observe which MOAs are clustering.
18.Return to the tab containing the feature value (rather than similarity matrices) and go to Tools > Marker Selection. Choose T-test as the metric, MOA as the field, class A as DMSO, and class B as the tubulin polymerization inhibitor. Leave the default values for Number of Markers and Permutations. This step reveals which features are driving the differences between these two groups (Fig. 3).
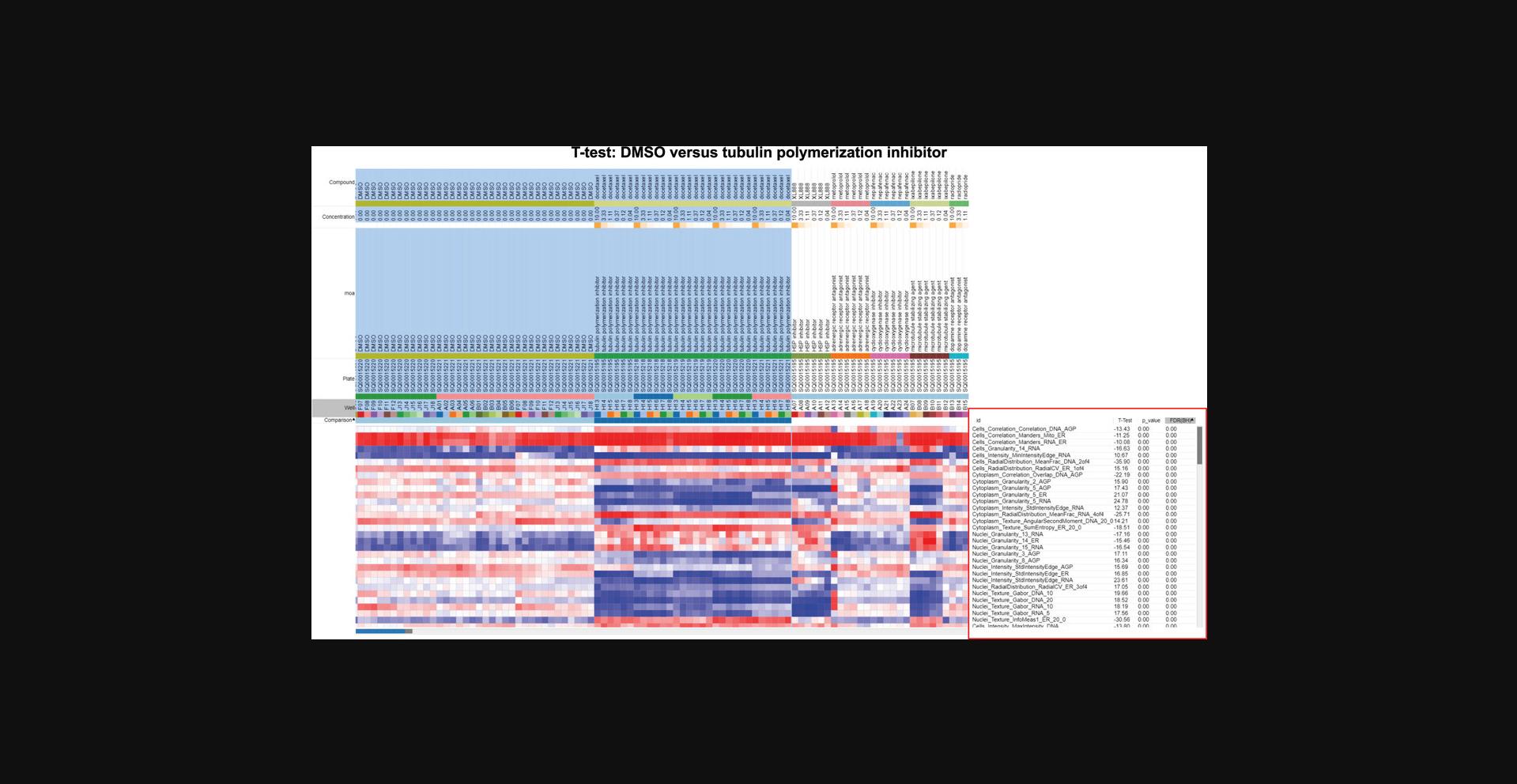
19.Sort the p value column by right-clicking on it. Explore the names of the features that determine the difference between DMSO and tubulin polymerization inhibitors. If you have a large number of features with a p value of 0.00, these will continue to be sorted alphabetically and not by strength; in this case, you can sort to find the highest and lowest T-test values, which should represent the strongest features.
20.Go back to the similarity matrix and go to File > Save Dataset. Write a name for your File, click OK to save as GCT version 1.3, and save the table, allowing it to be opened again in Morpheus when needed.
Basic Protocol 2: IMAGE AND SINGLE-CELL VISUALIZATION FOLLOWING PROFILE INTERPRETATION
With large datasets, it often becomes challenging to retrieve images of sites or single cells for visualization to perform quality control, validate a pipeline, and, most importantly, interpret any morphological changes detected in the profiles explored during the data analysis and exploration (visualized with heatmaps, UMAPs, etc.). Along with visualizing sample and feature correlations as in Basic Protocol 1, it is also important to think biologically about organelle distribution, morphological characteristics such as cell and nucleus shape, and intensities of each stain. Connecting the numbers (Pearson coefficients, T-tests, morphological feature values in profiles, etc.) with how the cells look in the images can help the user decipher a complex profile.
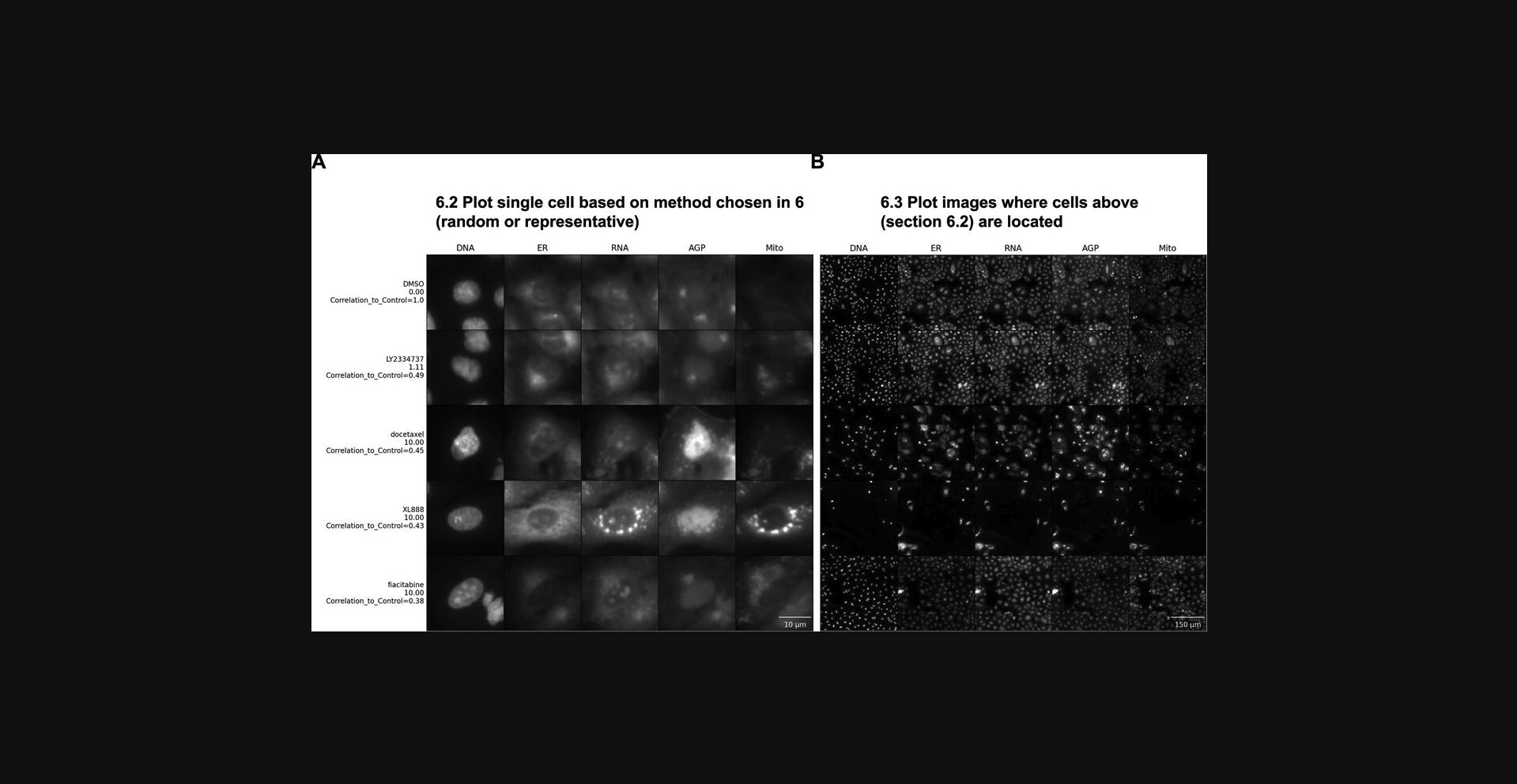
In this protocol, we describe how to use a script we created to retrieve random or representative images from the dataset and plot them together, allowing the user to choose which samples to observe and how to group and display them. While random images are often helpful, especially in cases of high heterogeneity, it can also be helpful to computationally determine which cells’ phenotypes are the most representative in a sample and compare them to control cells. This is not a trivial step, but can sometimes provide critical insight into morphological changes. In this protocol, we use Jupyter Notebook to derive representative cells by performing a clustering analysis on the morphological space of the population of single cells and sampling from the subpopulation closest to the center of the sample(s) of interest. This notebook can also be used to compute similarity matrices as in Morpheus; however, for large-scale experiments, we recommend examining the experiment using the per-well aggregated information as in Basic Protocol 1.Once a few treatments of interest are identified, single cells can be visualized using this protocol.
From the Jupyter Notebook, the user will obtain representative or random image sites and single cells, enabling comparison of the images with the correlation coefficient values obtained in the similarity matrix. By establishing the relationship between the images and heatmaps, the user can start hypothesizing about biological processes and morphological profiles that are significant, which could lead to more specific biological questions and assays. As in Basic Protocol 1, we provide some hints and interpretations for each step; for more detailed discussions of biological interpretations, see Understanding Results.
Materials
- Laptop or desktop computer with at least 2 GB RAM and a suitable web browser such as Google Chrome
- Internet access
- Gmail account if using Google Colab
- This protocol assumes the use of a web browser to run Google Colab. To run this protocol, open our Google Colab notebook (https://github.com/ciminilab/2023_Garcia-Fossa_Cruz_CurrentProtocols/blob/main/basic_protocol_2/notebook/Basic_Protocol_2.ipynb) and create a copy on your own Google Drive. To adapt this protocol to your own data, either download the Jupyter Notebook to your local computer and install the requirements based on the requirements.txt file or use Google Colab and mount your Google Drive (https://colab.research.google.com/notebooks/io.ipynb) to enable access to data you have stored in your Google Drive. In either case, you must adapt the pathnames and filenames within Section 2 of the Notebook to point to your dataset.
- Our dataset table is in a CSV format and contains the extracted features for single cells in a Cell Painting assay. In this assay, 1571 compounds were tested across six doses in A549 cells (Way, Chandrasekaran, et al., 2022). Here, we use the same dataset from Basic Protocol 1, but we require information about single cells, and each row of the table must have cell features and x -y locations within the image to enable single-cell image retrieval. We also provide all the images of where these single cells are located. For this purpose, we selected only a subset of samples within the dataset to minimize the memory requirements needed for users to explore the data. We performed normalization and feature selection with this dataset using pycytominer. The Jupyter Notebooks required to create this dataset from publicly available datasets (1_Samples_retrieval.ipynb and 2_Generate_Profiles.ipynb) are available on our GitHub under the basic_protocol_2/notebook folder. We also provide an alternate code in the sample retrieval notebook to allow the loading of entire plates when experiment size and memory permit.
- The Jupyter Notebook functions were written using Python 3.9 (Van Rossum & Drake, 2009). Data processing was performed using pycytominer tools for normalization, feature selection, and data annotation. Check pycytominer documentation (https://pycytominer.readthedocs.io/en/latest/) for details on how to change parameters and inputs depending on your dataset.
- The GitHub repository contains the following files relevant to Basic Protocol 2:
- util folder with .py files containing functions written to be used on this notebook. These functions are installed onto the notebook using pip install and then imported from utils.correlations import *.
- basic_protocol_2/Images folder, which contains the subset of images downloaded from https://github.com/broadinstitute/cellpainting-gallery. We provide PNG images that were compressed from the original TIFF images; PNG is a lossless format that requires less storage space.
- basic_protocol_2/data folder, which contains the BasicProtocols2_Example.zip with a CSV file. To use this notebook with your data, you could extract the features using CellProfiler and export the information to a spreadsheet that can be read in the Jupyter Notebook. Alternatively, if using a database file, you could transform it into a CSV file using our available Samples_retrieval.ipynb Jupyter Notebook. The notebook will perform annotation, normalization, and feature selection if you have not already run those steps. These steps can be bypassed if they have already been done (e.g., by notebook 2_Generate_Profiles.ipynb).
1.Open the Google Colab notebook Basic Protocol 2_Visualize cells and images.ipynb available in the link at https://github.com/ciminilab/2023_Garcia-Fossa_Cruz_CurrentProtocols/blob/main/basic_protocol_2/notebook/Basic_Protocol_2.ipynb. Be sure to access the notebook from our GitHub repository, allowing you to check for any recent updates.
2.Click the Copy to Drive button and the notebook will be available on your Google Drive in the Colab Notebooks folder.
3.Run the first three cells in the notebook Section 1 - Import Libraries by clicking on the start button at the top left (or hit Ctrl + Enter). The first line will clone the GitHub repository and install the functions; the second line will install the required libraries to run this notebook (this process takes ∼5 min) and import the libraries to allow their use inside the notebook. Run the lines of code in the order that they appear in the notebook.
4.Run only the first cell inside Section 2 - Define Inputs. This will define the inputs required to run the cells in the notebook. The script requires the filename and pathname to access the CSV table and read it as a DataFrame. It also needs the pathname for the images directory.
5.Run the cells inside Section 3a, which will import the dataset and perform annotation, normalization, and feature selection. The table contains all the features measured for every single cell, and also metadata information about compound MOAs, compound names, and concentrations tested. For more information about feature selection, see Critical Parameters.
6.Run the first three cells in Section 4 (through cell 4.1.1) and choose Metadata_Compound_Concentration for this demonstration. These options were generated based on the names of columns with the “Metadata_” prefix. This choice will impact the information visualized on the plots for the next steps. If the choice is Metadata_Compound_Concentration, you will see values such as DMSO 0.0, etc. When using new data, add the “Metadata_” prefix to any such columns before loading it into the notebook, as it will appear under this dropdown and be used for aggregation (Fig. 4A).
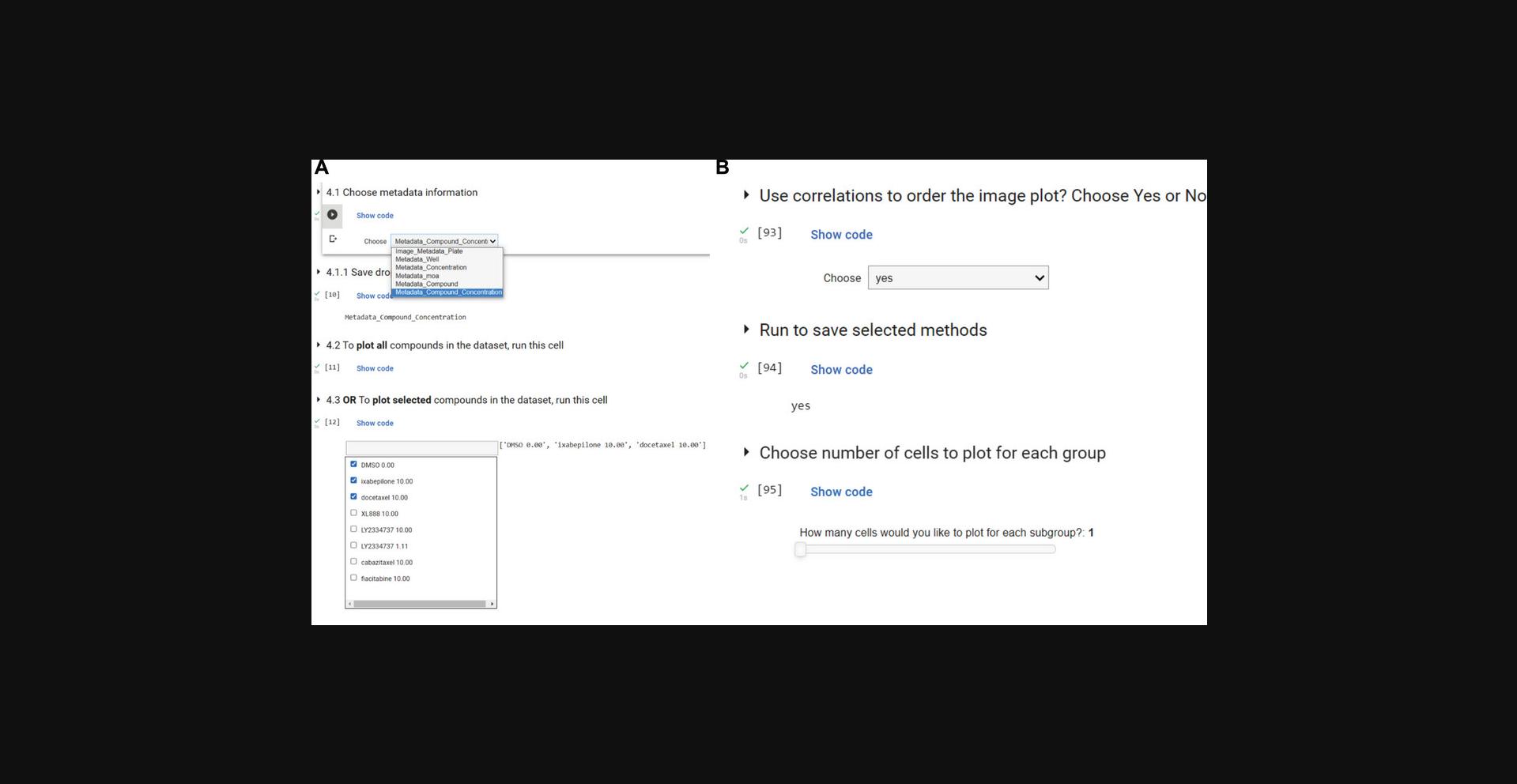
7.Run the cell in Section 4.2 to choose all the compounds available on the dataset to visualize. This step will select all the compounds in the dataset.
8.Run the cells in Section 5 to generate and graph the correlation between the compounds. Choose a column to be the labels for the correlation matrix using the dropdown, then use pycytominer to return a per-well aggregated DataFrame. A correlation matrix will be generated. There is an option to export the matrix as an image (type the name and press Enter/return).
9.Run the three cells in Section 5.1 to insert the correlation values calculated in the previous step inside the initial DataFrame as a new column. This function will get the chosen compound and find the correlation values for every other compound related to the first. Choose “DMSO 0.0” for comparison, because the aim for this dataset is to evaluate which compounds have morphological profiles more similar to the control.
10.Run all of the cells inside Section 5.2 and choose “DMSO 0.0”. This choice reflects the biological question of which compounds are closely correlated to the negative control (DMSO). However, this is a dynamic Jupyter Notebook where the user could be interested in other compounds or MOAs.
11.In Section 6 - Visualize Cells, run the first cell to choose whether to visualize randomly selected or representative single cells. Choose the random method to select random samples for each treatment/group you have; choose the representative method to select the most representative cell within each subgroup. Many cells in this section rely on correlation to the reference compound selected in Section 5.1; if you want to change reference compounds, rerun those cells before returning to Section 6 and running all cells here.
12.Run the next cell and select how many cells you would like to display from each subgroup and whether or not you would like the images shown in order of subgroup correlation to the reference compound.
13.Choose whether (a) each image should be rescaled to the minimum and maximum before being displayed or (b) the raw intensity values should be plotted. Raw intensities are typically more comparable across conditions (see below for caveats), but may be harder to see when the signal is dim and thus may require external rescaling after saving.
14.Insert the pixel size value. This is necessary to add a scale bar in your images. Type the value “0.29898” in this example to add the pixel size for this example dataset in μm/pixel. Each microscope and lens will have its own configuration.
15.Plot the selected single cells in random order by running the first cell of Section 6.1. This step allows a first view of the cells without the labels, so you can explore the images before knowing to which group the cells belong. Once you have explored the data, run the rest of the cells in Section 6.1 to append labels to see if your hypotheses were correct, to create an unshuffled version of the image, and to save the image to disk.
16.Run Section 6.2 to display the full images from which the single-cell crops have been pulled (Fig. 5B). Looking at the entire field of view (FOV) may provide insights into additional biological aspects.
COMMENTARY
Background Information
Image-based profiling typically starts with using fluorescent markers to stain different targets and/or compartments of the cell. In our example data for both protocols, we used Cell Painting data. Cell Painting is a morphological profiling assay that multiplexes six fluorescent dyes, imaged in five channels, to reveal eight relevant cellular components. The experiment's aim was to characterize chemical perturbations in cells by measuring morphological changes after cells were exposed to various treatments. Briefly, cells were plated in multiwell plates, perturbed with treatments to be tested, then stained, fixed, and imaged on a high-throughput microscope. Images were acquired for DNA, RNA, endoplasmic reticulum, mitochondria, and AGP (actin, Golgi, and plasma membrane).
Software such as CellProfiler (Stirling et al., 2021) makes it easy to obtain and extract information from these images, extracting thousands of morphological features distributed into categories relating to the compartment measured (nucleus, cell, cytoplasm) and types of metrics (size, shape, texture, intensity, granularity, and more) to produce a feature profile that enables the detection of subtle phenotypes. To facilitate understanding of the features, CellProfiler feature name outputs are organized as follows: [Compartment][FeatureGroup][Feature][Channel][Parameters]. Not all features have channel information; for example, shape features relate only to the outlines of the chosen cellular compartments. From a Cell Painting assay, Nuclei are identified by the DNA channel, Cells by the RNA or AGP channel, and Cytoplasm is defined as the cell excluding the nucleus object. FeatureGroups are associated with the measurements made on the compartments (e.g., AreaShape, Intensity, Texture, Granularity, and more). To understand how each module works to extract information from the images, check the latest documentation available for CellProfiler (https://broad.io/cellprofilermanual). You can check a list of all the features extracted from one particular analysis of a Cell Painting assay at https://github.com/carpenterlab/2022_Cimini_NatureProtocols/blob/main/CellProfiler_features.csv. Note that the names of the features will vary based on the parameters used to analyze the assay.
The essential steps after extraction of the features are aggregation, normalization, and feature selection. These are the steps we describe in our Jupyter Notebooks using pycytominer (Basic Protocol 1 support notebook and in the main notebook used for Basic Protocol 2). Profiles of cells treated with different experimental perturbations are then compared to identify the phenotypic impact of chemical or genetic perturbations, grouping compounds and/or genes into functional pathways and identifying signatures of disease. We demonstrate these last two steps using Morpheus software and scripts on Jupyter Notebooks in the protocols above.
Understanding the correlation coefficients calculated for the samples in both protocols is important for this protocol. A Pearson correlation coefficient is a way of representing the measurement of similarity, where it measures the strength of the linear relationship between two variables (in our case, between two wells across a large set of features or between two features across a large set of wells). A Pearson coefficient of 1 means a perfect positive correlation, 0 means no correlation, and –1 means a perfect negative correlation (Pearson & Galton, 1895). A similarity matrix is a way to assess the covariance in features between all pairs of columns or rows. In each square of the matrix, a Pearson correlation coefficient was calculated for all features in the dataset between each pair of samples. The squares at the intersection of those two samples are set as the value of that correlation coefficient, and so on for each pair of wells. This allows us to see at a high level how similar the overall phenotype is between any pairs of samples in our experiment, and therefore how phenotypically distinct our treatments are.
Critical Parameters and Troubleshooting
We reiterate that normalizing the features is fundamental before executing the steps in this paper. Normalization is usually performed on all of the features to fix range issues and allow comparison between features (Caicedo et al., 2017). Normalization is also recommended to increase the signal-to-noise ratio (Chandrasekaran, Ceulemans, Boyd, & Carpenter, 2021). Normalization performed on a plate level is recommended because this also corrects to some degree for plate-to-plate batch effects. Where sufficient negative controls exist, we recommend normalizing the features to the negative control. Check the profiling recipe for more information on how to process single-cell morphological profiles and how to normalize Cell Painting data for more information.
In data normalized to the negative controls, the negative control samples (or samples with otherwise weak phenotypes, here defined as a mean average precision across replicates of <0) will show limited similarity to one another and thus will show minimal clustering after step 16 of Basic Protocol 1 (hierarchical clustering). Somewhat unintuitively, this means that these samples will be spread across the entire dataset post-clustering. It is therefore expected, after hierarchical clustering and exploration (step 17 of Basic Protocol 1), to see one or a small number of “random” negative controls or weak perturbations clustering with a strong, consistent perturbation; this should not be taken as a sign that the strong perturbation in question is weak or similar to negative controls. Weak replicate correlation for any given sample can be checked in step 12 of Basic Protocol 1; if the replicate inconsistency looks possibly driven by technical issues (e.g., well position, Fig. 2B), one may consider performing another experiment to attempt to confirm if a profile is truly weak. In general, profiles with weak replicate correlation should not be used to draw biological conclusions, and hierarchically clustering results should always be checked for accidental spurious inclusion of weak profiles.
Proper reduction of the feature space is also an essential step to perform before analyzing new data in our protocols; this step will be automatically performed when following the profiling recipe (Chandrasekaran, Weisbart, Way, Carpenter, & Singh, 2022). If performing these steps on your own, a common starting point is to look for correlated features: when two features are too correlated, only one should be kept for further analysis. Since Pearson correlations are sensitive to large absolute feature values, we also recommend screening for unusual feature values; we provide guidance on performing this in Morpheus (see Basic Protocol 1, steps 3-6). Some feature reduction algorithms, such as support vector machines, give weights for each feature and remove the ones with fewer weights (Caicedo et al., 2017). We typically perform feature reduction in pycytominer, which provides six options for reducing the feature space based on (1) variance threshold (removing features that have relatively few unique feature values and/or a single value that is far more common than the rest of the feature values), (2) correlation threshold (removing features that are highly correlated to other features and thus redundant), (3) drop NA columns (removing features where a large number of values are missing), (4) drop outliers (removing features with aberrantly large absolute values), (5) noise (removing features that tend to have a high variance across replicates), and (6) blocklisting (removing features thought to not typically add useful biological information to Cell Painting profiles) (Way, 2019). Many of these feature removal methods have tunable parameters that ultimately guide the fraction of features removed; as such, it is critically important to check that the threshold values are appropriate for your data and adjust them when necessary.
Profiles should be assessed for their quality before data interpretation, to remove treatments with no apparent phenotype and, in some applications, to exclude compounds that are too toxic to the cells (Rezvani, Bigverdi, & Rohban, 2022). One method to perform profile quality assessment is to measure the precision with which one can correctly retrieve replicate wells. This approach was used in the example data we provide to check for the replicability of the profiles (for details see Way, Natoli, et al., 2022).
For troubleshooting of this method, problems, possible causes, and solutions are outlined in Table 1.
| Problem | Possible cause | Solution |
|---|---|---|
| All/almost all samples have a correlation value close to 1 (Morpheus after generating Similarity Matrix) | Features are not normalized | Check if the data were normalized (all features in range of 0-1) |
| Cells on Google Colab notebook cannot run | Notebook was not copied to user's Google Drive | Add a copy of the Notebook to your own Google Drive by clicking on Copy to Drive |
| User Warning: KMeans is known to have a memory leak on Windows with MKL (Math Kernel Library) when there are less chunks than available threads. You can avoid it by setting the environment variable OMP_NUM_THREADS=2. | Memory leak | Set the environment variable OMP_NUM_THREADS = x, with x being the value specified on your error output. Follow the solution in this thread on stack_over_flow. |
Understanding Results
When analyzing results, you may find that a profile of interest shows a dramatic difference from controls or other samples based on only a small number of similarly named features (such as a large number of features that fall within the nucleus or many changes in the texture of a particular stain), and the feature names have obvious meanings (e.g., nucleus area or integrated intensity of the mitochondria channel in the cytoplasm). In this scenario, interpretation may be straightforward, though you may need to look up the meaning of the feature names in the CellProfiler manual (https://broad.io/cellprofilermanual) to understand them better and discern their connection to the biological meaning. Some caution is warranted here; for example, DNA-damaging drugs could affect actin features because F-actin plays a role in DNA repair. Damage induced to the DNA induces nuclear actin formation (Belin, Lee, & Mullins, 2015), and these nuclear actin structures play a role in double-stranded break (DSB) repair, such as recruitment of proteins to enable repair of the heterochromatin through homologous recombination and assisting DSB movement in euchromatin repair (Caridi, Plessner, Grosse, & Chiolo, 2019). There may not be a straight line from a feature name to the biological function because cells are deeply interconnected systems and changes that start in a single genetic pathway can ripple throughout other pathways in the cell. Nevertheless, feature names can often create insights.
Instead of a few, easily interpretable features, you may find there are many dominant features in the profile and their collective meaning is not obvious. In such cases, an expert might be able to stare at the list and derive some meaning. For example, an expert might realize that many different stains showing increased correlation may actually be related to a decreased x -y cell size (because in a rounded cell, organelles are more likely to overlap one another on the x -y plane and may be either truly colocalized or merely spread across the z dimension). If you've looked at your feature list but need some backup, consider sharing your data on forum.image.sc so that experts can weigh in. An example of this can be found in the morphological profile induced by the microtubule inhibitor and microtubule-stabilizing agent in this dataset (cabazitaxel and ixabepilone, respectively). To understand the features that differentiate between our negative control (DMSO) and the microtubule perturbations, we performed marker selection using a T-test. Marker selection comes from genome analysis, but could be defined also as a feature selection. The model takes the features belonging to two classes as input and a T-test is calculated to assess marker features that discriminate between the two classes (DMSO vs. microtubule) (Gould et al., 2006). While individual T-tests performed in Morpheus do attempt to correct for sample number with a false discovery rate, it does not and cannot control for how many tests the user runs; these tests are therefore appropriate for gaining qualitative insight into the relative importance of various stains and/or feature classes in distinguishing a phenotype, but the values returned should not be directly reported, and any attempt to quantify these differences should be performed through standard statistical approaches. Our results show that many important features (Fig. 6A) belong to Granularity and Texture feature groups across a number of different stains, which makes sense in the context of induction of massive cytoskeletal disruption. Since microtubule disruption perturbs cell division, the presence of Nuclei_AreaShape_FormFactor (a measure of shape uniformity in which linear and/or irregular shapes have values near 0 and a perfect circle is 1) helps indicate that we are not looking at general cytoskeletal disruption, but specific disruption of the microtubules. This result highlights that the aggregate of different features is important for connecting profiles to perturbations.
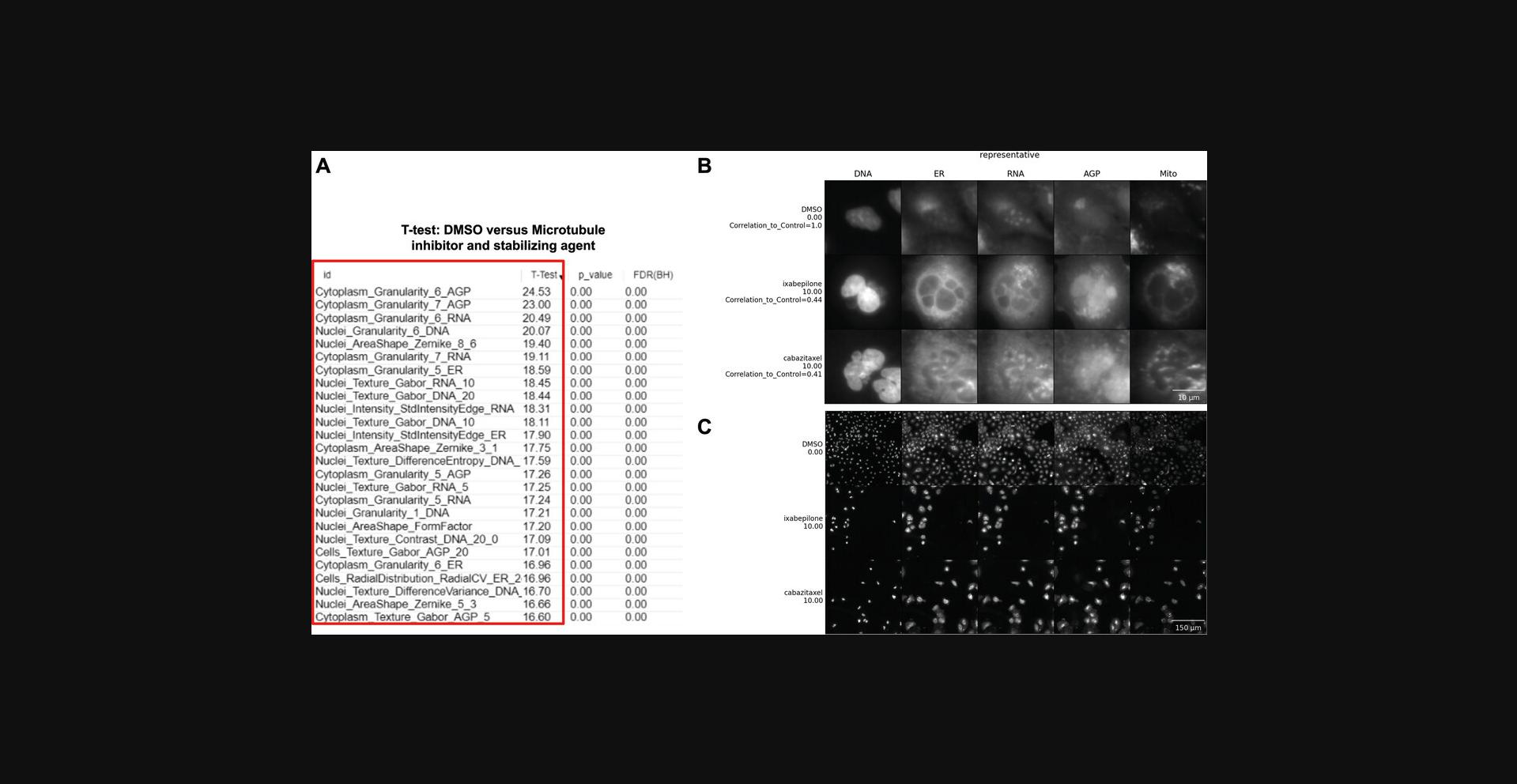
Examining example images directly alongside a list of important features can also help decipher a complex profile. An example where looking at features and images could help uncover the biological meaning of an event is during an assay to identify cells in different phases of the cell cycle using fluorescent markers such as DAPI to measure DNA content (Ferro et al., 2017). Based on significant changes in the feature space where the minor axis of the Nuclei and Cell area are low and DNA staining intensity is high, the user could look at single cells and realize these feature changes relate to cells that are going through metaphase. Basic Protocol 2 facilitates displaying single cells and images, which can otherwise be challenging to locate and access in large-scale experiments. In our example images of cells treated with two microtubule-related drugs, we observe that both drugs interfere with the cell cycle to produce similar morphologies, disrupting the overall appearance of every channel. As seen in Figure 6B, both treatments induce multinucleation (Fig. 6B, DNA column), as has been previously described for microtubule inhibitors (Azarenko, Smiyun, Mah, Wilson, & Jordan, 2014). Disruption of the cell cycle is also likely apparent in the lower overall cell count in treated vs. control cells (Fig. 6C). The Golgi localization and distribution are visually quite distinct compared to DMSO (Fig. 6B, AGP column), which could be related to the role of microtubules in vesicular trafficking and to their role in modeling the shape of organelles, including Golgi (Fourriere, Jimenez, Perez, & Boncompain, 2020; Thyberg & Moskalewski, 1985). We can therefore relate these morphological features and observations to the mechanism of actions of these drugs, providing a useful pattern to follow for investigators examining their own data and formulating their hypotheses. Sometimes, however, the most important differences are not visible to humans, and image-based profiling approaches have sometimes outperformed human expert image analysis for precisely such reasons (Gibson et al., 2015; Zhou et al., 2021).
Finally, we should note that, in some situations, following the procedures provided still does not allow you to make much headway in truly understanding the induced phenotype. If so, profile data can be used in other ways, e.g., by simply using the profile as a signature of the sample and trying to use drugs to revert this disease phenotype to a healthy-associated phenotype. If one has access to computational experts, one can also try to query their data against publicly available datasets (Rohban et al., 2022), though these approaches are currently still experimental. The interpretation of complex profiles is a challenge, but when successful can propel research in new directions to uncover exciting new mechanisms.
Time Considerations
For Basic Protocol 1, supposing that data tables were pre-processed for normalization and feature selection before input into Morpheus, the total time to explore the data is ∼1 hr. Basic Protocol 2 could take up to 2.5-3 hr if running the protocol with different settings and taking time to evaluate the images and create hypotheses.
Acknowledgments
We thank Rebecca Senft and Erin Weisbart for suggestions for the manuscript. We also thank Srinivas Niranj Chandrasekaran for helping with average precision concepts and data availability. Funding was provided by the National Institutes of Health (NIH COBA P41 GM135019 to BAC and AEC; MIRA R35 GM122547 to AEC). This project was made possible in part by grant number 2020-225720 to BAC from the Chan Zuckerberg Initiative DAF, an advised fund of the Silicon Valley Community Foundation. Funding was also provided by the São Paulo Research Foundation (FAPESP) #2022/01483-4, #2019/24033-1, and #2020/01218-3.
Author Contributions
Fernanda Garcia-Fossa : Software, data curation, writing (original draft, review, and editing); Mario Costa Cruz : Validation, writing (original draft, review, and editing); Marzieh Haghighi : Software, writing (review); Marcelo Bispo de Jesus : Funding acquisition, writing (review and editing); Shantanu Singh : Funding acquisition, writing (review and editing); Anne E. Carpenter : Conceptualization, supervision, funding acquisition, writing (original draft); Beth A. Cimini : Conceptualization, methodology, supervision, funding acquisition, writing (original draft, review, and editing).
Conflict of Interest
SS and AEC serve as scientific advisors for companies that use image-based profiling and Cell Painting (AEC: Recursion, SS: Waypoint Bio, Dewpoint Therapeutics) and receive honoraria for occasional talks at pharmaceutical and biotechnology companies.
Open Research
Data Availability Statement
The data that support the protocol are openly available at https://github.com/ciminilab/2023_Garcia-Fossa_Cruz_CurrentProtocols.
Literature Cited
- Azarenko, O., Smiyun, G., Mah, J., Wilson, L., & Jordan, M. A. (2014). Antiproliferative mechanism of action of the novel taxane cabazitaxel as compared with the parent compound docetaxel in MCF7 breast cancer cells. Molecular Cancer Therapeutics , 13, 2092–2103. doi: 10.1158/1535-7163.MCT-14-0265
- Belin, B. J., Lee, T., & Mullins, R. D. (2015). DNA damage induces nuclear actin filament assembly by Formin-2 and Spire-1/2 that promotes efficient DNA repair. [corrected]. eLife , 4, e07735. doi: 10.7554/eLife.07735
- Bray, M.-A., Singh, S., Han, H., Davis, C. T., Borgeson, B., Hartland, C., … Carpenter, A. E. (2016). Cell Painting, a high-content image-based assay for morphological profiling using multiplexed fluorescent dyes. Nature Protocols , 11, 1757–1774. doi: 10.1038/nprot.2016.105
- Caicedo, J. C., Cooper, S., Heigwer, F., Warchal, S., Qiu, P., Molnar, C., … Carpenter, A. E. (2017). Data-analysis strategies for image-based cell profiling. Nature Methods , 14, 849–863. doi: 10.1038/nmeth.4397
- Caridi, C. P., Plessner, M., Grosse, R., & Chiolo, I. (2019). Nuclear actin filaments in DNA repair dynamics. Nature Cell Biology , 21, 1068–1077. doi: 10.1038/s41556-019-0379-1
- Chandrasekaran, S. N., Ceulemans, H., Boyd, J. D., & Carpenter, A. E. (2021). Image-based profiling for drug discovery: Due for a machine-learning upgrade? Nature Reviews, Drug Discovery , 20, 145–159. doi: 10.1038/s41573-020-00117-w
- Chandrasekaran, S. N., Weisbart, E., Way, G., Carpenter, A., & Singh, S. (2022). Broad Institute imaging platform profiling recipe. Retrieved from https://github.com/cytomining/profiling-recipe
- Cimini, B. A., Chandrasekaran, S. N., Kost-Alimova, M., Miller, L., Goodale, A., Fritchman, B., … Carpenter, A. E. (2022). Optimizing the Cell Painting assay for image-based profiling. bioRxiv , 2022.07.13.499171. doi: 10.1101/2022.07.13.499171v1
- Ferro, A., Mestre, T., Carneiro, P., Sahumbaiev, I., Seruca, R., & Sanches, J. M. (2017). Blue intensity matters for cell cycle profiling in fluorescence DAPI-stained images. Laboratory Investigation , 97, 615–625. doi: 10.1038/labinvest.2017.13
- Fourriere, L., Jimenez, A. J., Perez, F., & Boncompain, G. (2020). The role of microtubules in secretory protein transport. Journal of Cell Science , 133, jcs237016. doi: 10.1242/jcs.237016
- Gibson, C. C., Zhu, W., Davis, C. T., Bowman-Kirigin, J. A., Chan, A. C., Ling, J., … Li, D. Y. (2015). Strategy for identifying repurposed drugs for the treatment of cerebral cavernous malformation. Circulation , 131, 289–299. doi: 10.1161/CIRCULATIONAHA.114.010403
- Gould, J., Getz, G., Monti, S., Reich, M., & Mesirov, J. P. (2006). Comparative gene marker selection suite. Bioinformatics , 22, 1924–1925. doi: 10.1093/bioinformatics/btl196
- Hirano, Y., Kinugasa, Y., Osakada, H., Shindo, T., Kubota, Y., Shibata, S., … Hiraoka, Y. (2020). Lem2 and Lnp1 maintain the membrane boundary between the nuclear envelope and endoplasmic reticulum. Communications Biology , 3, 276. doi: 10.1038/s42003-020-0999-9
- Kluyver, T., Ragan-Kelley, B., Pérez, F., Granger, B., Bussonnier, M., Frederic, J., … Jupyter Development Team. (2016). Jupyter Notebooks – a publishing format for reproducible computational workflows. In F. Loizides & B. Scmidt (Eds.), Positioning and power in academic publishing: Players, agents and agendas (pp. 87–90). IOS Press. doi: 10.3233/978-1-61499-649-1-87
- Pearson, K., & Galton, F. (1895). VII. Note on regression and inheritance in the case of two parents. Proceedings of the Royal Society of London , 58, 240–242.
- Pedregosa, F., Varoquaux, G., Gramfort, A., Michel, V., Thirion, B., Grisel, O., … Duchesnay, E. (2011). Scikit-learn: Machine learning in Python. Journal of Machine Learning Research , 12, 2825–2830.
- Reback, J., McKinney, W., jbrockmendel, Van den Bossche, J., Augspurger, T., Cloud, P., … Gorelli, M. (2020). pandas-dev/pandas: Pandas 1.2.0. Retrieved from https://zenodo.org/record/4394318
- Rezvani, A., Bigverdi, M., & Rohban, M. H. (2022). Image-based cell profiling enhancement via data cleaning methods. PLoS One , 17, e0267280. doi: 10.1371/journal.pone.0267280
- Rohban, M. H., Fuller, A. M., Tan, C., Goldstein, J. T., Syangtan, D., Gutnick, A., … Carpenter, A. E. (2022). Virtual screening for small-molecule pathway regulators by image-profile matching. Cell Systems , 13, 724–736.e9. doi: 10.1016/j.cels.2022.08.003
- Schindelin, J., Arganda-Carreras, I., Frise, E., Kaynig, V., Longair, M., Pietzsch, T., … Cardona, A. (2012). Fiji: An open-source platform for biological-image analysis. Nature Methods , 9, 676–682. doi: 10.1038/nmeth.2019
- Stirling, D. R., Swain-Bowden, M. J., Lucas, A. M., Carpenter, A. E., Cimini, B. A., & Goodman, A. (2021). CellProfiler 4: Improvements in speed, utility and usability. BMC Bioinformatics , 22, 433. doi: 10.1186/s12859-021-04344-9
- Thyberg, J., & Moskalewski, S. (1985). Microtubules and the organization of the Golgi complex. Experimental Cell Research , 159, 1–16. doi: 10.1016/S0014-4827(85)80032-X
- Van Rossum, G., & Drake, F. L. (2009). Python 3 reference manual. CreateSpace, Scotts Valley, CA.
- Way, G. P. (2019). Blocklist Features — Cell Profiler. Retrieved from https://figshare.com/articles/dataset/Blacklist_Features_-_Cell_Profiler/10255811
- Way, G. P., Chandrasekaran, S. N., Bornholdt, M., Fleming, S., Tsang, H., Adeboye, A., … Singh, S. (2022). Pycytominer: Data processing functions for profiling perturbations. Retrieved from https://github.com/cytomining/pycytominer [Accessed September 9, 2022]
- Way, G. P., Kost-Alimova, M., Shibue, T., Harrington, W. F., Gill, S., Piccioni, F., … Singh, S. (2021). Predicting cell health phenotypes using image-based morphology profiling. Molecular Biology of the Cell , 32, 995–1005. doi: 10.1091/mbc.E20-12-0784
- Way, G. P., Natoli, T., Adeboye, A., Litichevskiy, L., Yang, A., Lu, X., … Carpenter, A. E. (2022). Morphology and gene expression profiling provide complementary information for mapping cell state. Cell Systems , 13, 911–923.e9. doi: 10.1016/j.cels.2022.10.001
- Zhou, W., Yang, Y., Yu, C., Liu, J., Duan, X., Weng, Z., … Zhou, L. (2021). Ensembled deep learning model outperforms human experts in diagnosing biliary atresia from sonographic gallbladder images. Nature Communications , 12, 1259. doi: 10.1038/s41467-021-21466-z
Internet Resources
Versatile matrix visualization and analysis software.
Forum for image analysis questions and discussions.
Downloading and use of CellProfiler.
Beginner and advanced tutorials for CellProfiler.
Video tutorials for CellProfiler, Morpheus, and many other tools on the COBA YouTube channel.
Data processing functions for profiling perturbations. More information on how to use pycytominer, documentation, and workflows.
Citing Literature
Number of times cited according to CrossRef: 6
- Fernanda Garcia-Fossa, Marcelo Bispo de Jesus, Cationic solid lipid nanoparticles (SLN) complexed with plasmid DNA enhance prostate cancer cells (PC-3) migration, Nanotoxicology, 10.1080/17435390.2024.2307616, 18 , 1, (36-54), (2024).
- Laure Maneix, Polina Iakova, Charles G. Lee, Shannon E. Moree, Xuan Lu, Gandhar K. Datar, Cedric T. Hill, Eric Spooner, Jordon C. K. King, David B. Sykes, Borja Saez, Bruno Di Stefano, Xi Chen, Daniela S. Krause, Ergun Sahin, Francis T. F. Tsai, Margaret A. Goodell, Bradford C. Berk, David T. Scadden, André Catic, Cyclophilin A supports translation of intrinsically disordered proteins and affects haematopoietic stem cell ageing, Nature Cell Biology, 10.1038/s41556-024-01387-x, 26 , 4, (593-603), (2024).
- Fernanda Garcia-Fossa, Tuanny Leite Almeida, Rafaella Mascarelli Pereira, Thaís Moraes-Lacerda, Mariana Rodrigues Da Silva, Laura de Oliveira-Nascimento, Marcelo Bispo de Jesus, Assessment of nanotoxicology through in vitro techniques and image-based assays, Molecular Pharmaceutics and Nano Drug Delivery, 10.1016/B978-0-323-91924-1.00003-4, (311-340), (2024).
- Beth A. Cimini, Srinivas Niranj Chandrasekaran, Maria Kost-Alimova, Lisa Miller, Amy Goodale, Briana Fritchman, Patrick Byrne, Sakshi Garg, Nasim Jamali, David J. Logan, John B. Concannon, Charles-Hugues Lardeau, Elizabeth Mouchet, Shantanu Singh, Hamdah Shafqat Abbasi, Peter Aspesi, Justin D. Boyd, Tamara Gilbert, David Gnutt, Santosh Hariharan, Desiree Hernandez, Gisela Hormel, Karolina Juhani, Michelle Melanson, Lewis H. Mervin, Tiziana Monteverde, James E. Pilling, Adam Skepner, Susanne E. Swalley, Anita Vrcic, Erin Weisbart, Guy Williams, Shan Yu, Bolek Zapiec, Anne E. Carpenter, Optimizing the Cell Painting assay for image-based profiling, Nature Protocols, 10.1038/s41596-023-00840-9, 18 , 7, (1981-2013), (2023).
- Fabio Stossi, Pankaj K. Singh, Kazem Safari, Michela Marini, Demetrio Labate, Michael A. Mancini, High throughput microscopy and single cell phenotypic image-based analysis in toxicology and drug discovery, Biochemical Pharmacology, 10.1016/j.bcp.2023.115770, 216 , (115770), (2023).
- Callum Tromans‐Coia, Nasim Jamali, Hamdah Shafqat Abbasi, Kenneth A. Giuliano, Mai Hagimoto, Kevin Jan, Erika Kaneko, Stefan Letzsch, Alexander Schreiner, Jonathan Z. Sexton, Mahomi Suzuki, O. Joseph Trask, Mitsunari Yamaguchi, Fumiki Yanagawa, Michael Yang, Anne E. Carpenter, Beth A. Cimini, Assessing the performance of the Cell Painting assay across different imaging systems, Cytometry Part A, 10.1002/cyto.a.24786, 103 , 11, (915-926), (2023).

