Imaging Subcellular AMPK Activity Using an Excitation-Ratiometric AMPK Activity Reporter
Danielle L. Schmitt, Danielle L. Schmitt
Abstract
Adenosine monophosphate (AMP)-activated protein kinase (AMPK) is a master regulator of cellular metabolism, phosphorylating a variety of downstream targets throughout the cell. Subcellular AMPK activity results in regulation of glycolysis, lipid and protein biosynthesis, mitochondrial function, and gene expression. But how AMPK senses and responds to stimuli in a compartment-specific manner is not well understood, leaving an incomplete picture of compartmentalized AMPK activity. Key tools for studying subcellular AMPK activity are genetically encoded AMPK activity reporters (AMPKARs), which allow for the quantitative visualization of subcelluar AMPK activity. However, many AMPKARs suffer from poor dynamic range and sensitivity, limiting their application. I recently reported the development of a new excitation-ratiometric (ExRai) AMPKAR, a single-fluorophore AMPKAR with enhanced dynamic range for detection of subtle, subcellular AMPK activity. I used ExRai AMPKAR to study subcellular AMPK activity at several locations, including the lysosome and mitochondria, identifying new mechanisms for the regulation of AMPK activity. Here, I describe the use of ExRai AMPKAR to image subcellular AMPK activity in mouse embryonic fibroblasts using both widefield and confocal microscopy. I also describe the culture of mouse embryonic fibroblasts. Through the use of ExRai AMPKAR, subcellular AMPK activity can be illuminated to better understand how this central kinase regulates cellular metabolism. © 2023 The Author. Current Protocols published by Wiley Periodicals LLC.
Basic Protocol : Imaging subcellular AMPK activity using ExRai AMPKAR
Support Protocol 1 : Culturing of mouse embryonic fibroblasts for live-cell imaging
Support Protocol 2 : Live-cell imaging of ExRai AMPKAR using confocal microscopy
INTRODUCTION
Adenosine monophosphate (AMP)-activated protein kinase (AMPK) is a master regulator of cellular metabolism, with at least 100 identified downstream signaling targets (Hardie, 2022). In response to cellular stress or calcium signaling, AMPK controls a variety of metabolic processes, including glucose metabolism, mitochondrial function, lysosomal function, protein synthesis, and gene expression (Garcia & Shaw, 2017). As many AMPK targets occupy distinct subcellular locations, the spatiotemporal regulation and function of AMPK is an intense area of study. AMPK signaling complexes have been observed at several subcellular locations, including the lysosome and mitochondria. At the lysosome, AMPK has been found to be preferentially activated by glucose starvation, whereas more severe stress was needed to induce cytoplasmic and mitochondrial AMPK activity (Zong et al., 2019). At the mitochondria, AMPK has been found to regulate mitochondrial fission (Toyama et al., 2016), and selective inhibition of AMPK at the mitochondria resulted in reduction of exercise-induced mitophagy in skeletal muscle (Drake et al., 2021). Although these studies of spatial AMPK activity have provided significant insight into the subcellular role of AMPK in the cell, much of this work was done using approaches that are limited in their spatial and temporal resolution, and thus our understanding of spatiotemporal AMPK activity remains hampered.
A key tool for the study of spatiotemporal kinase activity are genetically encoded kinase activity reporters (KARs). KARs enable a sensitive, robust, and quantitative readout of kinase activity with high spatiotemporal resolution at the single-cell level (Schmitt et al., 2020). AMPK KARs (AMPKARs) that use changes in Förster resonance energy transfer (FRET) as the functional readout have previously been applied to study AMPK activity across scales (Hung et al., 2017; Konagaya et al., 2017; Sample et al., 2015; Tsou et al., 2011). Although FRET-based AMPKAR has been successfully used to measure subcellular AMPK activity, the poor dynamic range of the reporter has limited its use (Miyamoto et al., 2015). Recently, I reported the development of a single-fluorophore excitation-ratiometric (ExRai) AMPKAR (Schmitt et al., 2022). ExRai AMPKAR consists of circularly permutated enhanced green fluorescent protein (cpEGFP) flanked on the N-terminus by an AMPK-specific substrate peptide and on the C-terminus by an FHA1 phosphoamino acid–binding domain. AMPK phosphorylation of the substrate peptide induces binding by FHA1, resulting in a conformational change that shifts the peak excitation wavelength of ExRai AMPKAR (Fig. 1A). I found ExRai AMPKAR to have a significantly improved dynamic range over FRET-based AMPKAR, with a maximum ratio change about 3.5 times greater than FRET-based AMPKAR. This enabled the sensitive detection and study of subtle subcellular AMPK activity. Through the addition of signal peptides, ExRai AMPKAR can be targeted to various subcellular organelles for precise measurement of compartmentalized AMPK activity (Fig. 1B). For instance, using ExRai AMPKAR targeted to lysosomal surface, I found that lysosomal pools of AMPK are preferentially activated over mitochondrial or cytoplasmic pools, dependent on the presence of the upstream AMPK regulator liver kinase B1 (LKB1). I also used nuclear-localized ExRai AMPKAR to uncover a translocation-dependent mechanism for nuclear AMPK activity, which had remained elusive due to the limitations of FRET-based AMPKARs. Thus, ExRai AMPKAR is a sensitive reporter for subcellular AMPK activity that can be used to illuminate mechanisms for AMPK activity that have previously remained challenging to study.
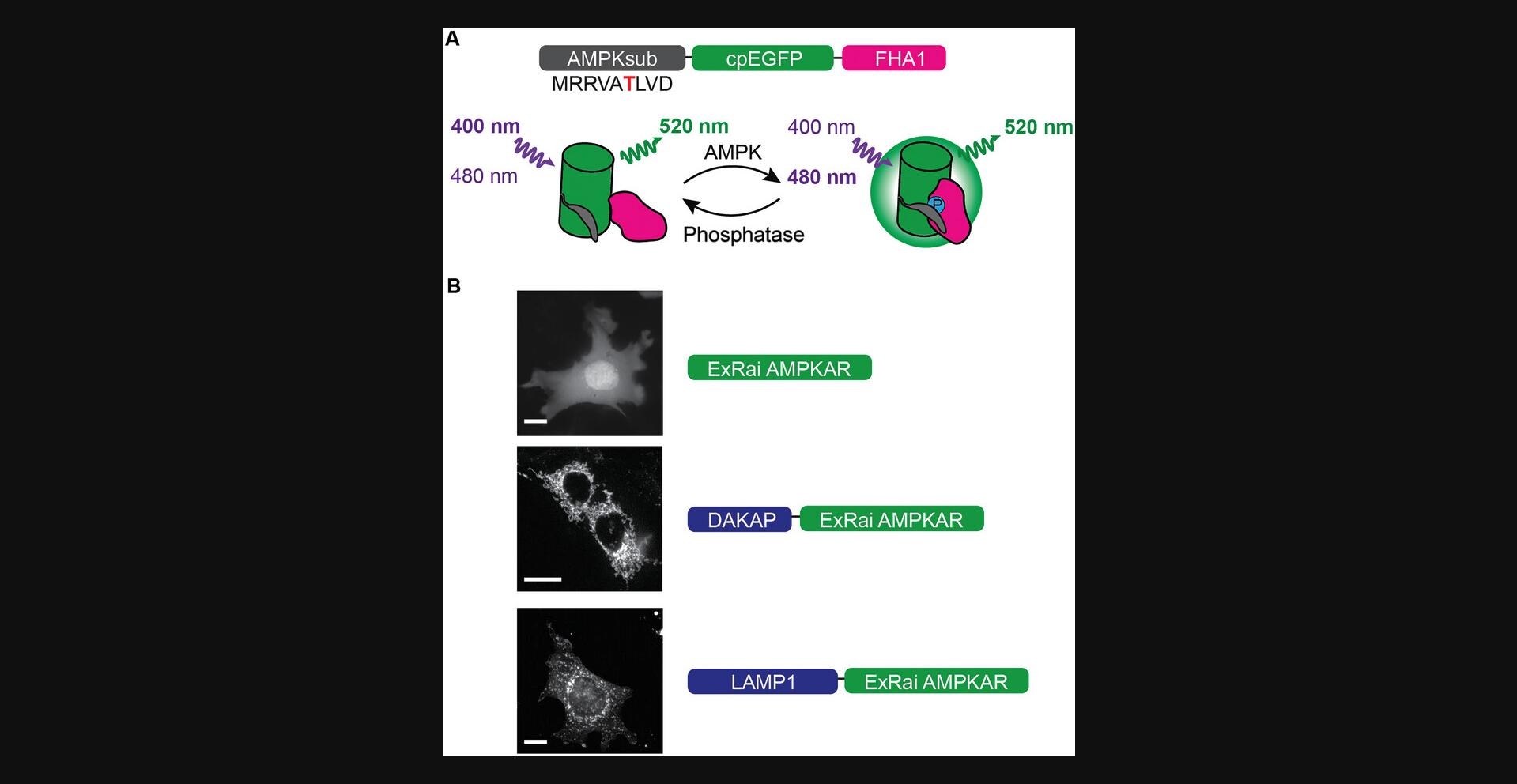
In this article, I describe a protocol to study subcellular AMPK activity using ExRai AMPKAR in living cells. I describe the procedure for imaging cellular stress-induced AMPK activity using 2-deoxyglucose (2-DG) and allosteric activation of AMPK using MK-8722 (Feng et al., 2017), including cell culture, transfection, preparation for imaging, image and data acquisition, and data analysis. Here, ExRai AMPKAR is expressed in mouse embryonic fibroblasts (MEFs), a cell line frequently used to study AMPK signaling (Egan et al., 2011; Shaw et al., 2004; Stein et al., 2019; Toyama et al., 2016). Thus, I provide Support Protocol 1, which describes maintaining MEFs in culture. As ExRai AMPKAR can be imaged using multiple imaging modalities, I provide Support Protocol 2, which describes confocal imaging of ExRai AMPKAR.
STRATEGIC PLANNING
ExRai AMPKAR can be used to measure subcellular AMPK activity in response to different stimuli, typically cellular stress or allosteric activation (Schmitt et al., 2022). When planning an imaging experiment with ExRai AMPKAR, careful consideration needs to be taken to ensure success of the experiment. The location of AMPK activity to be studied, stimulus, cell line, and imaging system need to be established.
ExRai AMPKAR has been successfully used to study AMPK activity in the cytoplasm, lysosome membrane, outer mitochondrial membrane, and nucleus. Measurement of AMPK activity at these or other locations requires development of subcellularly targeted ExRai AMPKAR incorporating the appropriate localization sequences or fusion of ExRai AMPKAR to proteins with the desired localization. For example, lysosomal membrane targeting of ExRai AMPKAR was accomplished by fusing full-length lysosomal-associated membrane protein 1 (LAMP1) to the N-terminus of ExRai AMPKAR (lyso-ExRai AMPKAR), and mitochondrial localization was achieved by adding an outer mitochondrial membrane localization sequence derived from dual-specific A kinase anchoring protein (mito-ExRai AMPKAR; AIQLRSLFPLALPGMLALLGWWWFFSRKKADP) to the N-terminus of ExRai AMPKAR (mito-ExRai AMPKAR; Fig. 1B; Schmitt et al., 2022). Although ExRai AMPKAR can be targeted to various subcellular locations, in this article I focus on imaging cytosolic, lysosomal, and mitochondrial AMPK activity.
Next, the stimulus and cell line used to study subcellular AMPK activity should be selected. Most studies of AMPK activity use cellular stress (e.g., 2-DG and inhibitors of oxidative phosphorylation) to activate AMPK or take advantage of a variety of allosteric AMPK activators, including MK-8722 and A-769662. ExRai AMPKAR has primarily been used to study acute activation of AMPK in response to stimuli (30 to 60 min). FRET-based AMPKARs have been used to study longer-term AMPK activity over the course of hours, which could be done using ExRai AMPKAR (Kosaisawe et al., 2021). Therefore, concentration and timing of treatment should be carefully thought out and optimized. An important consideration for studying subcellular AMPK activity is the cell line used and how this might impact studies. For example, many commonly used cancer cell lines, including HeLa and A549 lung cancer cells, lack the upstream kinase LKB1, which could influence results. The genetic background of each cell line should be carefully considered, including any necessary controls such as genetic knockout of AMPK. In addition, adherent cell lines are best for imaging, as they can be easily tracked over time. ExRai AMPKAR has been successfully imaged in MEFs, as well as Cos7, HEK293T, and HeLa cells (Schmitt et al., 2022).
Once cell lines for study have been selected, an important consideration is how the plasmid containing ExRai AMPKAR will be introduced to the cells. A variety of methods can be used, including chemical transfection, viral infection, and electroporation. Transfection efficiency can vary greatly depending on plasmid and cell type; therefore, optimization of transfection should be done, including the method and amount of DNA used in transfection. Here, I present transfection using a nonliposomal reagent; however, ExRai AMPKAR has also been successfully transfected into cells using liposome-based transfections and nucleofection.
An important consideration for designing experiments and imaging ExRai AMPKAR is the use of controls. Proper controls will validate measured AMPK activity and are essential for studying subcellular AMPK activity. One control to consider is ExRai AMPKAR T/A. This is a control construct where the target threonine residue phosphorylated by AMPK has been mutated to alanine, rendering this construct insensitive to AMPK activity. Other commonly used controls include vehicle controls, selective AMPK inhibitors like SBI-0206965 (Dite et al., 2018), or AMPKα1/2 knockout cell lines (Schmitt et al., 2022; Toyama et al., 2016). During experimental design, care should be taken to consider and use the most appropriate control conditions. In the data presented here, AMPKα1/2 knockout cells were used as controls.
Finally, before beginning the experiment, an appropriate imaging system must be in place to image ExRai AMPKAR. ExRai KARs have been successfully imaged using both widefield and confocal microscopy (Mehta et al., 2018; Zhang et al., 2020). In this article, I describe imaging ExRai AMPKAR using a widefield microscope with a standard set of excitation and emission filters. I also provide a support protocol for the use of confocal microscopy to image ExRai AMPKAR. For ratiometric imaging of ExRai AMPKAR, cells are sequentially imaged using 400- and 480-nm excitation filters or laser lines and a 520-nm emission filter for both excitation wavelengths. These wavelengths are commonly used to image DAPI and GFP, and only slight modification is needed to set up ExRai imaging. To minimize photobleaching and maintain an optimal signal-to-noise ratio, neutral-density (ND) filters are used in front of the light source, with short exposure times (50 to 100 ms) and images acquired every 30 s. Shorter or longer time intervals can be used depending on the application. When optimizing imaging settings, the appropriate objective lens should be used. For imaging smaller organelles, like lysosomes, a higher-powered magnification (e.g., 40× or 60×) should be used, whereas imaging large populations of cells with lower spatial resolution can be accomplished using a 20× objective lens with a high numerical aperture (N.A.). After consideration of experimental protocol, subcellular locations, cell lines, transfection method, controls, and microscope setup, imaging of subcellular AMPK activity can be accomplished.
CAUTION : Immortalized MEFs and many commonly used immortalized mammalian cell lines are considered a Biosafety Level 2 hazard. Follow all appropriate guidelines and regulations for the use and handling of pathogenic microorganisms.
Basic Protocol: IMAGING SUBCELLULAR AMPK ACTIVITY USING ExRai AMPKAR
This protocol allows users to image subcellular AMPK activity using targeted ExRai AMPKAR under widefield fluorescence microscopy. For imaging using confocal microscopy, please see Support Protocol 2.First, users prepare samples for imaging, which includes plating cells on imaging dishes and chemical transfection of cells with localized ExRai AMPKAR. Next, imaging of subcellular AMPK activity is accomplished using a widefield microscope to acquire images both before and after cells have been treated with 2-DG or an allosteric activator of AMPK (MK-8722) to induce AMPK activity. Finally, acquired images are analyzed to determine changes in excitation ratio after treatment with AMPK activators, the maximum ratio change for each location and stimulus, and the time to half maximum of reporter response. Successful completion of this protocol will result in measurement of subcellular AMPK activity induced by pharmacologic perturbation.
The Basic Protocol describes imaging ExRai AMPKAR localized to the cytoplasm, outer mitochondrial membrane, and lysosomal surface using cells grown in 35-mm imaging dishes, but this protocol can be adapted to image ExRai AMPKAR at locations and scales of interest.
Materials
-
MEFs (e.g., ATCC, cat. no. CRL-2991)
-
MEF culture medium (see recipe)
-
Opti-MEM I Reduced Serum Medium (e.g., Gibco, cat. no. 31985070)
-
Plasmid DNA encoding ExRai AMPKAR:
- Untargeted ExRai AMPKAR (e.g., Addgene, cat. no. 192446)
- Lysosomal outer membrane–targeted ExRai AMPKAR (lyso-ExRai AMPKAR; e.g., Addgene, cat. no. 192449)
- Mitochondrial outer membrane–targeted ExRai AMPKAR (mito-ExRai AMPKAR; e.g., Addgene, cat. no. 192448)
-
FuGENE HD (e.g., Promega, cat. no. E2311)
-
Hank's balanced salt solution (HBSS) imaging buffer (see recipe)
-
Fluorescence-free immersion oil (e.g., Immersol 518F; Carl Zeiss, cat. no. 444960)
-
2 M 2-DG (e.g., Sigma-Aldrich, cat. no. D6134-250MG) in 1× Dulbecco's phosphate-buffered saline (DPBS; e.g., Gibco, cat. no. 14190-144)
-
100 µM MK-8722 (e.g., Aobious, cat. no. AOB33226) in dimethyl sulfoxide (DMSO; e.g., Corning, cat. no. 25-950-CQC)
-
35-mm glass-bottom dishes, sterile (e.g., CellVis, cat. no. D35-14-1.5N)
-
Humidified incubator with 5% CO2 (e.g., Thermo Scientific Heracell VIOS 160i CO2 Incubator with Copper Chamber)
-
Tissue culture light microscope (e.g., Laxco LMI-3000 Series Routine Inverted Microscope)
-
Biological safety cabinet (e.g., Thermo Scientific 1300 Series A2)
-
Vortex mixer
-
1.7-ml microcentrifuge tubes, sterile (e.g., Corning, cat. no. 3207)
-
Inverted fluorescence microscope with appropriate objective lens, filter sets, camera, and image acquisition software:
- Zeiss AxioObserver Z1 microscope base equipped with definite focus
- Light source (e.g., HXP120; Carl Zeiss)
- Plan-apochromat 20×/0.8 N.A. and 40×/1.4 N.A. objective lens
- CMOS Orca Flash 4.0 camera
- Custom incubator chamber or stage top incubator to maintain 37°C environment
- Filter sets suitable for imaging dual GFP excitation ratio: ET405/40× and ET480/30× excitation filters, T505dczr dichroic, and ET535/50m emission
- External filter exchanger
-
Modeling clay
-
Computer running the following software:
- MATLAB (MathWorks) preconfigured to run MatScope (see Internet Resources) or other similar image acquisition software (e.g., NIS Elements, µ-manager, Zeiss Zen)
- Spreadsheet application (e.g., Microsoft Excel, Google Sheets)
- ImageJ or similar image analysis software
- Statistical analysis software (e.g., GraphPad Prism 8, R)
Cell transfection
Day 1 (30 min)
1.Seed a suspension of MEFs onto a 35-mm glass-bottom imaging dish, and culture cells in MEF culture medium (see Support Protocol 1 for details on passaging MEFs) to reach ∼70% confluency at the time of transfection. Incubate cells in a 37°C, 95% humidity, 5% CO2 incubator for 24 hr before transfection.
Day 2 (30 min)
2.Check confluency of imaging dishes containing MEFs under a light microscope. If cells have achieved at least 70% confluency on the glass bottom, proceed to step 3.Otherwise, exchange medium for fresh MEF culture medium, and incubate an additional 24 hr.
3.In a sterile biological safety cabinet, prepare transfection mix according to the recipe in Table 1.Briefly, add 100 µl Opti-MEM to each tube, and then add 0.5 µg plasmid DNA for each respective localized biosensor. Vortex quickly to mix. Then, add 1.5 µl FuGENE HD to the plasmid dilution. Mix by gently pipetting up and down. Incubate transfection mix for 15 min at room temperature.
| Reagent | Dish 1 | Dish 2 | Dish 3 | Dish 4 | Dish 5 | Dish 6 |
|---|---|---|---|---|---|---|
| Biosensor | ExRai AMPKAR | ExRai AMPKAR | Mito-ExRai AMPKAR | Mito-ExRai AMPKAR | Lyso-ExRai AMPKAR | Lyso-ExRai AMPKAR |
| Opti-MEM, μl | 100 | 100 | 100 | 100 | 100 | 100 |
| DNA, μg | 0.5 | 0.5 | 0.5 | 0.5 | 0.5 | 0.5 |
| Plasmid, μl | Variable | Variable | Variable | Variable | Variable | Variable |
| FuGENE HD transfection reagent, μl | 1.5 | 1.5 | 1.5 | 1.5 | 1.5 | 1.5 |
4.Place imaging dishes containing MEFs in biological safety cabinet. Add transfection mix dropwise to each dish, focusing on adding to cells adherent to the glass portion of the dish. Gently swirl to mix, and incubate overnight in a 37°C, 95% humidity, 5% CO2 atmosphere incubator.
Imaging preparation
Day 3 (30 min)
5.At ∼24 hr post-transfection, check imaging dishes for cell health.
6.In a biological safety cabinet, aspirate off culture medium from one 35-mm imaging dish, and wash cells with 2 ml of 1× HBSS imaging buffer prewarmed to 37°C, gently adding buffer to the side of the dish to avoid disturbing cells on the glass. Swirl gently, and aspirate off 1× HBSS. Repeat two times, and then add 2 ml of fresh 1× HBSS imaging buffer. Incubate cells at 37°C for 30 min.
7.While cells are incubating with 1× HBSS, turn on microscope power source, light source, definite focus, filter changer, camera, incubator, microscope base, and computer. Allow incubator to warm to 37°C. Open image acquisition software.
8.Configure a time course protocol for imaging ExRai biosensors. Ensure filter configuration is suitable for imaging ExRai AMPKAR. Set objective to the correct magnification (≥40× preferred for imaging localized ExRai AMPKAR) and imaging interval to 30 s for 30 min. For example:
- 400 nm excitation-emission: ET405/40× excitation filter, T505dcxr dichroic, and ET535/50m emission filter; 100-msec exposure.
- 480 nm excitation-emission: ET480/30× excitation filter, T505dcxr dichroic, and ET535/50m emission filter; 100-msec exposure.
The exposure and imaging interval should be modified for each imaging application and microscope setup. Exposure depends on camera sensitivity and light source. Too long of an exposure can increase cellular phototoxicity and photobleaching, whereas short exposure times result in noisy images with poor signal-to-noise ratios. Imaging intervals can be adjusted for imaging length (longer interval for longer imaging applications) or shortened to capture faster biological events. In my experience, 30- to 60-s intervals for 30 to 60 min is sufficient to capture changes in AMPK activity using ExRai AMPKAR.
9.Wet objective lens using a drop of immersion oil, and mount imaging dish on the stage. Use modeling clay to hold the imaging dish in place. Using 400-nm excitation-emission settings, bring cells into focus, and identify a region of the dish with several transfected cells. Once cells have been identified, set definite focus.
Data and image acquisition (30 min per dish)
10.Acquire a set of images of the field of view selected for imaging, and select regions of interest (ROIs) to monitor during imaging. Select at least three cells as ROIs. Select a background ROI, choosing a region of the field of view with no cells.
11.Begin experiment. Collect a time series of the field of view for each channel.
12.After acquiring ∼5 min of basal AMPK activity (≥10 image sets), pause acquisition to add stimulus to dish. Remove ∼500 µl imaging medium, and add to a microcentrifuge tube containing aliquoted stock solution of either 10 to 40 μl 2-DG (10 to 40 mM final ) or 10 μl MK-8722 (500 nM final). Gently pipet up and down to mix. Add dilution to the imaging dish dropwise around the edges of the dish. Once added, mix by gently pipetting four to five times, being careful to avoid directly pipetting on cells being imaged. Resume imaging for ∼25 min or until ExRai AMPKAR response reaches a maximum and does not increase further.
13.Repeat steps 9 to 12 until all dishes have been imaged. Upon completion of data acquisition, export data for analysis.
Image analysis (15 min per dish)
14.Open image analysis software. Load collected images, and select ROIs.
15.Review all selected ROIs over time to ensure ROIs track the cell or organelle faithfully, accounting for any changes in cell location (due to cell moving or bumping of dish during drug addition) or cell morphology. For the background ROI, ensure region remains free of floating cells or other artifacts. Extract fluorescence intensities (arbitrary units [a.u.]) from each ROI over each frame along with time each image was acquired, and compile in a spreadsheet (e.g., Excel or similar).
16.In the spreadsheet, normalize time to the time of drug addition by setting the time associated with drug addition as 0 min. For each time point, calculate background-subtracted intensity (I) of each channel and the excitation ratio using Equations 1 and 2.
17.Determine excitation ratio using Equation 3.
18.Normalize data to basal or pretreatment ExRai AMPKAR ratio (R) by averaging the first few data points to determine the basal ratio. Divide ratio at each time point by the basal ratio, and plot resulting normalized ratio. Plot the average of all single-cell normalized ratios and corresponding standard deviation using GraphPad Prism or similar statistical software (Fig. 2).
19.Determine maximum ratio change (∆Rmax/R0) using Equation 4, where Rmax is the maximum ratio achieved and R0 is ratio at t = 0 min.
20.Calculate time to half maximum (t1/2) for each single-cell time course. Identify ExRai AMPKAR ratio maximum, and then record the time at which the ratio was half of the maximum.
Support Protocol 1: CULTURING OF MOUSE EMBRYONIC FIBROBLASTS FOR LIVE-CELL IMAGING
Support Protocol 1 outlines the steps for successful culturing of MEFs and passaging for imaging experiments described in Basic Protocol. Adherent MEFs are collected from the previous culture dish using trypsin, diluted in culture medium, and a portion is placed in a new culture dish (cell passage). The remaining cells are used to seed glass-bottom imaging dishes for imaging ExRai AMPKAR. This protocol takes ∼30 min.
Materials
-
MEFs (e.g., ATCC, cat. no. CRL-2991)
-
MEF culture medium (see recipe)
-
70% (v/v) ethanol
-
TrypLE Express Enzyme (e.g., Gibco, cat. no. 12605010)
-
25-cm2 cell culture flasks (e.g., Fisher Scientific, cat. no. 101261C)
-
Humidified incubator with 5% CO2 (e.g., Thermo Scientific Heracell VIOS 160i CO2 Incubator with Copper Chamber)
-
Tissue culture light microscope (e.g., Laxco LMI-3000 Series Routine Inverted Microscope)
-
15-ml centrifuge tubes (e.g., USA Scientific, cat. no. 56188261)
-
Aerosol-tight centrifuge (e.g., Eppendorf Model 5810)
-
35-mm glass-bottom dishes (e.g., CellVis, cat. no. D35-14-1.5N)
1.Grow MEFs in at least a 25-cm2 cell culture flask in 5 ml MEF culture medium in a 37°C incubator with 95% humidity and 5% CO2. Passage cells once they are ∼90% confluent, as observed under a tissue culture light microscope. At this time, examine MEFs to ensure they exhibit characteristic fibroblast morphology (e.g., elongated shape).
2.Before passage, maintain a sterile environment using 70% ethanol to sterilize medium aliquots, dishes, tubes, and serological pipets before placing in biological safety cabinet.
3.Gently aspirate culture medium from MEFs, taking care to aspirate from the side of the flask. Add 1 ml TrypLE Express Enzyme, and gently rock dish to cover cells. Incubate at room temperature for 3 to 5 min.
4.Add 2 ml prewarmed MEF culture medium to the dish, and suspend cells by gently pipetting up and down. Wash dish with the suspension to ensure all cells are collected, and transfer cell suspension to a 15-ml centrifuge tube. Centrifuge cell suspension 3 to 5 min at 500 × g , room temperature.
5.While the cell suspension is in the centrifuge, prepare a fresh maintenance flask and glass-bottom imaging dishes. Label maintenance flask with initials, date, cell line, and passage number. Label imaging dishes with initials, date, cell line, and dish number. Add prewarmed MEF culture medium to each dish.
6.Remove pelleted cells from centrifuge, and remove or aspirate supernatant. Resuspend cell pellet in 5 ml prewarmed MEF culture medium by gently by pipetting up and down. Aliquot cell suspension to the flask (500 µl) and imaging dishes (100 µl). Swirl and rock gently to evenly distribute cells. Return passaged cells to 37°C incubator with 95% humidity and 5% CO2 environment until ready for transfection or passage.
Support Protocol 2: LIVE-CELL IMAGING OF ExRai AMPKAR USING CONFOCAL MICROSCOPY
Imaging of ExRai AMPKAR and other ExRai-based biosensors can be done using multiple modalities, including confocal microscopy (Mehta et al., 2018; Zhang et al., 2020). Confocal microscopy provides better spatial resolution over widefield microscopy through rejection of out-of-focus light and enables imaging of thick specimens (Elliott, 2020). These properties could be advantageous when studying subcellular AMPK activity, making confocal microscopy a beneficial imaging modality. For confocal microscopy, sample preparation is the same, with changes only made to how data are collected. The confocal microscope is programmed with the correct optical setup, and then ROIs are acquired and time series collected similar to in the Basic Protocol. Data analysis is done similar to in the Basic Protocol.
Additional Materials (also see Basic Protocol)
- Inverted confocal microscope with appropriate objective lens, laser lines, filter sets, camera, and image acquisition software:
- Nikon Ti2 spinning-disk confocal microscope equipped with PerfectFocus
- S Fluor 40×/1.3 N.A. oil objective
- Photometrics Prime 95B sCMOS camera
- 405- and 488-nm lasers, 525/50 nm emission filter, 488 dichroic
- Stage-top incubator set at 37°C and 5% CO2 (e.g., Okolab)
Sample preparation
Days 1 and 2
1.Perform Basic Protocol steps 1 to 4 for seeding imaging dishes with MEFs and transfection.
Imaging preparation (30 min)
Day 3
2.Perform Basic Protocol steps 5 and 6 for preparing cells for imaging.
3.While cells incubate in 1× HBSS, prepare microscope for imaging. Turn on microscope, and allow lasers to equilibrate and incubator to reach 37°C.
4.Program a protocol for ExRai AMPKAR imaging. Ensure optical configuration is suitable for imaging ExRai AMPKAR. Set objective to the correct magnification (≥40× for imaging localized ExRai AMPKAR) and imaging interval to 30 s for 30 min. For example:
- 400 nm excitation-emission: 405 nm excitation laser, 488 dichroic, and 525/50 nm emission filter; 200-ms exposure.
- 480 nm excitation-emission: 488 nm excitation laser, 488 dichroic, and 525/50 nm emission filter; 200-ms exposure.
The exposure and imaging interval should be modified for each imaging application and microscope setup. Exposure depends on laser power and camera sensitivity. Some ExRai-based KARs have been imaged using different power for the 488- and 405-nm lasers, with exposure times between 50 and 200 ms (Zhang et al., 2020). For example, laser power across laser lines can be standardized to 1 µW, as others have reported for ratiometric imaging (Yaginuma & Okada, 2021). This ensures the same amount of light is produced by both lasers. Images are set up to be acquired sequentially.
5.Wet objective lens using a drop of immersion oil, and mount imaging dish on the stage. Use 405 nm excitation-emission settings to bring cells into focus and set PerfectFocus.
6.Identify a region of the cell with at least three transfected cells with good cell morphology and correct localization of ExRai AMPKAR.
Data and image acquisition (30 min per dish)
7.Acquire an image of the selected field of view, and select ROIs to monitor during imaging.
8.Begin experiment. Collect a time series of the field of view for each channel for ∼5 min.
9.Pause acquisition to add AMPK activators to the dish. Remove ∼500 µl imaging medium, and add to a microcentrifuge tube containing aliquoted stock solution of either 10 to 40 μl 2-DG (10 to 40 mM final) or 10μl MK-8722 (500 nM final). Gently pipet up and down to mix. Add dilution to the imaging dish dropwise around the edges of the dish. Once added, mix by gently pipetting four to five times, being careful to avoid directly pipetting on cells being imaged.
10.Resume imaging for ∼25 min or until ExRai AMPKAR response reaches a maximum and does not continue to increase.
11.Repeat steps 5 to 10 until all dishes have been imaged. Upon completion of data acquisition, export data for analysis.
Image analysis (15 min per dish)
12.Perform Basic Protocol steps 14 to 19.
REAGENTS AND SOLUTIONS
HBSS imaging buffer
- 1× HBSS (e.g., Fisher Scientific, cat. no. 14065056)
- 20 mM HEPES (e.g., Fisher Scientific, cat. no. SH3023701)
- 2.0 g/L D-glucose (e.g., VWR, cat. no. BT132735-500G)
- Mix well and adjust pH to 7.4 with NaOH (e.g., Fisher Scientific, cat. no. S318-500)
- Filter sterilize using a 0.22-µm membrane filter (e.g., Fisher Scientific, cat. no. 09761112)
- Store at 4°C for up to 6 months protected from light
- Warm 50-ml aliquots to 37°C before using for imaging
- Prepare and store under sterile environment.
Prepare 1× HBSS from 10× HBSS and 20 mM HEPES from 1 M HEPES using water with 18.2-MΩ-cm resistivity under sterile conditions.
MEF culture medium
- Dulbecco's modified Eagle medium (DMEM) containing 4.5 g/L glucose, pyruvate, and L-glutamine (e.g., Gibco, cat. no. 11995-065)
- 10% (v/v) fetal bovine serum (e.g., Gibco, cat. no. 26140-079)
- 1% (v/v) penicillin-streptomycin (e.g., Gibco, cat. no. 15140-122)
- Mix well and prepare 50-ml aliquots
- Store at 4°C for up to 3 months protected from light in a sterile environment
- Prewarm to 37°C before using for cell culture
Prepare and store under sterile environment.
COMMENTARY
Background Information
AMPK is a central regulator of cellular metabolism, involved in maintaining cellular energy homeostasis. AMPK has a diversity of targets throughout the cell, and it is only more recently being understood how subcellular AMPK activities are regulated and impact cell function. For example, lysosomal pools of AMPK have been found to be activated in response to mild stress, whereas more severe stress was required for activation of other pools of AMPK (Zong et al., 2019). Pools of AMPK at the mitochondria respond to localized energetic stress and are involved in mitochondrial quality control (Drake et al., 2021). Although these studies have uncovered the compartmentalized nature of AMPK activity, these findings relied on bulk techniques, which are both spatially and temporally limited and can obscure heterogeneous signaling events in a cell population.
To study compartmentalized AMPK activity in real time with high spatiotemporal resolution, AMPKARs have been developed. The first AMPKAR was based on FRET and enabled visualization of AMPK activity in real time in single living cells (Tsou et al., 2011). However, this FRET-based AMPKAR exhibited limited dynamic range and was not suitable for studying subcellular AMPK activity. Subsequent generations of FRET-based AMPKARs improved upon the dynamic range by introducing flexible linkers and using enhanced fluorescent proteins (Hung et al., 2017; Konagaya et al., 2017; Sample et al., 2015). These improved AMPKARs supported the study of AMPK activity in heterogeneous cell populations and for intravital imaging (Konagaya et al., 2017; Kosaisawe et al., 2021). Introduction of targeting motifs subsequently allowed for study of subcellular AMPK activity (Miyamoto et al., 2015). Bimolecular AMPKARs were developed to improve dynamic range of AMPK activity sensing at the plasma membrane (Depry et al., 2015). Despite these advances, FRET-based AMPKARs have a couple disadvantages. First, FRET-based biosensors suffer from poor dynamic range due to only small changes in FRET being measured, despite efforts made to improve FRET-based reporters (Bajar et al., 2016; Komatsu et al., 2011; Lam et al., 2012). Subtle changes in cellular signaling or biochemical activity might be missed using FRET-based reporters. For example, using FRET-based AMPKAR, nuclear AMPK activity could not be observed, notwithstanding improvements made to the dynamic range of the biosensor (Miyamoto et al., 2015; Sample et al., 2015; Tsou et al., 2011). Second, FRET-based reporters use two fluorescent proteins, resulting in limited availability for multiplexing. As a result observation of multiple cellular activities at once cannot be easily accomplished with FRET-based KARs (Piljic & Schultz, 2008).
To overcome these disadvantages, single-fluorophore ExRai KARs have been developed (Chen et al., 2021; Mehta et al., 2018; Zhang et al., 2020). ExRai KARs exhibit significantly enhanced dynamic range and use only one fluorescent protein, making these reporters advantageous for multiplexed experiments. I found ExRai AMPKAR to have a significantly enhanced dynamic range, signal-to-noise ratio, and assay fitness score over FRET-based AMPKAR (Schmitt et al., 2022). Importantly, through inclusion of localization sequences, ExRai AMPKAR can detect subtle changes in local AMPK signaling. For instance, ExRai AMPKAR could be used to detect nuclear AMPK activity, which was not possible using FRET-based reporters. I subsequently used ExRai AMPKAR to uncover a translocation-based mechanism for nuclear AMPK activity. Although not shown with ExRai AMPKAR, other ExRai KARs have been successfully used in multiplexed experiments, demonstrating the use of these single fluorescent protein–based biosensors for observing multiple biochemical activities in a single cell (Mehta et al., 2018; Zhang et al., 2020). Therefore, the high dynamic range and good assay fitness of ExRai AMPKAR finally enabled the detection of subtle AMPK signaling events, which were missed using other techniques.
Critical Parameters
Imaging conditions
In this protocol, cells are imaged in 1× HBSS and incubated for 30 min in 1× HBSS before imaging. I observed an unstable, drifting baseline reading of ExRai AMPKAR when imaged immediately after medium exchange. Allowing cells to adjust for 30 min from full medium to 1× HBSS imaging buffer ensures the cells have recovered from any stress caused by the medium exchange and results in a stable baseline reading. This 30-min incubation period and subsequent imaging should be done at 37°C to avoid unnecessary cell stress. Some more sensitive cell lines may require imaging in a more complete medium, like FluoroBrite DMEM-based imaging buffer.
Selection of cells
The selection of cells to image is critical for the success of any live-cell imaging experiment. Within a single field of view, cells will vary in ExRai AMPKAR expression and cell health. For reproducible results, cells that appear healthy and have good ExRai AMPKAR expression should be selected. For MEFs, cells should appear fibroblast like and not rounded up. Cells should have a fluorescence intensity above background. Selection of very dim cells will result in data with poor signal-to-noise ratio. Localized reporters should appear correctly localized to the desired subcellular location. When first performing the experiment, make sure basal measurements of ExRai AMPKAR are steady and do not significantly drift >5% of initial value. This unstable baseline reading could confound interpretation of collected data, or cells will appear to not respond to stimulation.
Use of control experiments and validation
Control experiments should be carefully designed and performed alongside experimental trials. These control experiments validate ExRai AMPKAR results and ensure proper data interpretation. Control experiments to consider are vehicle controls, the use of AMPK KO cell lines, pharmacologic inhibitors of AMPK, and ExRai AMPKAR T/A (Schmitt et al., 2022). When testing new AMPK-activating or inhibiting conditions, western blotting for phosphorylated AMPKα1/2 and/or a validated downstream target of AMPK like acetyl Co-A carboxylase can be done to ensure the experimental conditions activate or inhibit AMPK activity.
Troubleshooting
Common problems with imaging ExRai AMPKAR, possible causes, and solutions can be found in Table 2.
| Problem | Possible cause | Potential solution |
|---|---|---|
| Dim cells or minimal transfection | Poor transfection | Optimize transfection procedure for each cell line considering DNA amount, DNA:transfection reagent ratio, type of transfection reagent, and transfection medium, using Opti-MEM instead of full growth medium |
| Optimize imaging time post-transfection (18-48 hr) | ||
| Consider viral transduction (see Strategic Planning) | ||
| Unstable baseline | Incorrect filter set | Verify correct selection of microscope filters |
| No equilibration time in imaging buffer | Allow cells to equilibrate in imaging buffer for ∼30 min before imaging, or image in more complete medium (see Critical Parameters) | |
| Dramatic changes in cell morphology | Optimize imaging conditions (image in medium like FluoroBrite DMEM-based buffer) | |
| Photobleaching | Reduce exposure time, and decrease lamp intensity | |
| Minimal responsea or slow response | Incorrect drug addition | Ensure medium is mixed well when drug is added; ensure consistency in adding drug between trials or use a perfusion system |
| Incorrect drug concentration | Optimize concentration of drug to be used (i.e., use a higher or lower dose) | |
| Validate concentrations used successfully activate AMPK via western blotting for phosphorylated AMPK and downstream targets | ||
| Check solubility of drugs used | ||
| Low expression of reporter | Optimize transfection of the biosensor (see above) | |
| Mislocalized biosensor | Extreme overexpression of reporter | Optimize transfection of biosensor, focusing on decreasing amount of DNA used; optimize time of imaging post-transfection |
| Localization tag selected inappropriate for cell line | Change targeting sequence on biosensor, selecting targeting sequences previously shown to localize correctly in selected cell line |
- AMPK, adenosine monophosphate–activated protein kinase; DMEM, Dulbecco's modified Eagle medium.
- a Ratio change <10% relative to baseline.
Understanding Results
Effective imaging of subcellular AMPK activity using ExRai AMPKAR should result in the 400 nm excitation-emission decreasing and 480 nm excitation-emission increasing upon stimulation of AMPK with either 2-DG or MK-8722. The ratio (480/400) will increase when AMPK is activated and decrease when AMPK is deactivated. Control experiments could include vehicle (DPBS or DMSO) or ExRai AMPKAR T/A. These controls should exhibit minimal change in excitation-emission from either channel.
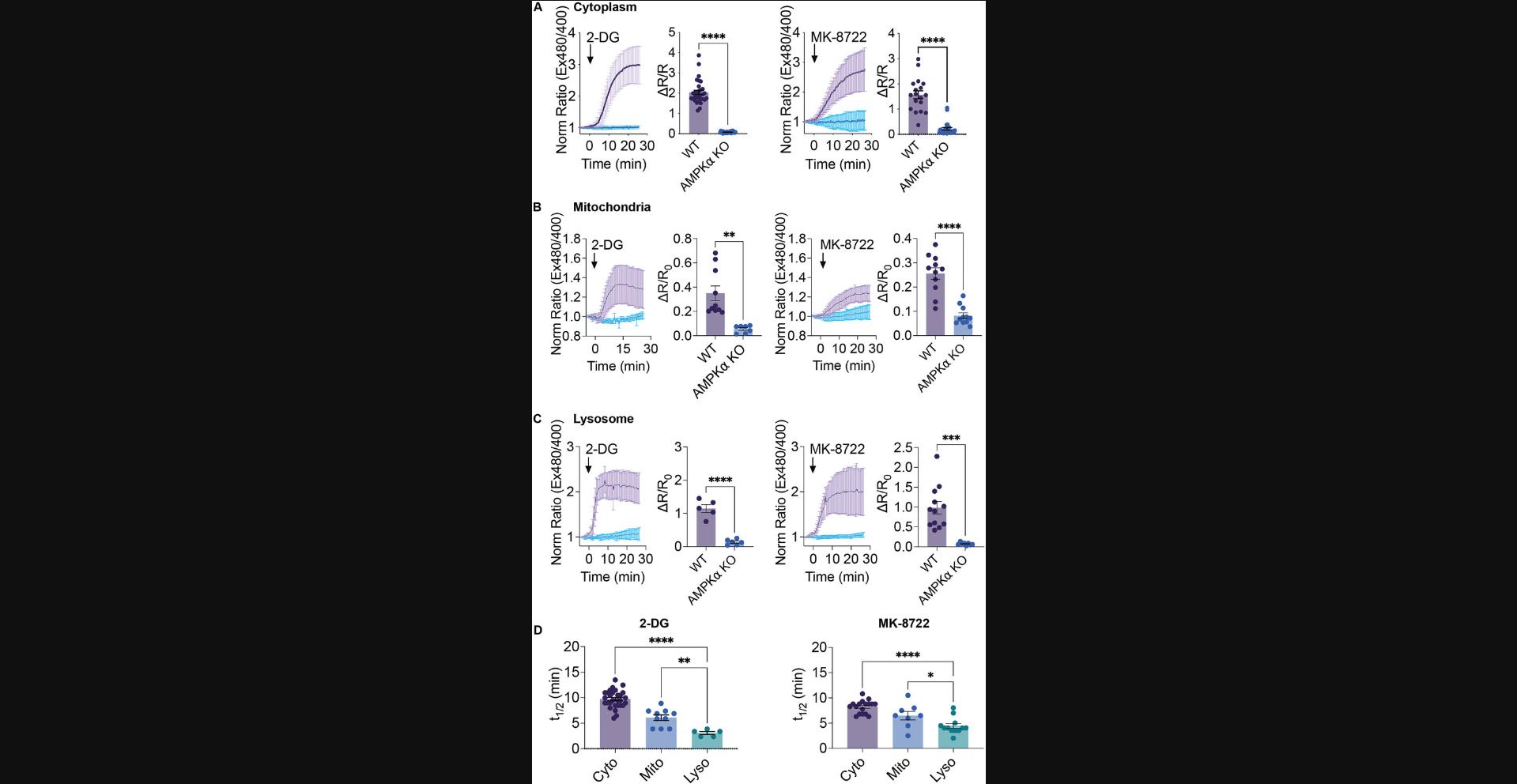
I suggest imaging at least two technical replicates per experiment per day and independently replicating the experiment at least three times using different cell preparations. Following image analysis and generation of single-cell ExRai AMPKAR responses (see Basic Protocol steps 14 to 20), data can be pooled and averaged across cells, standard deviation for each time point determined, and results plotted over time (Fig. 2). The pooled and single-cell data should be carefully considered, and any trends seen in the data should be noted. For example, extreme heterogeneity in response, biphasic changes, or other changes in response should be noted. The ∆R/R0 can also be plotted and mean and standard error of the mean determined. From these data, change in AMPK activity over time and magnitude of change in AMPK activity can be determined. The equation ∆R/R0 can be used to compare AMPK activity across conditions, including comparing treatment conditions to control, using an unpaired Student's t -test or Welch's unequal variance t -test for data where sample sizes and variance are not equal between data sets. When comparing more than three data sets, use a one-way ANOVA followed by an appropriate multiple comparisons test (e.g., Dunnett's multiple comparison test or Tukey's multiple comparison test). Since subcellular targeting can impact biosensor dynamic range, I do not recommend comparison of the maximum ratio change across locations (Schmitt et al., 2022). In Figure 2, I compare levels of activity of AMPK induced by treatment with either 2-DG or MK-8722 at each location to control.
To compare response kinetics across locations, t1/2 can be used. Mean t1/2 can be determined, as can standard error of the mean. The same statistical tests as those described above should be used. As shown in Figure 2D, comparisons of t1/2 across locations reveals significant spatial differences in the induction of AMPK activity by both 2-DG and MK-8722. I find lysosomal activity is induced first, followed by the mitochondria and cytoplasm.
Time Considerations
Imaging experiments in general take up to 1 week to complete, including time to prepare cells. Before sample preparation, MEFs take 2 to 3 days to reach ∼90% confluency for passage (see Support Protocol 1). On day 1, cells are plated on imaging dishes, which takes ∼30 min. On day 2, cells are transfected, which takes ∼30 min. On day 3, cells are prepared for imaging (∼30 min), imaging is performed (∼30 min), and data are analyzed (∼15 min), which takes ∼1.5 hr. The timing for each step has also been included throughout the protocols.
Acknowledgments
This work was supported by the University of California Los Angeles. I thank Sohum Mehta for his helpful discussion and support.
Author Contribution
Danielle L. Schmitt : conceptualization, data curation, resources, writing–original draft, writing–review and editing.
Conflict of Interest
The author declares no conflicts of interest.
Open Research
Data Availability Statement
The data, tools, and material (or their source) that support the protocol are available from the corresponding author upon reasonable request.
Literature Cited
- Bajar, B. T., Wang, E. S., Lam, A. J., Kim, B. B., Jacobs, C. L., Howe, E. S., Davidson, M. W., Lin, M. Z., & Chu, J. (2016). Improving brightness and photostability of green and red fluorescent proteins for live cell imaging and FRET reporting. Scientific Reports , 6, 1–12. https://doi.org/10.1038/srep20889
- Chen, M., Sun, T., Zhong, Y., Zhou, X., & Zhang, J. (2021). A highly sensitive fluorescent Akt biosensor reveals lysosome-selective regulation of lipid second messengers and kinase activity. ACS Central Science , 7(12), 2009–2020. https://doi.org/10.1021/acscentsci.1c00919
- Depry, C., Mehta, S., Li, R., & Zhang, J. (2015). Visualization of compartmentalized kinase activity dynamics using adaptable BimKARs. Chemistry & Biology, 22(11), 1470–1479. https://doi.org/10.1016/j.chembiol.2015.10.004
- Dite, T. A., Langendorf, C. G., Hoque, A., Galic, S., Rebello, R. J., Ovens, A. J., Lindqvist, L. M., Ngoei, K. R. W., Ling, N. X. Y., Furic, L., Kemp, B. E., Scott, J. W., & Oakhill, J. S. (2018). AMP-activated protein kinase selectively inhibited by the type II inhibitor SBI-0206965. Journal of Biological Chemistry , 293(23), 8874–8885. https://doi.org/10.1074/jbc.ra118.003547
- Drake, J. C., Wilson, R. J., Laker, R. C., Guan, Y., Spaulding, H. R., Nichenko, A. S., Shen, W., Shang, H., Dorn, M. V., Huang, K., Zhang, M., Bandara, A. B., Brisendine, M. H., Kashatus, J. A., Sharma, P. R., Young, A., Gautam, J., Cao, R., Wallrabe, H., … Yan, Z. (2021). Mitochondria-localized AMPK responds to local energetics and contributes to exercise and energetic stress-induced mitophagy. Proceedings of the National Academy of Sciences of the United States of America , 118(37), e2025932118. https://doi.org/10.1073/pnas.2025932118
- Egan, D. F., Shackelford, D. B., Mihaylova, M. M., Gelino, S., Kohnz, R. A., Mair, W., Vasquez, D. S., Joshi, A., Gwinn, D. M., Taylor, R., Asara, J. M., Fitzpatrick, J., Dillin, A., Viollet, B., Kundu, M., Hansen, M., & Shaw, R. J. (2011). Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science , 331(6016), 456–461. https://doi.org/10.1126/science.1196371
- Elliott, A. D. (2020). Confocal microscopy: Principles and modern practices. Current Protocols in Cytometry , 92, e68. https://doi.org/10.1002/cpcy.68
- Feng, D., Biftu, T., Romero, F. A., Kekec, A., Dropinski, J., Kassick, A., Xu, S., Kurtz, M. M., Gollapudi, A., Shao, Q., Yang, X., Lu, K., Zhou, G., Kemp, D., Myers, R. W., Guan, H.-P., Trujillo, M. E., Li, C., Weber, A., & Sebhat, I. K. (2017). Discovery of MK-8722: A systemic, direct pan-activator of AMP-activated protein kinase. ACS Medicinal Chemistry Letters , 9(1), 39–44. https://doi.org/10.1021/acsmedchemlett.7b00417
- Garcia, D., & Shaw, R. J. (2017). AMPK: Mechanisms of cellular energy sensing and restoration of metabolic balance. Molecular Cell , 66(6), 789–800. https://doi.org/10.1016/j.molcel.2017.05.032
- Hardie, D. G. (2022). AMP-activated protein kinase — a journey from 1 to 100 downstream targets. Biochemical Journal , 479(22), 2327–2343. https://doi.org/10.1042/bcj20220255
- Hung, Y. P., Teragawa, C., Kosaisawe, N., Gillies, T. E., Pargett, M., Minguet, M., Distor, K., Rocha-Gregg, B. L., Coloff, J. L., Keibler, M. A., Stephanopoulos, G., Yellen, G., Brugge, J. S., & Albeck, J. G. (2017). Akt regulation of glycolysis mediates bioenergetic stability in epithelial cells. eLife , 6, 598. https://doi.org/10.7554/elife.27293
- Komatsu, N., Aoki, K., Yamada, M., Yukinaga, H., Fujita, Y., Kamioka, Y., & Matsuda, M. (2011). Development of an optimized backbone of FRET biosensors for kinases and GTPases. Molecular Biology of the Cell , 22(23), 4647–4656. https://doi.org/10.1091/mbc.e11-01-0072
- Konagaya, Y., Terai, K., Hirao, Y., Takakura, K., Imajo, M., Kamioka, Y., Sasaoka, N., Kakizuka, A., Sumiyama, K., Asano, T., & Matsuda, M. (2017). A highly sensitive FRET biosensor for AMPK exhibits heterogeneous AMPK responses among cells and organs. Cell Reports , 21(9), 2628–2638. https://doi.org/10.1016/j.celrep.2017.10.113
- Kosaisawe, N., Sparta, B., Pargett, M., Teragawa, C. K., & Albeck, J. G. (2021). Transient phases of OXPHOS inhibitor resistance reveal underlying metabolic heterogeneity in single cells. Cell Metabolism , 33(3), 649–665.e8. https://doi.org/10.1016/j.cmet.2021.01.014
- Lam, A. J., St-Pierre, F., Gong, Y., Marshall, J. D., Cranfill, P. J., Baird, M. A., McKeown, M. R., Wiedenmann, J., Davidson, M. W., Schnitzer, M. J., Tsien, R. Y., & Lin, M. Z. (2012). Improving FRET dynamic range with bright green and red fluorescent proteins. Nature Methods , 9(10), 1005–1012. https://doi.org/10.1038/nmeth.2171
- Mehta, S., Zhang, Y., Roth, R. H., Zhang, J., Mo, A., Tenner, B., Huganir, R. L., & Zhang, J. (2018). Single-fluorophore biosensors for sensitive and multiplexed detection of signalling activities. Nature Cell Biology , 20(10), 1215–1225. https://doi.org/10.1038/s41556-018-0200-6
- Miyamoto, T., Rho, E., Sample, V., Akano, H., Magari, M., Ueno, T., Gorshkov, K., Chen, M., Tokumitsu, H., Zhang, J., & Inoue, T. (2015). Compartmentalized AMPK signaling illuminated by genetically encoded molecular sensors and actuators. Cell Reports , 11(4), 657–670. https://doi.org/10.1016/j.celrep.2015.03.057
- Piljic, A., & Schultz, C. (2008). Simultaneous recording of multiple cellular events by FRET. ACS Chemical Biology , 3(3), 156–160. https://doi.org/10.1021/cb700247q
- Sample, V., Ramamurthy, S., Gorshkov, K., Ronnett, G. V., & Zhang, J. (2015). Polarized activities of AMPK and BRSK in primary hippocampal neurons. Molecular Biology of the Cell , 26(10), 1935–1946. https://doi.org/10.1091/mbc.e14-02-0764
- Schmitt, D. L., Curtis, S. D., Lyons, A. C., Zhang, J., Chen, M., He, C. Y., Mehta, S., Shaw, R. J., & Zhang, J. (2022). Spatial regulation of AMPK signaling revealed by a sensitive kinase activity reporter. Nature Communications , 13(1), 3856. https://doi.org/10.1038/s41467-022-31190-x
- Schmitt, D. L., Mehta, S., & Zhang, J. (2020). Illuminating the kinome: Visualizing real-time kinase activity in biological systems using genetically encoded fluorescent protein-based biosensors. Current Opinion in Chemical Biology , 54, 63–69. https://doi.org/10.1016/j.cbpa.2019.11.005
- Shaw, R. J., Kosmatka, M., Bardeesy, N., Hurley, R. L., Witters, L. A., DePinho, R. A., & Cantley, L. C. (2004). The tumor suppressor LKB1 kinase directly activates AMP-activated kinase and regulates apoptosis in response to energy stress. Proceedings of the National Academy of Sciences of the United States of America , 101(10), 3329–3335. https://doi.org/10.1073/pnas.0308061100
- Stein, B. D., Calzolari, D., Hellberg, K., Hu, Y. S., He, L., Hung, C.-M., Toyama, E. Q., Ross, D. S., Lillemeier, B. F., Cantley, L. C., Yates, J. R., & Shaw, R. J. (2019). Quantitative in vivo proteomics of metformin response in liver reveals AMPK-dependent and -independent signaling networks. Cell Reports , 29(10), 3331–3348.e7. https://doi.org/10.1016/j.celrep.2019.10.117
- Toyama, E. Q., Herzig, S., Courchet, J., Lewis, T. L., Loson, O. C., Hellberg, K., Young, N. P., Chen, H., Polleux, F., Chan, D. C., & Shaw, R. J. (2016). AMP-activated protein kinase mediates mitochondrial fission in response to energy stress. Science , 351(6270), 275–281. https://doi.org/10.1126/science.aab4138
- Tsou, P., Zheng, B., Hsu, C.-H., Sasaki, A. T., & Cantley, L. C. (2011). A fluorescent reporter of AMPK activity and cellular energy stress. Cell Metabolism , 13(4), 476–486. https://doi.org/10.1016/j.cmet.2011.03.006
- Yaginuma, H., & Okada, Y. (2021). Live cell imaging of metabolic heterogeneity by quantitative fluorescent ATP indicator protein, QUEEN-37C. BioRxiv , [preprint]. https://doi.org/10.1101/2021.10.08.463131
- Zhang, J., Liu, B., Hong, I., Mo, A., Roth, R. H., Tenner, B., Lin, W., Zhang, J. Z., Molina, R. S., Drobizhev, M., Hughes, T. E., Tian, L., Huganir, R. L., Mehta, S., & Zhang, J. (2020). An ultrasensitive biosensor for high-resolution kinase activity imaging in awake mice. Nature Chemical Biology , 118, 11707–11724. https://doi.org/10.1038/s41589-020-00660-y
- Zong, Y., Zhang, C.-S., Li, M., Wang, W., Wang, Z., Hawley, S. A., Ma, T., Feng, J.-W., Tian, X., Qi, Q., Wu, Y.-Q., Zhang, C., Ye, Z., Lin, S.- Y., Piao, H.-L., Hardie, D. G., & Lin, S.-C. (2019). Hierarchical activation of compartmentalized pools of AMPK depends on severity of nutrient or energy stress. Cell Research , 29(6), 460–473. https://doi.org/10.1038/s41422-019-0163-6
Key References
- Schmitt et al. (2022). See above.
Describes the development and application of ExRai AMPKAR.
Internet Resources
MatScope Suite: open-sources MATLAB-based imaging program that interfaces with µ-Manager for automated image acquisition and that was used to collect and analyze the data in this protocol.

