Identifying Cell-Type-Specific Metabolic Signatures Using Transcriptome and Proteome Analyses
Nadja Gebert, Nadja Gebert, Shahadat Rahman, Shahadat Rahman, Caroline A. Lewis, Caroline A. Lewis, Alessandro Ori, Alessandro Ori, Chia-Wei Cheng, Chia-Wei Cheng
Abstract
Studies in various tissues have revealed a central role of metabolic pathways in regulating adult stem cell function in tissue regeneration and tumor initiation. The unique metabolic dependences or preferences of adult stem cells, therefore, are emerging as a new category of therapeutic target. Recently, advanced methods including high-resolution metabolomics, proteomics, and transcriptomics have been developed to address the growing interest in stem cell metabolism. A practical framework integrating the omics analyses is needed to systematically perform metabolic characterization in a cell-type-specific manner. Here, we leverage recent advances in transcriptomics and proteomics research to identify cell-type-specific metabolic features by reconstructing cell identity using genes and the encoded enzymes involved in major metabolic pathways. We provide protocols for cell isolation, transcriptome and proteome analyses, and metabolite profiling and measurement. The workflow for mapping cell-type-specific metabolic signatures presented here, although initially developed for intestinal crypt cells, can be easily implemented for cell populations in other tissues, and is highly compatible with most public datasets. © 2021 The Authors. Current Protocols published by Wiley Periodicals LLC.
This article was corrected on 4 August 2022. See the end of the full text for details.
Basic Protocol 1 : Intestinal crypt isolation and cell population purification
Basic Protocol 2 : Transcriptome analyses for cell-type-specific metabolic gene expression
Basic Protocol 3 : Proteome analyses for cell-type-specific metabolic enzyme levels
Basic Protocol 4 : Metabolite profiling and measurement
INTRODUCTION
The capacity to adjust metabolic pathways in response to nutrient availability and energy status is pivotal for cell survival and proliferation. It is generally assumed that unless they are genetically aberrant, normal cells are equipped with common metabolic machinery (i.e., transporters and enzymes) to cope with the changing nutritional environment. Cell metabolism, therefore, is often described as the metabolic reactions in one generic cell, without a specified cell origin or identity.
Recent studies in stem cell biology suggest a different picture, in which adult stem cells are metabolically different from their differentiated daughter cells in the same tissue (Shapira & Christofk, 2020). In the hematopoietic system, metabolic differences between stem cells and multipotent progenitors can be identified by means of the 160 metabolites detected in 10,000 flow-sorted cells, using methods involving hydrophilic liquid interaction chromatography and high-sensitivity Orbitrap mass spectrometry (DeVilbiss et al., 2021). However, it remains difficult to systematically compare metabolite levels between stem cells and other rare cell populations in solid tissues, such as intestine, mostly due to the additional dissociation process that is required prior to the flow cytometry−based cell purification.
In the small intestine, stem cells residing in the crypts of Lieberkühn play a critical role in epithelial tissue renewal, homeostasis, and cancer development (Calibasi-Kocal et al., 2021). The balance between self-renewal and differentiation of intestinal stem cells (ISCs) is critically regulated by metabolic pathways. Several metabolic enzymes and metabolite transporters that mediate ISC glucose metabolism have been identified, including lactate dehydrogenase (LDH), which converts glucose-derived pyruvate into lactate in the niche Paneth cells (Rodriguez-Colman et al., 2017), mitochondrial pyruvate carrier 1 (MPC1), which transports pyruvate into mitochondria in ISCs (Schell et al., 2017), and pyruvate dehydrogenase (PDH), which converts pyruvate into acetyl-CoA, thereby increasing the influx of acetyl-CoA from glycolysis into the TCA cycle (Bensard et al., 2020). These metabolic enzymes act separately in stem cells and niche cells while coordinately regulating glycolytic and oxidative phosphorylation (OxPhos) fluxes that critically influence the proliferation and tumorigenic potential of ISCs (Wang, Odle, & Liu, 2021a), highlighting a significant regulatory role for the metabolic differences between cell types.
We have recently determined that enzymes mediating lipid metabolism may function as metabolic cell-fate determinants. Specifically, we found that ISC expressed high levels of the ketogenic enzyme Hmgcs2 (3-hydroxy-3-methylglutaryl-CoA synthase 2) to produce the ketone body β-hydroxybutyrate, derived from acetyl CoA produced by fatty acid oxidation (FAO). This metabolic feature of ISCs is required for ISC self-renewal, lineage decision, and adaptation to diet. Here, we will describe the pipeline for experimental design and analysis that we developed to identify the cell-type-specific metabolic signatures using transcriptome (Cheng et al., 2019; Haber et al., 2017) and proteome analyses (Gebert et al., 2020; Fig. 1). We will also describe how we use liquid chromatography–mass spectrometry (LC–MS) and target-specific assays to validate the individual metabolites.
NOTE : All protocols involving animals must be reviewed and approved by the appropriate Animal Care and Use Committee and must follow regulations for the care and use of laboratory animals.
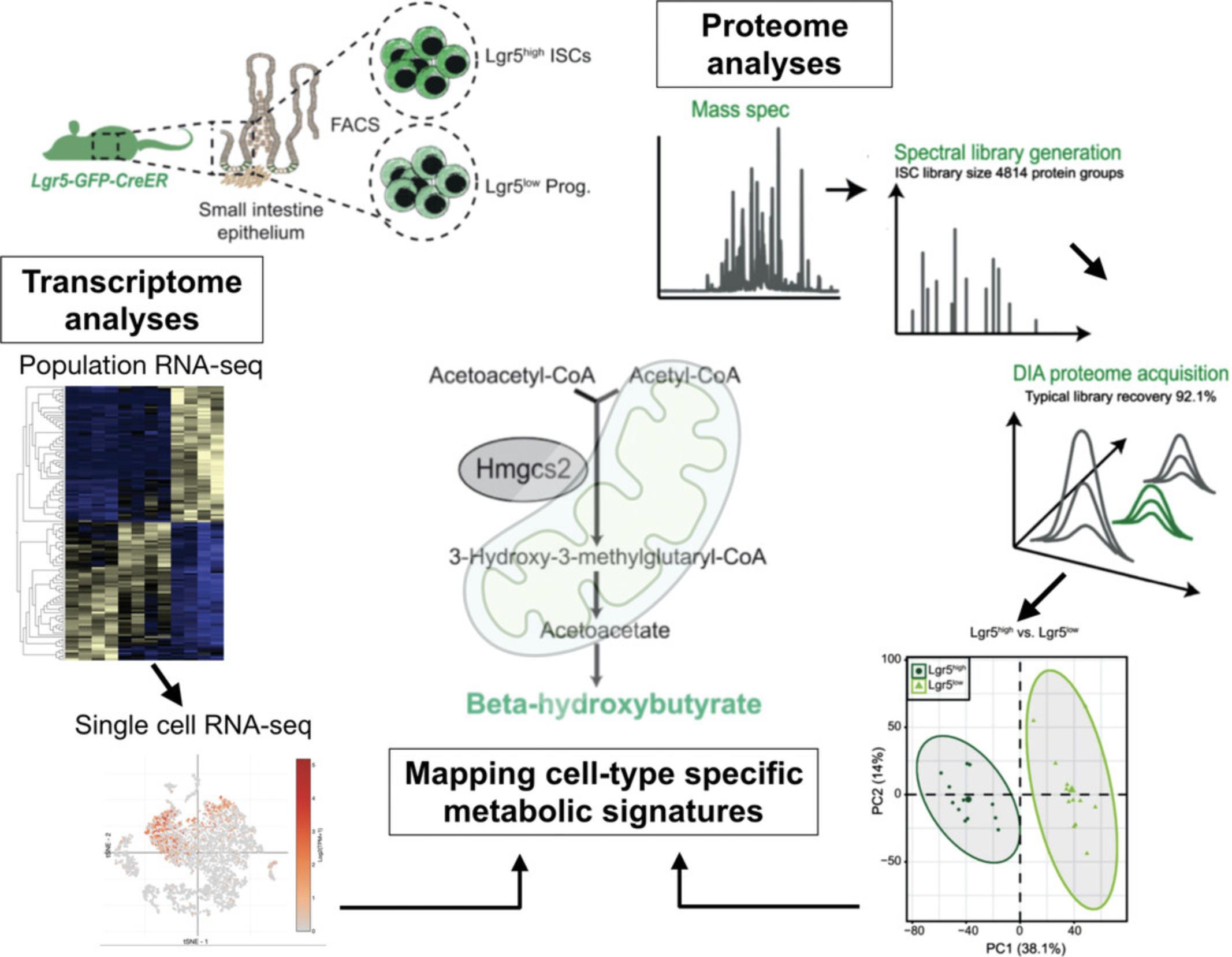
Basic Protocol 1: INTESTINAL CRYPT ISOLATION AND CELL POPULATION PURIFICATION
Cell populations at the bottoms of intestinal crypts serve as an ideal model for studying the inherent metabolic features of cells. They share the same nutritional environment while displaying distinct metabolic features (Baulies, Angelis, & Li, 2020; Urbauer, Rath, & Haller, 2020; Wang, Odle, & Liu, 2021b). Three major cell types enriched in the intestinal crypts are self-renewing ISCs, transient amplifying (TA) progenitor cells, and terminally differentiated niche Paneth cells. To enhance the efficiency of purification, crypt isolation is a critical step prior to flow-cytometry-based cell sorting. Using a GFP reporter for Lgr5 (leucine-rich-repeat-containing G-protein-coupled receptor 5, also known as Gpr49), a marker gene exclusively expressed in cycling columnar ISCs (Barker et al., 2007), the stem cells and progenitor cells can be distinguished based on GFP intensity. In combination with surface proteins such as c-kit and CD24 that mark secretory Paneth cells, one can purify the three basic crypt cell populations with one FACS panel: Lgr5-GFPhigh ISCs, Lgr5-GFPlow progenitors, and c-kit+CD24+ Paneth cells (Cheng, Yilmaz, & Mihaylova, 2020).
Materials
-
Mouse (adult, older than 8 weeks)
-
70% ethanol in spray bottle
-
500 ml phosphate-buffered saline (PBS; PBS Tablets; Gibco, cat. no. 18912014, to be dissolved in 500 ml deionized water; keep on ice)
-
0.5 M disodium EDTA (Promega, cat. no. V4231)
-
Tweezers
-
Dissecting scissors
-
Oral gavage needle on 20-ml syringe
-
Ice bath
-
Large petri dish (at least 22 mm)
-
50-ml conical tubes (e.g., Corning Falcon)
-
50 ml conical tubes
-
Large beaker
-
Nutating mixer/rocker
-
70-µm mesh
-
Additional reagents and equipment for euthanasia of mice (see Current Protocols article: Donovan & Brown, 2006)
1.Euthanize mouse by cervical dislocation (see Current Protocols article: Donovan & Brown, 2006).
2.Place on large lab towel and spray abdomen of mouse with 70% ethanol.
3.Beginning at the base of the abdomen, cut open peritoneal cavity and locate stomach, usually beneath the liver.
4.Clip beginning of small intestine adjacent to stomach and remove it from the mouse by pulling until you reach the caecum and colon, readjusting grip every 3-5 cm. Be sure to remove any fat or blood vessels attached to the intestine.
5.Place small intestine in petri dish containing ice-cold PBS immediately.
6.Keep ice-cold PBS in a separate 50-ml tube to draw from, and use an oral gavage needle to flush intestine with cold PBS twice into large beaker. First flush from the proximal end (usually lighter in color), then from the distal end.
7.Open up the intestine (split lengthwise so that it is flat rather than tubular) using the syringe by inserting the gavage needle into the intestine and gently pulling down the intestine perpendicular to the tip of the needle, using the needle to cut the intestine.
8.Lay the tissue flat in a petri dish filled with PBS to clean. Gently wipe off excess mucus and lumen contents.
9.Cut the tissue into 3-5 fragments.
10.Place the tissue into a conical tube with 50 ml cold PBS.
11.Add 1 ml of 0.5 M disodium EDTA to tube with 50 ml PBS and intestine (10 mM EDTA final).
12.Incubate in cold room on nutating mixer/rocker for ∼45 min.
13.Invert tube 3-5 times and wash by pouring out fluid and refilling the tube with ice-cold PBS to 30 ml.
14.Shake vigorously in an elliptical motion for 3-5 min until crypts detach from the intestine.
15.To remove villi from solution, filter the 30 ml of collected crypts through 70-µm mesh into another clean 50-ml tube.
16.Add another 30 ml PBS to the tube containing the intestine.
17.Repeat steps 13 and 15.Collect up to 6 fractions (∼150 ml).
18.Spin down the collected crypts 5 min at 300 × g , 4°C.
19.Remove the PBS and re-suspend the pellet (crypts) with reagents for downstream applications.
Basic Protocol 2: TRANSCRIPTOME ANALYSES FOR CELL-TYPE-SPECIFIC METABOLIC GENE EXPRESSION
A fundamental question of stem cell biology is what intrinsic features bestow stem cells with unique properties. Transcriptome analysis, which offers the capability to simultaneously discover new transcripts and to quantify gene expression, has been a significant step toward answering this question. Accompanied by advances in technologies that enable the quantification of gene expression in rare cell populations, new computational analyses have been developed to further grant access to the more holistically described cell states and identities (Morris, 2019). In this protocol, we will explain how we re-define crypt cell identity with cell-type-specific metabolic gene expression patterns using population RNA-seq and single-cell RNA-seq data generated from murine intestinal crypt cells.
Materials
-
Intestinal crypt pellets (see Basic Protocol 1, step 19)
-
TrypLE (Life Technologies 12604021)
-
S-MEM (Life Technologies 11380-037)
-
0.4% BSA: 200 mg of BSA (Sigma-Aldrich A4161) in 50 ml of PBS
-
15-ml centrifuge tubes (e.g., Corning Falcon)
-
Low-binding 1.5-ml microcentrifuge tubes
-
Centrifuge with temperature control (Eppendorf Microcentrifuge 5430 R)
-
FACSAria3 flow sorter: 4-laser, 11-color sorter running BDFACS Diva software on XP
-
Chromium Single Cell Platform (10X Genomics; Cheng et al., 2019)
-
Qlucore Omics Explorer Version 3.7(21)
-
R 3.4.0
-
DESeq2
-
Seurat v3 (Macosko et al., 2015)
-
dittoSeq package
-
Additional reagents and equipment for antibody staining and FACS analysis (Cheng et al. 2020)
Prepare for fluorescence-activated cell sorting (FACS) for RNA sequencing
1.To prepare samples for cell sorting by flow cytometry, resuspend intestinal crypt pellets isolated from step 19 of Basic Protocol 1 in 500 μl TrypLE per sample, and transfer to individual 15-ml Falcon centrifuge tubes.
2.Dissociate crypts by incubating with TrypLE for 1 min at 32°C.
3.To stop the dissociation, add 10 ml ice-cold S-MEM and invert the tube twice to wash the cell pellets.
4.Centrifuge 5 min at 300 × g , 4°C.
5.Remove supernatant and process by antibody staining and FACS analysis as previously described (Cheng et al., 2020).
6.Sort cells by FACS.
-
Using the FACSAria3 cell sorter for bulk RNA-seq, sort 25,000 live cells (7AAD−) per population (i.e., Epcam+/CD24-/GFPhighLgr5+ ISCs, Epcam+/CD24-/GFPlowprogenitor cells, and Epcam+/GFP-/CD24+/c-Kit+/SSChighPaneth cells) into the assigned collection tubes (low-binding tubes containing 400 μl TRIzol™Reagent).
-
Following the completion of each sort, vortex and place the collected sample on ice until 200,000 cells have been sorted for each cell population.
-
Perform RNA purification and bulk RNA sequencing as previously described (Cheng et al.,2020).
-
For single-cell RNA-seq, sort 50,000 live (7AAD-) epithelial cells (Epcam+) into a 1.5-ml microcentrifuge tube containing 50 μl of 0.4% BSA-PBS and store on ice.
-
Proceed to the Chromium Single Cell Platform (Cheng et al.,2019).
Identify cell-type-specific metabolic gene clusters using bulk RNA-seq data
7.Extract data of 2948 genes encoding mammalian metabolic enzymes using Qlucore Omics Explorer (Birsoy et al., 2015; Corcoran, Grady, Pisitkun, Parulekar, & Knepper, 2017).
8.Filter out lowly expressed genes and genes with small profile variance to reduce noise. In the current example, we choose to retain genes with the variance (σ/σ max) greater than 9e-7.In addition, perform multi-group comparison of the cell types by applying ANOVA using Qlucore Omics Explorer 3.7 or Likelihood Ratio Test (LRT) using R package DESeq2.Set q -value (i.e., p -value adjusted for multiple testing by means of false discovery rate, FDR) as selection factor, for example, as q < 0.005.
9.Normalize Z -score (mean = 0, var = 1) and visualize gene-expression levels with heatmap. To highlight the differences between genes, make Z -score by samples (force each sample to have zero mean and standard deviation = 1). Center and normalize Z -scores; it also helps to have an intuitive idea of the relative variation.
10.Reorder genes with hierarchical clustering. Depicting the data matrix in this way puts genes (rows) with similar expression patterns across cell types (columns) close to each other and helps to find co-varying genes. Three major clusters of genes are identified based on the cell-type-specific expression patterns (Fig. 3A). Also see Readme file in the folder containing files generated using Qlucore Omics Explorer 3.7, available at https://gitlab.leibniz-fli.de/ori-lab/metabolic-signatures-in-stem-cells/.
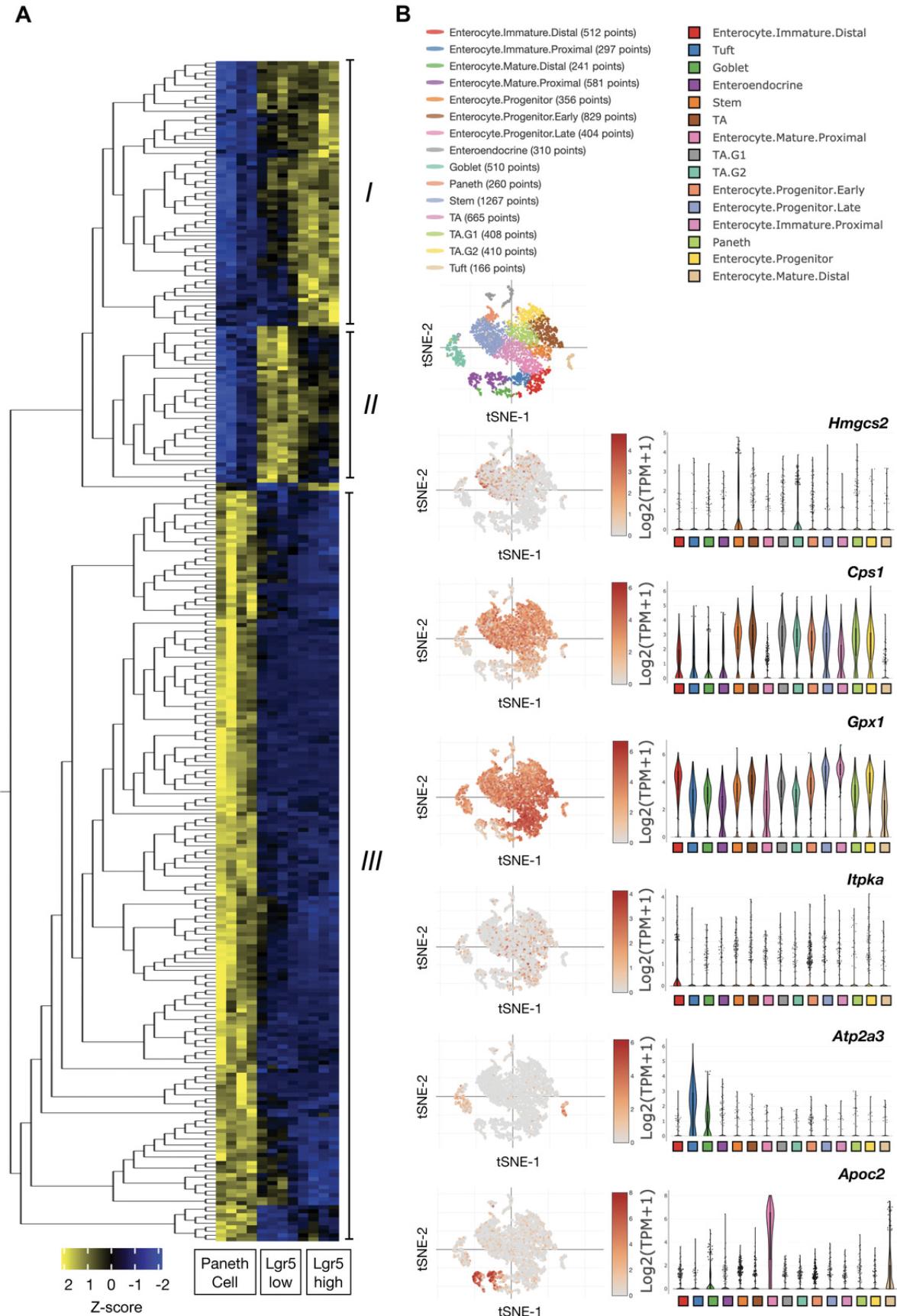
Delineate the cell metabolism−identity patterning using scRNA-seq
11.Cluster the cells using R package Seurat v3 (Macosko et al., 2015).
12.Run non-linear dimensional reduction and visualize cell cluster by t -distributed stochastic neighborhood embedding Epical cells clustered (t-SNE) or Uniform Manifold Approximation and Projection (UMAP), using RunTSNE() or RunUAMP() functions, respectively.
13.Annotate cell identity post hoc based on the expression of literature-curated marker genes. Plot Heatmap shows the relative expression level (row-wise Z scores) of genes (rows) in cell-type-specific signatures across the individual intestinal epithelial cells (column) in the full-length scRNA-seq data. This step can be achieved using R , dittoHeatmap command from dittoSeq package, and the annot.by parameter.
14.Revisit expression of the targeted genes identified in step 10 (e.g., Hmgcs2, Cps1, Gpx1, ATP2a3, and Apoc2) to examine the cell metabolism−to−cell identity patterning and heterogeneity. Examples shown in Figure 3C are generated using the single-cell portal (https://singlecell.broadinstitute.org/single_cell).
Basic Protocol 3: PROTEOME ANALYSES FOR CELL-TYPE-SPECIFIC METABOLIC ENZYME LEVELS
The homeostasis of the proteome, also known as proteostasis, is pivotal for the maintenance of stem cell function, and its perturbation has been shown to contribute to loss of regenerative capacity in different stem cell compartments, for example during aging (Schuler, Gebert, & Ori, 2020). Mass spectrometry−based proteomics enables the quantification of thousands of proteins in cells and tissues. However, the analysis of purified stem cells has proven challenging due to the limited number of cells that can be obtained from a single animal. Importantly, direct measurement of enzyme levels makes it possible to take into account both transcriptional and post-transcriptional mechanisms (e.g., changes in protein synthesis or degradation) that determine the amount of available proteins in different cell populations (Liu, Beyer, & Aebersold, 2016). The workflow of sample preparation presented here allows efficient preparation of peptides from a small number of cells (down to ∼50,000) obtained by FACS. This protocol describes the appropriate collection of ISC populations and preparation as well as analysis of peptides for the acquisition of the proteomics dataset by LC-MS/MS. The same protocol can be adapted to any other cell population obtained by FACS, e.g., muscle stem cells (Schuler et al., 2021).
Materials
-
Intestinal crypts from step 17 of Basic Protocol 1
-
Crypt digestion buffer (see recipe; prepare 3 ml/sample)
-
Phosphate-buffered saline (PBS; PBS Tablets; Gibco, cat. no. 18912014, to be dissolved in 500 ml deionized water; keep on ice)
-
Y-27632 dihydrochloride monohydrate (Abcam ab120129)
-
2× proteomics lysis buffer (see recipe)
-
1 M dithiothreitol (DTT; Roth 6908.3)
-
200 mM iodoacetamide (Sigma I1149); prepare fresh directly before use and keep in the dark (cannot be stored)
-
Acetone (Biosolve 0001037801BS), ice-cold
-
HPLC water (Sigma 270733)
-
Protein digestion buffer (see recipe)
-
LysC stock solution (see recipe)
-
Trypsin stock solution (see recipe)
-
10% trifluoroacetic acid (Biosolve 0020234131BS)
-
Acetonitrile (Biosolve 0001204102BS)
-
Formic acid (Roth 4724.3)
-
HRM kit peptides (Biognosys AG 42896)
-
50-ml conical tubes (e.g., Corning Falcon)
-
1.5-ml microcentrifuge tubes (Eppendorf 0030120.086)
-
37°C water bath
-
Centrifuge with temperature control (Eppendorf 5804R)
-
70-μm mesh filters
-
Bioruptor PlusTM (Diagenode)
-
Thermomixer (Eppendorf 460-0223E and 460-0220)
-
pH paper (Roth KEE1.1)
-
Waters Oasis® HLB μElution Plate, 30 μm (Waters 186001828BA)
-
Extraction Plate Manifold for Oasis 96-Well Plates (Waters 186001831)
-
Vacuum pump (Gast Manufacturing Inc. DOA P504 PN)
-
Speed vacuum centrifuge (Eppendorf Concentrator Plus)
-
PCR tubes (Ratiolab 8610040)
-
MS vials (VDS Optilab 93909134)
-
nanoAcquity UPLC system (Waters)
-
Trapping column (nanoAcquity Symmetry C18, 5 μm, 180 μm × 20 mm; Waters)
-
Analytical column (nanoAcquity BEH C18, 1.7 μm, 75 μm × 250 mm; Waters)
-
Orbitrap Fusion Lumos Tribrid Mass Spectrometer (ThermoFisher Scientific)
The outlet of the column needs to be coupled directly to the Orbitrap Fusion Lumos (ThermoFisher Scientific) using the Proxeon nanospray source.
- SpectronautTM v.14
- R v3.6.3
- R Studio Server v1.2.5042
Isolating targeted cell populations for proteome analysis
In order to avoid contamination of the sample, nitrile gloves should be used throughout all the protocol steps. Sample tubes should be resistant to centrifugation at high speed (20,800 × g) and acetone at indicated steps.
1.Isolate crypts according to Basic Protocol 1, steps 1-17 (collect 3 fractions, combine fraction 2 and 3, discard fraction 1).
2.For dissociating cells to single-cell suspensions, resuspend crypt pellet (∼400,000 crypts) in 1 ml crypt digestion buffer.
3.Add an additional 2 ml crypt digestion buffer.
4.Incubate in a water bath at 37°C for 10 min.
5.Vortex 1 min.
6.Incubate in a water bath at 37°C for 10 min.
7.Fill up to 50 ml with cold PBS containing 80 μM Y-27632 dihydrochloride monohydrate.
8.Centrifuge 5 min at 1000 × g , 4°C.
9.Remove the buffer and resuspend cells in 1 ml PBS containing 80 µM Y-27632 dihydrochloride monohydrate.
10.Filter through a 70-μm mesh directly before sorting.
11.Sort two populations of GFP-positive cells (GFPhigh and GFPlow) cells into 1.5-ml tubes containing 50 μl 2× proteomics lysis buffer:
12.After sorting, spin down samples at room temperature and, if necessary, adjust the volume of lysis buffer to the volume of sheath fluid collected from the sorter by adding 2× proteomics lysis buffer.
13.After sorting, keep the sample on ice until all samples are sorted and frozen, and store at −80°C until protein digestion and mass spectrometry analysis.
Protein digestion and peptide purification for proteomics
14.Thaw ISC lysates and solubilize by sonication in a Bioruptor PlusTM for 10 cycles (1 min on and 30 s off), with high intensity, at 20°C.
15.Heat lysates for 10 min at 95°C in a Thermomixer.
16.After heating at 95°C, sonicate lysates again as in step 14.
17.Clarify lysates by centrifugation for 5 min at 20,800 × g , 4°C, in a centrifuge with temperature control.
18.Transfer the supernatant containing proteins to a new 1.5-ml tube.
19.Reduce cysteines by adding 1 M DTT to 10 mM and incubating at 37°C in a Thermomixer.
20.Alkylate cysteine by addition of 200 mM iodoacetamide to a final concentration of 15 mM and incubate for 30 min in the dark.
21.Precipitate proteins by adding 8× the sample volume of ice-cold acetone and incubate overnight at -20°C.
22.Pellet the protein by centrifugation for 1 hr at 20,800 × g , 4°C.
23.Remove the supernatant.
24.Wash the pellet twice with ice cold 80% acetone (v/v in HPLC water).
25.Air-dry the pellets in a fume hood.
26.When dried, dissolve the pellet in protein digestion buffer at a concentration of 0.25-0.5 µg/μl:
27.To facilitate the resuspension of the protein pellet, subject the samples tothree rounds of sonication (1 min on and 30 s off) with high intensity at 20°C using the Bioruptor PlusTM.
28.Digest proteins for 4 hr at 37°C using 1:100 (enzyme/protein) LysC.
29.Dilute samples to 1.5 M urea, 50 mM HEPES (see protein digestion buffer recipe) with HPLC water and digest with 1:100 (enzyme/protein) trypsin for 16 hr at 37°C.
30.Acidify digested peptide solutions with 10% trifluoroacetic acid. Check the pH using 0.5 μl sample and pH paper. If the pH is higher than 2, add more trifluoroacetic acid.
31.Desalt the peptides with Waters Oasis® HLB μElution Plate 30 μm:
-
Place a waste collection tray in an Extraction Plate Manifold for Oasis 96-Well Plates.
-
Condition the columns by applying 3 × 100 μl solvent B (80% acetonitrile/0.05% formic acid in HPLC water) in presence of a low vacuum of 5 mbar created by a vacuum pump.
-
Equilibrate the columns by applying 2 × 100 μl solvent A (0.05% formic acid) in the presence of a low vacuum.
-
Add 100 μl of solvent A without vacuum.
-
Add each sample to one well containing solvent A without vacuum.
-
Load the sample on the column by applying a low vacuum.
-
Wash the samples by adding 3 × 100 μl solvent A in presence of a low vacuum.
-
After the last wash, dry the columns by applying a strong vacuum (up to 10 mbar).
-
Remove the waste collection tray and place PCR tubes for collection of samples.
-
Pipet 50 μl of solvent B and apply low vacuum until all samples are eluted.
-
Dry eluates in a speed vacuum centrifuge.
32.Dissolve the dried peptides in 5% acetonitrile/0.1% formic acid to a peptide concentration of 1 μg/μl.
33.Transfer 10 μl to an MS vial.
34.Prior to analysis by LC-MS/MS, spike into each sample 0.5 μl of HRM kit peptides reconstituted according to the manufacturer's instruction and then diluted 1:4 using 5% acetonitrile/0.1% formic acid.
Label-free LC-MS analysis using Data Independent Acquisition (DIA)
For direct LC-MS analysis using DIA, a volume that contains 1-2 µg of peptides needs to be injected. The separation gradient is nonlinear, according to Table 1, at a constant flow of 0.3 μl/min.
| Time | %B |
|---|---|
| 0.0 | 0 |
| 1.0 | 5 |
| 4.0 | 6 |
| 9.0 | 7 |
| 18.0 | 9 |
| 77.0 | 21 |
| 93.0 | 24 |
| 101.0 | 26 |
| 106.0 | 28 |
| 110.0 | 29 |
| 113.0 | 31 |
| 115.0 | 32 |
| 116.0 | 33 |
| 117.0 | 34 |
| 119.0 | 36 |
| 120.0 | 40 |
| 121.0 | 50 |
| 124.0 | 50 |
| 125.0 | 85 |
| 129.0 | 85 |
| 130.0 | 0 |
| 145.0 | 0 |
- a
Non-linear gradient for the nanoLC during an LC-MS run (step 22). The table shows the runtime (in min) and the corresponding percentage of mobile phase B (%B) on the column.
Buffers for nanoLC during LC-MS runs: mobile phase A is 0.1% (v/v) formic acid in water, mobile phase B is 0.1% (v/v) formic acid in acetonitrile.
35.DIA data acquisition:
-
Use the nanoAcquity UPLC system fitted with a trapping (nanoAcquity Symmetry C18, 5 μm, 180 μm × 20 mm) and an analytical column (nanoAcquity BEH C18, 1.7 μm, 75 μm × 250 mm). The outlet of the column needs to be coupled directly to an Orbitrap Fusion Lumos using the Proxeon nanospray source.
-
Load 1-2 μg of reconstituted peptides.
-
Separate peptides using the gradient described in Table1.
-
Acquire MS data using the parameters described in Table2.
| Entry | Parameter | Setting |
|---|---|---|
| MS OT (scan experiment 1) | Detector Type: | Orbitrap |
| 0-145 min | Resolution | 120,000 |
| Mass range | Normal | |
| Use quadrupole isolation | Yes | |
| Scan Range (m/z) | 350-1650 | |
| RF lens | 30% | |
| AGC target | 5.0e5 | |
| Maximum injection Time | 20 ms | |
| Microscans | 1 | |
| Data Type | Profile | |
| Polarity | Positive | |
| tMS2 OT HCD (scan experiment 2) | MSn level | 2 |
| 0-145 min | Isolation mode | Quadrupole |
| MS Isolation windows (m/z) | See Table 3 | |
| Activation type | HCD | |
| HCD collision energy (%) | 30 | |
| Detector Type | Orbitrap | |
| Orbitrap resolution | 30,000 | |
| Scan range (m/z) | 200-2000 | |
| AGC target | 1.0e6 | |
| Maximum Injection Time | 70 ms | |
| Microscans | 1 | |
| Data type | Profile | |
| Inject ions for All Available Parallelizable Time | Yes | |
| Loop control | All |
| m/z | Isolation window (m/z) |
|---|---|
| 376.5 | 53 |
| 412.5 | 21 |
| 431 | 18 |
| 448 | 18 |
| 464.5 | 17 |
| 480.5 | 17 |
| 496.5 | 17 |
| 512 | 16 |
| 527 | 16 |
| 542.5 | 17 |
| 558 | 16 |
| 573.5 | 17 |
| 589.5 | 17 |
| 605.5 | 17 |
| 621.5 | 17 |
| 637.5 | 17 |
| 654 | 18 |
| 671 | 18 |
| 688 | 18 |
| 706 | 20 |
| 725 | 20 |
| 744 | 20 |
| 764 | 22 |
| 785.5 | 23 |
| 808.5 | 25 |
| 833 | 26 |
| 859 | 28 |
| 887.5 | 31 |
| 919.5 | 35 |
| 956 | 40 |
| 1000 | 50 |
| 1056.5 | 65 |
| 1138.5 | 101 |
| 1419 | 462 |
Proteomics raw data analysis
36.Generate a library by searching DIA raw data using the “directDIA” option in SpectronautTM v.14 using default settings.
| Entry (node in method editor) | Parameter | Setting |
|---|---|---|
| MS OT | Detector Type: | Orbitrap |
| Resolution | 60,000 | |
| Mass range | Normal | |
| Use quadrupole isolation | Yes | |
| Scan Range (m/z) | 350-1650 | |
| RF lens | 40% | |
| AGC target | 2.0e5 | |
| Maximum injection Time | 50 ms | |
| Microscans | 1 | |
| Data Type | Profile | |
| Polarity | Positive | |
| MIPS | Monoisotopic peak determination | Peptide |
| Intensity | Filter type, Intensity threshold | 5.0e4 |
| Charge state | Include charge state(s) | 2-7 |
| Dynamic exclusion | Exclude after n times | 1 |
| Exclusion duration (s) | Depends on LC runtime: 15 s for 1 hr run, 60 s for 2.5 hr | |
| Mass Tolerance | ppm | |
| Low | 10 | |
| High | 10 | |
| Exclude isotopes | Yes | |
| ddMS2 IT HCD | Isolation mode | Quadrupole |
| Isolation Window (m/z) | 1.4 | |
| Activation type | HCD | |
| HCD collision energy (%) | 30 | |
| Detector type | Orbitrap | |
| Resolution | 15,000 | |
| First mass (m/z) | 120 | |
| AGC target | 2.0e5 | |
| Maximum Injection Time | 22 ms | |
| Microscans | 1 | |
| Data type | Profile | |
| Data Dependent Properties | Select: “Cycle time” | 3 s |
37.Search DIA raw data against the generated spectral library using default settings.
38.Perform relative quantification in SpectronautTM for each pairwise comparison using the replicate samples from each condition.
39.Export the data (candidates table) and protein quantity data reports.
Proteomics data analysis using R
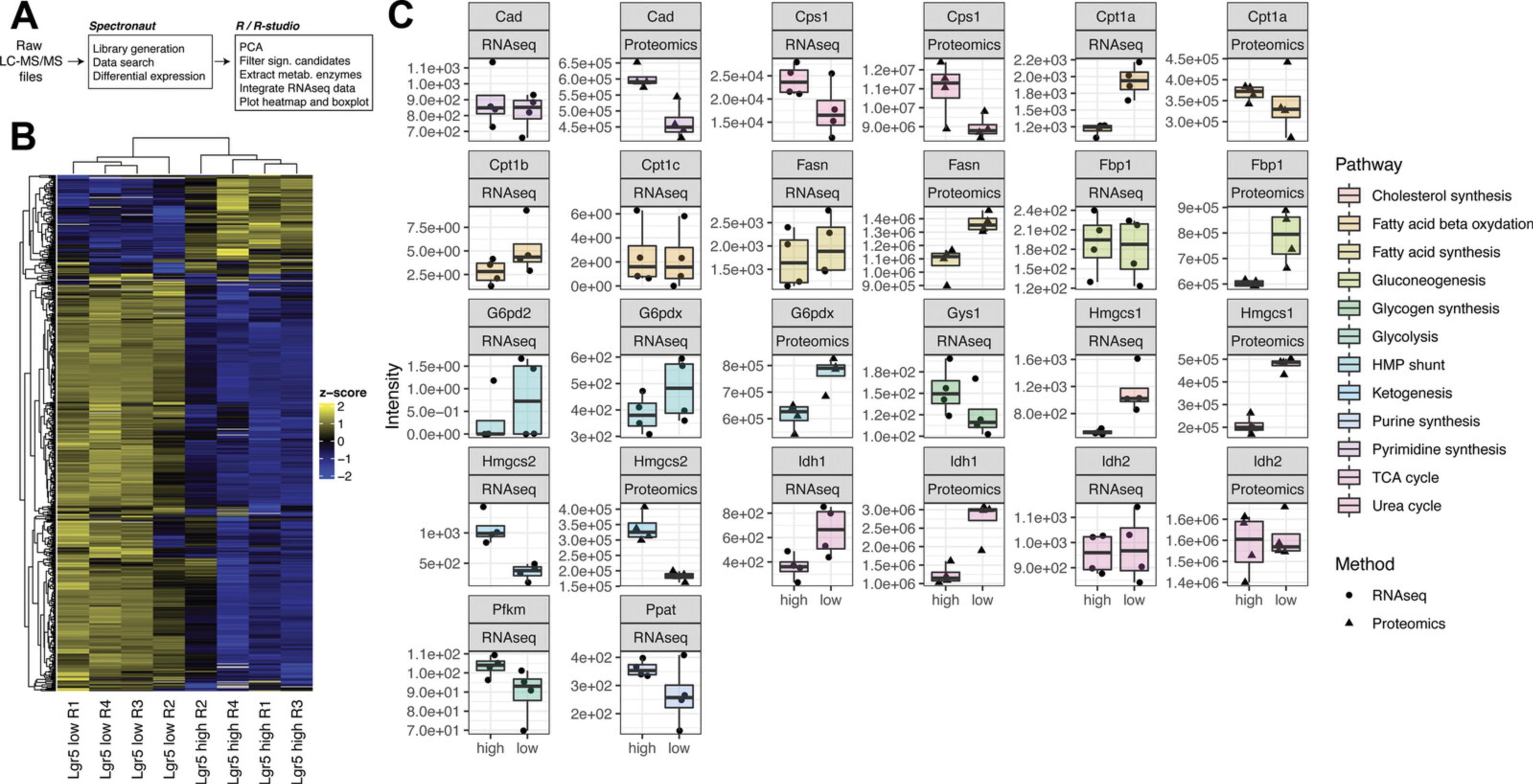
Downstream data analyses and visualization can be performed using R v3.6.3 in R Studio Server v1.2.5042 using a similar strategy to the one for population RNA-seq (Fig. 4A).
40.Perform principal component analysis (PCA) using all the quantified proteins to compare samples.
-
Extract protein intensities (“PG.quantity” from Spectronaut report table), log2 transform, perform PCA using the functionprcomp, and visualize results using the functionfviz_pca_ind, which is part of the packagefactoextra (https://cran.r-project.org/package=factoextra).
-
Samples from different cell populations should form clearly distinguishable clusters on the PCA space (Fig.1, proteome analyses). Possible outlier samples can also be identified with this procedure and removed for downstream analysis.
41.Analyze the abundance of metabolic enzymes quantified.
-
Extract protein intensities from the Spectronaut report table for the 2948 genes encoding mammalian metabolic enzymes (Birsoy et al.,2015).
-
Filter for proteins showing differential abundance between cell types according to the Spectronaut candidates table. Here, proteins are retained ifqvalue is smaller than 0.05 and the absolute log2 fold change (“Absolute.AVG.Log2.Ratio”) is greater than 0.3.
In our dataset, we quantified 393 metabolic enzymes with q < 0.05 and absolute log2 fold change >0.3.
- Normalize data viaZ-score (mean = 0, var = 1) and visualize protein levels with heatmap. To highlight the differences between proteins, makeZ-score by sample (force each sample to have zero mean and standard deviation = 1).
In general, centering and normalizing Z-scores also help in getting an intuitive idea of the relative variation.
- Reorder genes with hierarchical clustering. Depicting the data matrix in this way puts genes (rows) with similar expression patterns across cell types (columns) close to each other and helps to find co-varying proteins. In Figure4B, two major clusters of proteins are identified based on the cell-type-specific expression patterns.
Z-score-based analysis is powerful for identifying robust differences between cell types; however, in order to estimate the effect size of protein abundance differences between samples, complementary approaches should be used. In particular, proteomics data allow one to estimate absolute amounts of proteins and, thereby, to compare directly the abundance of metabolic enzymes in different sample types.
- Protein abundance estimates can be derived from the intensities of the peptides identified for each protein. Default settings in Spectronaut utilize a Top3 strategy for peptide summarization, whereby the mean value of the three most confidently identified peptides for each protein is used to derive the protein abundance value. Since this strategy considers the same number of peptides for each protein, it does not require further normalization to take into account differences in protein sizes. Other summarization strategies, i.e., based on summed peptide intensities, require additional normalization steps to derive comparable estimates of protein absolute abundances (see for example iBAQ values; Schwanhausser et al.,2011).
In order to correctly estimate abundances of related (homologous) proteins, only proteotypic peptides (peptides unique to a given gene product) should be used. In addition, it is recommended to consider only proteins for which at least two proteotypic peptides were identified, as abundance estimates based on single peptides can be less accurate due to different physical/chemical properties of individual peptides.
-
In Figure4C, absolute levels of key rate-limiting metabolic enzymes are compared between GFPhighand GFPlowISC samples. Illustrative R code and input files are available at:https://gitlab.leibniz-fli.de/ori-lab/metabolic-signatures-in-stem-cells/.
Basic Protocol 4: METABOLITE PROFILING AND MEASUREMENT
In contrast to RNA and proteins, metabolites do not follow a fixed structural template and accordingly have diverse physical properties. As a result, no single analytical tool can measure all metabolites. One advantage of the current framework is that once the cell-type-specific metabolic genes or enzymes are identified, the targeted metabolic pathways can be further investigated in combination with metabolic gene ablation. Here, we use Hmgcs2 and ketogenesis (i.e., ketone body metabolite production), which we identified as ISC-specific metabolic signature, as an example to describe the validation process using HRAM (high-resolution accurate-mass) liquid chromatography–mass spectrometry (LC–MS) (”a” steps below) and NAD+/NADH assay (”b” steps below) for measuring ketone body β-hydroxybutyrate (β-OHB). The former provides overall changes of relative metabolite levels in response to the target gene deletion. The latter is designed specifically for determining levels of the targeted metabolite β-OHB, which is based upon the oxidation of D-3-hydroxybutyrate to acetoacetate by the enzyme 3-hydroxybutyrate dehydrogenase (McMurray, Blanchflower, & Rice, 1984).
Materials
- LC/MS OPTIMA−grade methanol (Fisher Scientific A4564)
- Metabolomics Amino Acid Mix Standard (Cambridge Isotope Laboratories, Inc. MSK-A2-1.2)
CIL has since brought out a new Canonical Amino Acid Standard Mix (MSK-CAA-1) which contains all 20 proteogenic amino acids labeled with 13C and 15N; we recommend using this as an internal standard option instead.
-
Blood bank saline, 0.85% NaCl (Azer Scientific 16005-092)
-
LC/MS OPTIMA−grade water (Fisher Scientific W64)
-
LC/MS grade chloroform with ethanol as a stabilizer (Fisher Scientific C606SK1)
-
LC/MS OPTIMA grade acetonitrile (Fisher Scientific A9554)
-
Ammonium carbonate (Sigma 379999)
-
Ammonium hydroxide (Sigma 338818)
-
100 mM Tris·HCl, pH 8.5
-
β-Hydoxybutyrate Standard (Sigma H0265)
-
β-Hydoxybutyrate Enzyme Solution (Caymen 700193)
-
β-Hydroxybutyrate Colorimetric Detector (Caymen 700194)
-
Glycine (Sigma G7126)
-
Aspartate (Sigma A7219)
-
1.5-ml microcentrifuge tubes (Axygen MCT-150-C)
-
Centrifuge with temperature control (Eppendorf 5804R)
-
Nitrogen evaporator (MultiVap, Organomation)
-
Polypropylene autosampler vials with fused insert (ThermoFisher 03340620)
-
Autosampler caps with PTFE/silicone septa (ThermoFisher 03376481)
-
Dionex Ultimate 3000 UHPLC (ThermoFisher Scientific)
-
QExactive hybrid mass spectrometer equipped with a HESI II probe (ThermoFisher Scientific)
-
SeQuant® ZIC-pHILIC analytical column (5 μm, 150 × 2.1 mm) fitted with a SeQuant® ZIC-pHILIC guard column (5 μm, 20 × 2.1 mm)
-
Microsoft Excel
-
TraceFinder 4.1 (ThermoFisher Scientific)
-
96-well plates for colorimetric assays
NOTE : If absolute quantification of metabolites is required, we recommend including the labeled metabolite of interest as an internal standard if possible, and running a standard curve alongside the experimental samples. Determining cell number and volume/ tissue weight is also required to calculate absolute concentration.
Polar metabolite extraction and detection using HRAM LC/MS
Chloroform-methanol extraction
Before you begin, prepare methanol containing 500 nM Metabolomics Amino Acid Mix Standard (methanol with internal standards) and chill to −20°C (this can be stored long-term at −20°C). Chill appropriate amount of chloroform to −20°C and LC/MS grade water to 4°C. The following protocol should be performed on ice and as quickly as possible to quench metabolism in the intestinal crypts.
1a. Harvest intestinal crypts from WT and Hmgcs2-intestinal KO mice as previously described (Cheng et al., 2020; also see Basic Protocol 1).
2a. Wash crypt pellets once with cold 0.85% NaCl.
3a. Add 600 μl LC/MS-grade methanol with internal standards.
4a. Add 300 μl LC/MS-grade water.
5a. Pipette up and down to mix.
6a. Add 400 μl chloroform to each sample.
7a. Vortex for 10 min at 4°C
8a. Spin 10 min at 21,100 × g , 4°C, to initiate phase separation.
9a. Carefully collect the two layers separately, avoiding the precipitate at the interface of the two layers (top layer is polar; bottom layer is organic/lipids). Evaporate to dryness under a stream of nitrogen.
10a. Store dried polar and organic extracts at −80°C until analysis.
Preparation of samples for LC-MS analysis of polar metabolites
11a. Resuspend dried polar extracts in 100 μl LC/MS-grade water (on ice)
12a. Vortex for 10 min at 4°C to ensure complete resuspension and even mixing
13a. Spin for 10 min at 21,100 × g , 4°C, to remove any remaining debris/precipitate
14a. Transfer 90 μl to a new 1.5-ml microcentrifuge tube (on ice)
15a. Combine equal volumes of each sample to create a 1× pooled sample (this will be used for QC).
16a. Take 3 μl of your pooled sample and add 7 μl LC/MS-grade water. This will be your 0.3× dilution. Transfer to an LC/MS autosampler vial.
17a. Take 1 μl of your pooled sample and add 9 μl LC/MS-grade water. This will be your 0.1× dilution. Transfer to an LC/MS autosampler vial.
18a. Transfer at least 40 μl of your 1× pooled sample to an LC/MS autosampler vial.
19a. Transfer 20 μl of sample to an LC/MS autosampler vial. Store any remaining sample (from the original 90 μl resuspension) at −80°C as a backup in case the initial run fails.
20a. Transfer LC/MS vials to the autosampler of the LC system
LC-MS analysis of polar metabolites
For LC-MS analysis of polar metabolites inject 2 μl of each sample. Metabolites are separated on a Dionex Ultimate 3000 UHPLC using a linear gradient according to Table 5 with a constant flow rate of 0.15 ml/min.
| Time | %B |
|---|---|
| 0.0 | 80 |
| 20.0 | 20 |
| 20.5 | 80 |
| 28.0 | 80 |
- a
Linear gradient for the UHPLC during an LC-MS run (step 21a). The table shows the runtime (in min) and the corresponding percentage of mobile phase B (%B) on the column.
Other LC systems may be used. However, the technology transfer may need to be determined empirically.
- Mobile Phase A: 20 mM ammonium carbonate with 0.1% ammonium hydroxide
- Mobile Phase B: acetonitrile
The following MS parameters are specific to the QExactive quadrupole-orbitrap hybrid mass spectrometer (ThermoFisher Scientific). While alternative mass spectrometers may be used, their parameters will need to be adjusted according to the specific instrument being utilized.
21a. Data acquisition:
- Use the Dionex UltiMate 3000 UHPLC system coupled to a QExactive hybrid mass spectrometer equipped with a HESI II probe.
- Prior to analysis, calibrate the mass spectrometer using the standard calibration mix if calibration has not been performed within the last 7 days, or a mass check fails. Perform a custom calibration of the lower end masses by spiking the standard calibration mix with glycine (positive ionization mode) and aspartate (negative ionization mode).
- Inject 2 μl sample onto a SeQuant® ZIC-pHILIC analytical column (5 μm, 150 × 2.1 mm) fitted with a SeQuant® ZIC-pHILIC guard column (5 μm, 20 × 2.1 mm).
Note that injection volume may need to be determined empirically depending on the cell/tissue type. Choose an injection volume that detects metabolites in the linear range (use the linear dilution series to determine the linear range of each metabolite).
- Set the column oven to 25°C and the autosampler tray to 4°C.
- Use Excel to randomize the order in which your samples will be injected and inject the pooled QC sample every 8-10 samples, including the 0.3× and 0.1× dilution series at the end of your sequence. As part of this linear dilution series, include a larger 2× QC pool injection. In addition, acquire MS/MS data from the QC pool in both positive and negative ionization mode.
- Separate metabolites using the gradient described in Table 5.
- Acquire MS data using the parameters described in Tables 6-8.
Processing and analysis of LC-MS data to determine relative quantification of polar metabolites
22a. Use TraceFinder 4.1 to integrate raw peak areas for metabolites of interest and internal standards by extracting ion chromatograms from the full scan data (or tSIM data in the example of β-OHB)
- For metabolite identification, match retention time and MS/MS spectra against in-house retention time and MS/MS spectral libraries previously established using identical LC-MS conditions and authentic chemical standards.
The in-house retention time library is especially important for the integration of β-hydroxybutyrate due to close-eluting isomers of α-hydroxybutyrate, β-isohydroxybutyrate, and γ-hydroxybutyrate.
| Entry | Parameter | Setting |
|---|---|---|
| Full MS | Polarity | Positive |
| 0-16.5 min | Resolution | 70,000 |
| AGC Target | 1e6 | |
| Max IT | 20 ms | |
| Microscans | 1 | |
| Data Type | Profile | |
| Scan Range | 70-1000 m/z | |
| Full MS | Polarity | Negative |
| 0-16.5 min | Resolution | 70,000 |
| AGC Target | 1e6 | |
| Max IT | 20 ms | |
| Microscans | 1 | |
| Data Type | Profile | |
| Scan Range | 70-1000 m/z | |
| tSIM | Polarity | Negative |
| 3-7 min | Inclusion list | On: 120.03230 m/z |
| Resolution | 70,000 | |
| AGC Target | 1e5 | |
| Max IT | 250 ms | |
| Isolation Window | 1.2 m/z | |
| Microscans | 1 | |
| Data Type | Profile |
| Entry | Parameter | Setting |
|---|---|---|
| Full MS | Polarity | Positive |
| 0-16.5 min | Default charge state | 1 |
| Exclusion | Ona | |
| Resolution | 70,000 | |
| AGC Target | 1e6 | |
| Max IT | 50 ms | |
| Microscans | 1 | |
| Data Type | Profile | |
| Scan Range | 70-1000 m/z | |
| ddMS2 | Resolution | 17,500 |
| AGC Target | 2e5 | |
| Maximum IT | 100 ms | |
| Loop Count | 10 | |
| TopN | 10 | |
| Isolation Window | 1.0 m/z | |
| dd properties | (N)CE/Stepped (N)CE | 15, 30, 45 |
| Exclude Isotopes | On | |
| Dynamic Exclusion | 5.0 s |
- a
Exclusion list for positive mode should be generated prior to experiment using a mock extraction run on identical Full Scan settings. Include the top 100 most abundant ions.
| Entry | Parameter | Setting |
|---|---|---|
| Full MS | Polarity | Negative |
| 0-16.5 min | Default charge state | 1 |
| Exclusion | Ona | |
| Resolution | 70,000 | |
| AGC Target | 1e6 | |
| Max IT | 50 ms | |
| Microscans | 1 | |
| Data Type | Profile | |
| Scan Range | 70-1000 m/z | |
| ddMS2 | Resolution | 17,500 |
| AGC Target | 2e5 | |
| Maximum IT | 100 ms | |
| Loop Count | 10 | |
| TopN | 10 | |
| Isolation Window | 1.0 m/z | |
| (N)CE/Stepped (N)CE | 15, 30, 45 | |
| dd properties | Exclude Isotopes | On |
| Dynamic Exclusion | 5.0 s |
- a
Exclusion list for negative mode should be generated prior to experiment using a mock extraction run on identical Full Scan settings. Include the top 100 most abundant ions.
23a. Normalize raw peak areas by dividing the raw peak area for each metabolite by the raw peak area of the corresponding internal standard (selected based on similarity of retention time and/or chemical similarity). If performing metabolite profiling to measure a particular metabolite, consider purchasing a stable isotope−labeled version of that metabolite if available.
24a. Further normalize these values to tissue weight (mg) or cell number, if the extraction volume was not adjusted for biological input prior to analysis by LC-MS.
25a. Generate QC metrics by calculating the coefficient of variation (CV) for the multiple pooled QC injections. Reject metabolites with a CV >0.25, as these metabolites are not being detected in a reliable fashion.
26a. Use the pooled dilution series (average of pooled 1× QC injections, 0.3× QC pool injection, and 0.1× QC pool injection) to calculate the linear correlation coefficient for each metabolite. Reject metabolites that have an R value of <0.975, as these metabolites are not being detected in a linear fashion.
27a. Use the normalized values to calculate relative log2 fold change across different conditions and to perform statistical analysis. Many open-source platforms exist that can be used for this purpose, including Metaboanalyst (Xia, Psychogios, Young, & Wishart, 2009).
NAD+/NADH assay for β-OHB determination
1b. Resuspend the crypt cells from Basic Protocol 1 (step 19) or directly sort cell populations from Basic Protocol 2 (step 6) in 50 μl 100 mM Tris·HCl, pH 8.5.
2b. Homogenize the cell suspension on ice.
3b. Centrifuge cell suspension 10 min at 10,000 × g , 4°C.
4b. Transfer the supernatant to another tube on ice
5b. Prepare standards by serially diluting β-hydroxybutyrate solution (50 μl each, from 100 to 1 μM) using 100 mM Tris·HCl.
6b. Load standard duplicates and samples into 96-well microplate for colorimetric assay. Use 100 mM Tris·HCl as the blank for calibration.
7b. Prepare developer solution by mixing 100 μl of the β-HB Colorimetric Detector to the vial of reconstituted β-HB Enzyme Solution. Store the developer solution on ice.
8b. Add 50 μl developer solution to each well and mix the reagents by pipetting to initiate the enzymatic reaction
9b. Incubate the plate in dark, at 25°C, up to 2 hr for lysates from crypts or up to 6 hr for lysates from sorted cells.
10b. Read the absorbance at 445-455 nm.
11b. Calculate the average absorbance of each standard and sample.
12b. Subtract the absorbance value of standard A (0 mM) from itself and all other values (both standards and samples).
13b. Calculate the values of the β-HB samples using the equation obtained from the linear regression of the standard curve by substituting the corrected absorbance values for each sample into the equation.
14b. Normalize to sample volume (measured prior to lysis) or by number of cells.
REAGENTS AND SOLUTIONS
Crypt digestion buffer
- 70% (v/v) TrypLE
- 0.4 U/μl DNase I (Sigma 4716728001)
- 5 mM MgCl2 (Roth KK36.2)
- 80 μM Y-27632 dihydrochloride monohydrate (Abcam ab120129)
- Prepare fresh before use
LysC stock solution
Prepare a stock solution of LysC (Wako Chemicals GmbH 125-05061) at a concentration of 0.5 µg/μl by reconstituting lyophilized enzyme (20 µg vial) in 40 μl. LysC stock solution can be stored at −20°C for up to 1 year. Avoid multiple cycles of freeze-thawing.
Protein digestion buffer
- Prepare in HPLC water (Sigma 270733):
- 3 M urea (Sigma, U6504)
- 100 mM HEPES (Sigma, H3375), pH 8
- Prepare fresh before use
Proteomics lysis buffer, 2×
- Prepare in MilliQ water:
- 4% sodium dodecyl sulfate (SDS; Sigma 75746)
- 200 mM HEPES (Sigma H3375), pH 8 (adjust with NaOH)
- 20 mM dithiothreitol (DTT; Roth 6908.3)
- Store up to 1 week at −20°C
Since SDS might precipitate at low temperature, do not keep the proteomics lysis buffer on ice!
Trypsin stock solution
Prepare a trypsin stock solution at a concentration of 0.5 µg/μl by reconstituting lyophilized enzyme (Promega, V5111; 20 µg vial) in 40 μl of provided trypsin buffer (supplied with the enzyme). Trypsin stock solution can be stored at −80°C for up to 1 year. Avoid multiple cycles of freeze-thawing.
COMMENTARY
Background Information
The concept of cell identity has evolved with methods that allow detecting cell features for classification. Although transcriptome and proteome analyses are primary approaches for studying cell identity, they have not been utilized for discovering cell-type-specific metabolic features until very recently (Cheng et al., 2019; Gebert et al., 2020). In this protocol, we use intestinal crypts as an example to describe the essential steps involved in tissue dissociation and cell population purification (Basic Protocol 1). We explain how to cross-reference transcriptome (Basic Protocol 2) and proteome (Basic Protocol 3) data and use these data as an exploratory tool to identify inherent metabolic features in different cell types. We use ketogenic HMGCS2 and ketone body metabolites as examples to describe how to further investigate the metabolic activities by metabolite profiling (Basic Protocol 4).
The integrative framework offers several advantages in identifying the cell-type-specific metabolic features. First, the transcriptome and proteome-based analyses allow immediate discovery of the cell marker−to−metabolism association and the more advanced pathway analyses linking the differentially expressed metabolic enzymes to the cell-fate determination signaling. Also, genetic or enzymatic perturbations can be directly applied to the screening pipeline for downstream functional validation. Significantly, this approach narrows down the pool of metabolite candidates, which eases the technical hurdles of measuring a wide breadth of small molecules in rare cell populations or transient cell states. Finally, compared to the metabolome datasets, many more transcriptome and proteome datasets containing cell-type-specific information are publicly available. Therefore, biologists are much more encouraged to explore the cell-type-specific metabolic signatures in the tissues or diseases of interest using transcriptomics and proteomics approaches.
Data Interpretation
The differential expression of metabolic genes/proteins identified in Basic Protocols 2 and 3 (Fig. 3A and 4A) reveals the inherent metabolic differences interlinked with the defined cell types. When mapping the expression pattern of the identified genes/proteins onto tSNE plots (Fig. 3B), one can further visualize heterogeneity of metabolic features within the stem cell cluster and more holistically explore the relationship between the metabolic features and cell states and identities at single-cell resolution. For example, Hmgcs2 enriching for Lgr5high stem cells (Fig. 3A, cluster I) is expressed moderately in transient amplifying progenitor cells at G2/M phase (TA-G2) but sparsely in TAs at G1/S phase (TA-G1) (Fig. 3B). Cps1 expression is fairly high in stem and progenitor clusters, but very low in the differentiated cells (i.e., enterocytes, goblet, Paneth, tuft, and enteroendocrine cells). Expression of Gpx1 enriching for the Lgr5low TAs (Fig. 3A, cluster II) is particularly up-regulated in TAs and early progenitors, but significantly down-regulated in the terminally differentiated cells. Similarly, Itpka (Fig. 3A, cluster II) expression is almost exclusive to TAs and progenitors. Finally, the result of scRNA-seq indicates that Atp2a3 and Apoc2 identified through the enrichment in CD24+ Paneth cells (Fig. 3A, cluster III) are not specific to the Paneth cell cluster. In fact, the data shows that genes highlighted in cluster III (Fig. 3A) mostly are markers for the terminally differentiated cells. Atp2a3, for example, marks the goblet and Tuft cells, and Apoc2 is almost exclusively expressed in the enterocytes. Overall, these data indicate a great diversity of metabolic gene and protein expression patterns across cell types. They also highlight the utility of transcriptome and proteome analyses for systematically exploring the relationship between cell metabolism and identity.
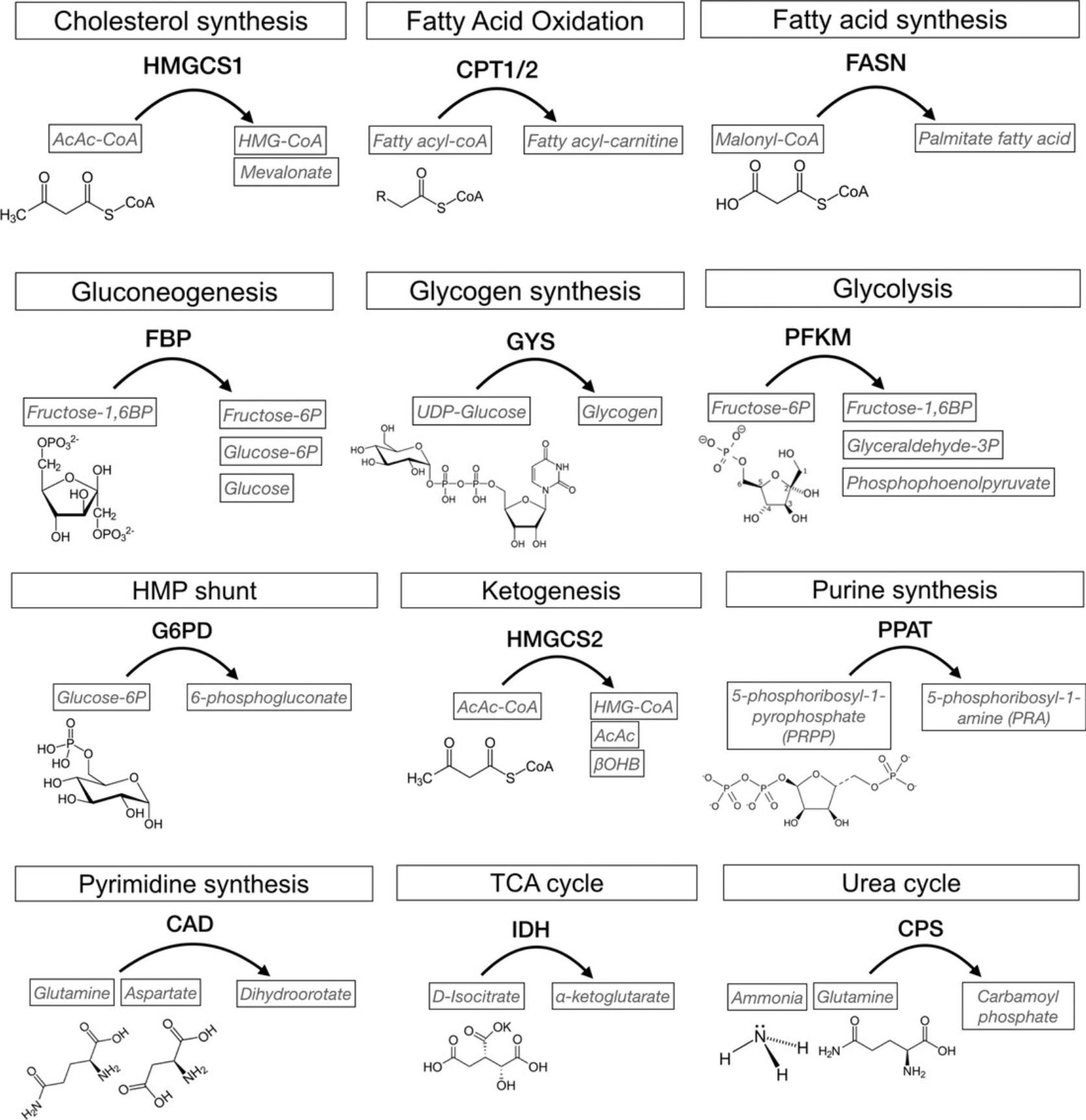
It is worth mentioning that differential metabolic gene expression is not always consistent with protein levels. In Figure 4C, we examine the gene expression and protein levels of the rate-limiting enzymes of twelve major metabolic pathways in Lgr5high stem cells and Lgr5low progenitors. The differential expressions of Cps1 for urea cycle, Hmgcs1 for cholesterol synthesis, Hmgcs2 for ketogenesis, and Idh1 for TCA cycles are consistent at both RNA and protein levels. Cpt1a for fatty acid oxidation is higher in the progenitor at RNA levels but not at protein levels, which is the opposite of Fasn and Fbp1 expression. Cpt1b and Cpt1c (isoforms of Cpt1a), Gys1 for glycogen synthesis, Pfkm for glycolysis, and Ppat for purine synthesis are not detectable at protein levels. The consistency between RNA and protein profiles of the targeted genes can also serve as the selection criterion for robust inherent metabolic features. Given that rate-limiting enzymes catalyze specific reactions at a relatively lower velocity and in a relatively substrate-independent manner, one can further investigate the metabolic activity based on the levels of corresponding metabolites (Fig. 5). As described in Basic Protocol 4, to investigate the metabolic activity of HMGCS2, one can measure the levels of acetoacetate (AcAc) β-hydroxybutyrate (β-OHB; Fig. 5, ketogenesis) using HRAM LC/MS (Basic Protocol 4, ‘a’ steps) and β-OHB specifically NAD+/NADH assay (Basic Protocol 4, ‘b’ steps).
Alternative Methods
Although we are able to distinguish 153 polar metabolites, with 108 metabolites detected in intestinal crypts, using the HRAM LC-MS method described in the ‘a’ steps of Basic Protocol 4, we encourage readers to modify the extraction protocol or to consider alternative methods optimized for the metabolites of interest. Gas chromatography−mass spectrometry (GC-MS)−based metabolite profiling, for instance, is ideal for identifying and quantitating small molecular metabolites (<650 Da), such as small acids, alcohols, sugars, and sterols (see Current Protocols article: Fiehn, 2016). In addition to MS-based methods, nuclear magnetic resonance (NMR) technologies have also been used for metabolite profiling (Perez-Ramirez & Christofk, 2021). The NMR-based techniques offer a better resolution for the difficult-to-distinguish metabolites. However, they have lower sensitivity compared to MS, and are therefore more suitable for abundant metabolites or large cell populations in the tissue (Perez-Ramirez & Christofk, 2021). Because we incorporate the metabolite profiling and measurement here for target validation instead of data exploration, readers can also take advantage of the target-driven approach using assays developed for specific metabolites. For example, the NAD+/NADH assay described in the ‘b’ steps of Basic Protocol 4 is specialized for measuring levels of β-OHB. There are also commercially available bioluminescent assays to detect the levels of glucose, glutamine, glutamate, and lactate at nanomolar (nM) concentrations from cell lysates. Also, for rate-limiting enzymes that catalyze specific metabolites (Fig. 5), metabolic flux analysis using stable isotope-labeled tracers will allow a dynamic profile of the targeted metabolic activity. Stable-isotope measurements are increasingly used to study cell metabolism in vivo (Fernandez-Garcia, Altea-Manzano, Pranzini, & Fendt, 2020). These are beneficial when the metabolic gene/protein of interest has a well-established primary substrate and can be efficiently delivered to the targeted tissue.
Critical Parameters
Although enzyme availability helps determine cell-type-exclusive metabolic ability, it cannot be used to quantify the levels of metabolic activity. For example, a low-expressing enzyme may display high enzyme activity, which cannot be detected by gene or protein expression levels. Additional investigation combining gene deletion, inhibitor treatments, and metabolic profiling will be necessary to complete the characterization for the identified cell-type-specific metabolic signatures. Another critical point is that, although less susceptible than the metabolome, RNA and protein profiling are not always resistant to the tissue dissociation processes. Recent studies have reported significant artifacts, including the unexpected stress-induced gene expression and the selection bias favoring stress-resistant cell types (Butler, Hoffman, Smibert, Papalexi, & Satija, 2018; Denisenko et al., 2020). Finally, the instruments and settings for mass spectrometry analyses indicated in Basic Protocols 3 and 4 are based on the current setup in our laboratories, and they are meant to be of an illustrative nature. Instruments from other vendors or alternative acquisition methods might be employed for the quantitative mass spectrometry analyses.
Acknowledgments
This work was supported by funding from NIH/R00 (DK123407) to C.-W.C. and S.R., and the Else Kröner Fresenius Stiftung (award number: 2019_A79) to A.O. The Fritz Lipmann Institute (FLI) is a member of the Leibniz Association and is financially supported by the Federal Government of Germany and the State of Thuringia.
Author Contributions
Nadja Gebert : formal analysis, methodology, writing original draft, writing review an editing; Shahadat Rahman : methodology, validation, writing review and editing; Caroline A. Lewis : methodology, writing original draft, writing review and editing; Alessandro Ori : formal analysis, investigation, methodology, supervision, writing original draft, writing review and editing; Chia-Wei Cheng : conceptualization, funding acquisition, methodology, supervision, validation, writing original draft, writing review and editing
Conflict of Interest
The authors declare no conflict of interest.
Open Research
Data Availability Statement
The data that support the findings of this study are openly available at https://gitlab.leibniz-fli.de/ori-lab/metabolic-signatures-in-stem-cells/.
Literature Cited
- Barker, N., van Es, J. H., Kuipers, J., Kujala, P., van den Born, M., Cozijnsen, M., … Clevers, H. (2007). Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature , 449(7165), 1003–1007. doi: 10.1038/nature06196.
- Baulies, A., Angelis, N., & Li, V. S. W. (2020). Hallmarks of intestinal stem cells. Development , 147(15), dev182675. doi: 10.1242/dev.182675.
- Bensard, C. L., Wisidagama, D. R., Olson, K. A., Berg, J. A., Krah, N. M., Schell, J. C., … Rutter, J. (2020). Regulation of tumor initiation by the mitochondrial pyruvate carrier. Cell Metabolism , 31(2), 284–300.e287. doi: 10.1016/j.cmet.2019.11.002.
- Birsoy, K., Wang, T., Chen, W. W., Freinkman, E., Abu-Remaileh, M., & Sabatini, D. M. (2015). An essential role of the mitochondrial electron transport chain in cell proliferation is to enable aspartate synthesis. Cell , 162(3), 540–551. doi: 10.1016/j.cell.2015.07.016.
- Bruderer, R., Muntel, J., Muller, S., Bernhardt, O. M., Gandhi, T., Cominetti, O., … Reiter, L. (2019). Analysis of 1508 plasma samples by capillary-flow data-independent acquisition profiles proteomics of weight loss and maintenance. Molecular and Cellular Proteomics , 18(6), 1242–1254. doi: 10.1074/mcp.RA118.001288.
- Butler, A., Hoffman, P., Smibert, P., Papalexi, E., & Satija, R. (2018). Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nature Biotechnology , 36(5), 411–420. doi: 10.1038/nbt.4096.
- Calibasi-Kocal, G., Mashinchian, O., Basbinar, Y., Ellidokuz, E., Cheng, C. W., & Yilmaz, O. H. (2021). Nutritional control of intestinal stem cells in homeostasis and tumorigenesis. Trends in Endocrinology and Metabolism , 32(1), 20–35. doi: 10.1016/j.tem.2020.11.003.
- Cheng, C. W., Biton, M., Haber, A. L., Gunduz, N., Eng, G., Gaynor, L. T., … Yilmaz, O. H. (2019). Ketone body signaling mediates intestinal stem cell homeostasis and adaptation to diet. Cell , 178(5), 1115–1131.e1115. doi: 10.1016/j.cell.2019.07.048.
- Cheng, C. W., Yilmaz, O. H., & Mihaylova, M. M. (2020). Strategies for measuring induction of fatty acid oxidation in intestinal stem and progenitor cells. Methods in Molecular Biology , 2171, 53–64. doi: 10.1007/978-1-0716-0747-3_4.
- Corcoran, C. C., Grady, C. R., Pisitkun, T., Parulekar, J., & Knepper, M. A. (2017). From 20th century metabolic wall charts to 21st century systems biology: Database of mammalian metabolic enzymes. American Journal of Physiology, Renal Physiology , 312(3), F533–F542. doi: 10.1152/ajprenal.00601.2016.
- Demichev, V., Messner, C. B., Vernardis, S. I., Lilley, K. S., & Ralser, M. (2020). DIA-NN: Neural networks and interference correction enable deep proteome coverage in high throughput. Nature Methods , 17(1), 41–44. doi: 10.1038/s41592-019-0638-x.
- Denisenko, E., Guo, B. B., Jones, M., Hou, R., de Kock, L., Lassmann, T., … Forrest, A. R. R. (2020). Systematic assessment of tissue dissociation and storage biases in single-cell and single-nucleus RNA-seq workflows. Genome Biology , 21(1), 130. doi: 10.1186/s13059-020-02048-6.
- DeVilbiss, A. W., Zhao, Z., Martin-Sandoval, M. S., Ubellacker, J. M., Tasdogan, A., Agathocleous, M., … Morrison, S. J. (2021). Metabolomic profiling of rare cell populations isolated by flow cytometry from tissues. Elife , 10, e61980. doi: 10.7554/eLife.61980.
- Donovan, J., & Brown, P. (2006). Euthanasia. Current Protocols in Immunology , 73, 1.8.1–1.8.4. doi: 10.1002/0471142735.im0108s73.
- Fernandez-Garcia, J., Altea-Manzano, P., Pranzini, E., & Fendt, S. M. (2020). Stable isotopes for tracing mammalian-cell metabolism in vivo. Trends in Biochemical Sciences , 45(3), 185–201. doi: 10.1016/j.tibs.2019.12.002.
- Fiehn, O. (2016). Metabolomics by gas chromatography-mass spectrometry: Combined targeted and untargeted profiling. Current Protocols in Molecular Biology , 114, 30.34.31–30.34.32. doi: 10.1002/0471142727.mb3004s114.
- Gebert, N., Cheng, C. W., Kirkpatrick, J. M., Di Fraia, D., Yun, J., Schadel, P., … Ori, A. (2020). Region-specific proteome changes of the intestinal epithelium during aging and dietary restriction. Cell Reports , 31(4), 107565. doi: 10.1016/j.celrep.2020.107565.
- Haber, A. L., Biton, M., Rogel, N., Herbst, R. H., Shekhar, K., Smillie, C., … Regev, A. (2017). A single-cell survey of the small intestinal epithelium. Nature , 551(7680), 333–339. doi: 10.1038/nature24489.
- Liu, Y., Beyer, A., & Aebersold, R. (2016). On the dependency of cellular protein levels on mRNA abundance. Cell , 165(3), 535–550. doi: 10.1016/j.cell.2016.03.014.
- Macosko, E. Z., Basu, A., Satija, R., Nemesh, J., Shekhar, K., Goldman, M., … McCarroll, S. A. (2015). Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell , 161(5), 1202–1214. doi: 10.1016/j.cell.2015.05.002.
- McMurray, C. H., Blanchflower, W. J., & Rice, D. A. (1984). Automated kinetic method for D-3-hydroxybutyrate in plasma or serum. Clinical Chemistry , 30(3), 421–425.
- Morris, S. A. (2019). The evolving concept of cell identity in the single cell era. Development , 146(12), dev169748. doi: 10.1242/dev.169748.
- Perez-Ramirez, C. A., & Christofk, H. R. (2021). Challenges in studying stem cell metabolism. Cell Stem Cell , 28(3), 409–423. doi: 10.1016/j.stem.2021.02.016.
- Pino, L. K., Searle, B. C., Bollinger, J. G., Nunn, B., MacLean, B., & MacCoss, M. J. (2020). The Skyline ecosystem: Informatics for quantitative mass spectrometry proteomics. Mass Spectrometry Reviews , 39(3), 229–244. doi: 10.1002/mas.21540.
- Rodriguez-Colman, M. J., Schewe, M., Meerlo, M., Stigter, E., Gerrits, J., Pras-Raves, M., … Burgering, B. M. (2017). Interplay between metabolic identities in the intestinal crypt supports stem cell function. Nature , 543(7645), 424–427. doi: 10.1038/nature21673.
- Rost, H. L., Aebersold, R., & Schubert, O. T. (2017). Automated SWATH data analysis using targeted extraction of ion chromatograms. Methods in Molecular Biology , 1550, 289–307. doi: 10.1007/978-1-4939-6747-6_20.
- Schell, J. C., Wisidagama, D. R., Bensard, C., Zhao, H., Wei, P., Tanner, J., … Rutter, J. (2017). Control of intestinal stem cell function and proliferation by mitochondrial pyruvate metabolism. Nature Cell Biology , 19(9), 1027–1036. doi: 10.1038/ncb3593.
- Schuler, S. C., Gebert, N., & Ori, A. (2020). Stem cell aging: The upcoming era of proteins and metabolites. Mechanisms of Ageing and Development , 190, 111288. doi: 10.1016/j.mad.2020.111288.
- Schuler, S. C., Kirkpatrick, J. M., Schmidt, M., Santinha, D., Koch, P., Di Sanzo, S., … von Maltzahn, J. (2021). Extensive remodeling of the extracellular matrix during aging contributes to age-dependent impairments of muscle stem cell functionality. Cell Reports , 35(10), 109223. doi: 10.1016/j.celrep.2021.109223.
- Schwanhausser, B., Busse, D., Li, N., Dittmar, G., Schuchhardt, J., Wolf, J., … Selbach, M. (2011). Global quantification of mammalian gene expression control. Nature , 473(7347), 337–342. doi: 10.1038/nature10098.
- Shapira, S. N., & Christofk, H. R. (2020). Metabolic regulation of tissue stem cells. Trends in Cell Biology , 30(7), 566–576. doi: 10.1016/j.tcb.2020.04.004.
- Tran, N. H., Qiao, R., Xin, L., Chen, X., Liu, C., Zhang, X., … Li, M. (2019). Deep learning enables de novo peptide sequencing from data-independent-acquisition mass spectrometry. Nature Methods , 16(1), 63–66. doi: 10.1038/s41592-018-0260-3.
- Urbauer, E., Rath, E., & Haller, D. (2020). Mitochondrial metabolism in the intestinal stem cell niche-sensing and signaling in health and disease. Frontiers in Cell and Developmental Biology , 8, 602814. doi: 10.3389/fcell.2020.602814.
- Wang, D., Odle, J., & Liu, Y. (2021a). Metabolic regulation of intestinal stem cell homeostasis. Trends in Cell Biology , 31(5), 325–327. doi: 10.1016/j.tcb.2021.02.001.
- Wang, D., Odle, J., & Liu, Y. (2021b). Metabolic regulation of intestinal stem cell homeostasis. Trends in Cell Biology , 31(5), 325–327. doi: 10.1016/j.tcb.2021.02.001.
- Xia, J., Psychogios, N., Young, N. and Wishart, D.S. (2009). MetaboAnalyst: a web server for metabolomic data analysis and interpretation. Nucleic Acids Research , 37, W652–660.
Corrections
In this publication, a note about the use of human or animal subjects has been added.
The current version online now includes this information and may be considered the authoritative version of record.
Citing Literature
Number of times cited according to CrossRef: 1
- Shahadat Rahman, Xi Lan, Christopher Terranova, Rayan El-Kholdi, Omer H. Yilmaz, Chia-Wei Cheng, Epigenetic and Transcriptional Dynamics of Notch Program in Intestinal Differentiation, Intestinal Differentiated Cells, 10.1007/978-1-0716-3076-1_7, (77-88), (2023).

