How to Illuminate the Dark Proteome Using the Multi-omic OpenProt Resource
Marie A. Brunet, Marie A. Brunet, Amina M. Lekehal, Amina M. Lekehal, Xavier Roucou, Xavier Roucou
Abstract
Ten of thousands of open reading frames (ORFs) are hidden within genomes. These alternative ORFs, or small ORFs, have eluded annotations because they are either small or within unsuspected locations. They are found in untranslated regions or overlap a known coding sequence in messenger RNA and anywhere in a “non-coding” RNA. Serendipitous discoveries have highlighted these ORFs’ importance in biological functions and pathways. With their discovery came the need for deeper ORF annotation and large-scale mining of public repositories to gather supporting experimental evidence. OpenProt, accessible at https://openprot.org/, is the first proteogenomic resource enforcing a polycistronic model of annotation across an exhaustive transcriptome for 10 species. Moreover, OpenProt reports experimental evidence cumulated across a re-analysis of 114 mass spectrometry and 87 ribosome profiling datasets. The multi-omics OpenProt resource also includes the identification of predicted functional domains and evaluation of conservation for all predicted ORFs. The OpenProt web server provides two query interfaces and one genome browser. The query interfaces allow for exploration of the coding potential of genes or transcripts of interest as well as custom downloads of all information contained in OpenProt. © 2020 The Authors.
Basic Protocol 1 : Using the Search interface
Basic Protocol 2 : Using the Downloads interface
INTRODUCTION
Historically, open reading frames (ORFs) shorter than 100 codons were discarded from genome annotations unless previously characterized, as they were deemed too short to be functional (Cheng et al., 2011). This length criterion, alongside requirement of an ATG start codon and the restriction of a single coding sequence per transcript, has considerably shaped and limited the exploration of the proteome (Brunet, Levesque, Hunting, Cohen, & Roucou, 2018; Hellens, Brown, Chisnall, Waterhouse, & Macknight, 2016; Olexiouk & Menschaert, 2016; Orr, Mao, Storz, & Qian, 2019). Deeper ORF annotation is key to functional proteomic discoveries and a better understanding of physiological and pathological mechanisms (Brunet et al., 2018; Ma et al., 2014; Menschaert et al., 2013; Samandi et al., 2017). With the development of ribosome profiling (Ingolia, 2014), a technique detecting ribosome-protected fragments (footprints) originating from translating ribosomes, all translation events across the genome can potentially be captured (Ingolia, 2016). This observation led the community to use ribosome profiling to capture a deeper ORF landscape and identify small ORF (sORF) candidates for functional characterization (Andreev et al., 2015a, 2015b; Bazzini et al., 2014; Chen et al., 2020; Menschaert et al., 2013). Several repositories of sORFs have been published, all relying on ribosome profiling data for ORF annotation (Hao et al., 2017; Olexiouk, Van Criekinge, & Menschaert, 2018; Xie et al., 2016). Despite being an undeniable resource for novel ORF identification, the ribosome profiling technique still presents biases that can hinder detection of functional yet non-annotated ORFs, including ORFs in low-abundance transcripts or in repetitive regions (Brar & Weissman, 2015; Brunet et al., 2018; Ingolia, 2014; Ingolia, Ghaemmaghami, Newman, & Weissman, 2009; Raj et al., 2016).
At OpenProt (Brunet et al., 2019), we computationally predict all possible alternative ORFs (altORFs) from an exhaustive transcriptome. All known transcripts are retrieved from both Ensembl and NCBI RefSeq annotations (O'Leary et al., 2016; Yates et al., 2020), and after in silico translation, all ORFs starting with an ATG and longer than 30 codons are listed. The predicted proteins are then divided into three categories (Table 1): known proteins are called refProts, non-annotated proteins similar to a known protein in the same gene are called novel isoforms, and non-annotated proteins with no significant similarity to a known protein in the same gene are called altProts. Such a computational strategy allows for annotation of an exhaustive set of ORFs, yet it also certainly results in false positives. Thus, OpenProt evaluates protein conservation and mines ribosome profiling and proteomics datasets to cumulate experimental evidence for all predicted proteins (Barrett et al., 2013; Deutsch et al., 2020; Perez-Riverol et al., 2019). All evidence is listed in the OpenProt resource, allowing the user an in-depth review of evidence for any protein supported by OpenProt. OpenProt currently supports 10 species and explores 114 proteomics and 87 ribosome profiling datasets. For a complete overview of the OpenProt resource, including the computational and analytic methods, we refer the user to the original publication (Brunet et al., 2019). The web server also contains a detailed help section with tutorials and frequently asked questions (https://openprot.org/p/help).
| OpenProt name | Description | Accession number |
|---|---|---|
| RefProt | Known protein present in current annotations (Ensembl and/or RefSeq) and/or UniProtKB |
ENSP*** NP_*** or XP_*** UniProt accession |
| Isoform | Non-annotated protein with high homology to a known protein from the same gene | II_*** |
| AltProt | Non-annotated protein with no significant homology to a known protein from the same gene | IP_*** |
Basic Protocol 1 described here guides the novice user in how to explore ORFs using the Search interface. This protocol is designed to provide a rapid view of the coding potential and translation products of genes of interest. Basic Protocol 2 describes how to download custom data from the OpenProt resource. Guidelines for investigation of a specific altORF are provided afterward, alongside discussion on critical parameters.
Basic Protocol 1: USING THE SEARCH INTERFACE
This protocol details the use of the Search interface to query specific genes, transcripts, or proteins. The interface is optimized to accommodate many questions that a researcher may have. For example, a researcher may want to know if a specific gene contains novel ORFs with supporting experimental evidence or whether a given transcript may contain several ORFs. The protocol will guide novice users in how to exploit the Search interface of the OpenProt resource. First, the protocol describes how to navigate to the Search interface from the homepage. Then, it details the features available on the interface to tailor the results to any query. Finally, the protocol explains how to investigate a specific ORF of interest.
Necessary Resources
- OpenProt is accessible via all major web browsers supporting JavaScript, such as Safari, Firefox, Chrome, or Internet Explorer. All pages can be viewed on mobile phones, but the interfaces have been optimized for display on computers or tablets. The Search interface is designed for exploration of specific genes, transcripts, and/or proteins. The interface is accessible via the homepage by clicking on the “Search” tab or can be accessed directly at https://openprot.org/p/altorfDbView. The Downloads interface is designed for custom downloads of any data in the OpenProt resource (see Basic Protocol 2). The interface is accessible via the homepage by clicking on the “Downloads” tab or can be accessed directly at https://openprot.org/p/download. Each protein annotated in OpenProt has a dedicated page containing a genome browser and all supporting information.
Navigating from the homepage to the Search interface
1.Visit OpenProt homepage (https://openprot.org/).
2.Hover cursor over each tab of the navigation bar at the top of the page to highlight it and click to navigate to Search interface (see steps 3 to 5) or another desired page (for Downloads, see Basic Protocol 2).

| Navigation tab | Description |
|---|---|
| Home | The Home tab navigates to the OpenProt homepage. The page contains general information about the web server, the reasons to use the OpenProt resource, links to detailed tutorials, and an overview of the concept and methods behind OpenProt. |
| Browse | The Browse tab navigates to a genome browser for each species, with customizable tracks, allowing visualization of all ORFs present in OpenProt. |
| Search | The Search tab navigates to the OpenProt query interface. |
| Downloads | The Downloads tab navigates to a query interface for custom downloads. |
| About | The About tab navigates to a page containing general information about the resource, the developers and funding agencies, and OpenProt publications. |
| Help | The Help tab navigates to a page containing detailed tutorials and frequently asked questions (FAQs) about OpenProt. |
Exploring the Search interface
3.Use main filters to define a search.

| Category | Name | #a | Description | Notes |
|---|---|---|---|---|
| Main filters (circles) | Species | 1 | List of species supported by OpenProt |
Supported species: - All species - Homo sapiens (Hs) - Pan troglodytes (Pt) - Mus musculus (Mm) - Rattus norvegicus (Rn) - Bos taurus (Bt) - Ovis aries (Oa) - Danio rerio (Dr) - Drosophila melanogaster (Dm) - Caenorhabditis elegans (Ce) - Saccharomyces cerevisiae S288c (Sc) |
| Assembly | 2 | List of supported genome assemblies |
Supported assemblies: - Hs: GRCh38.p5 - Pt: CHIMP2.1.4 - Mm: GRCm38.p4 - Rn: Rnor_6.0 - Bt: UMD_3.1 - Oa: Oar_v3.1 - Dr: GRCz10 - Dm: Release 6 plus ISO1 MT - Ce: WBcel235 - Sc: R64 |
|
| Annotation | 3 | List of supported annotations |
Supported annotations: - Hs: GRCh38.p7/GRCh38.83 - Pt: CHIMP2.1.4/CHIMP2.1.4.87 - Mm: GRCm38.p4/GRCm38.84 - Rt: Rnor_6.0/Rnor_6.0.84 - Bt: UMD_3.1/UMD_3.1.86 - Oa: Oar_v3.1/Oar_v3.1.89 - Dr: GRCz10/GRCz10.84 - Dm: BDGP6/BDGP6.84 - Ce: WBcel235/WBcel235.84 - Sc: R64/R64.83 |
|
| Gene | 4 | Query box to input genes of interest | Accepted formats are Entrez, Ensembl, and/or RefSeq gene accessions. | |
| Transcript | 5 | Query box to input transcripts of interest | Accepted formats are Ensembl and/or RefSeq transcript accessions. | |
| Protein | 6 | Query box to input proteins of interest | Accepted formats are Ensembl, RefSeq, and/or UniProt protein accessions. | |
| Filters (squares): any combination of… | Experimental evidence | A | Selection of proteins with experimental evidence | Experimental evidence is MS and/or Ribo-seq. |
| MS | B | Selection of proteins detected by MS | The list of re-analyzed MS datasets is available under the help section | |
| Ribo-seq | C | Selection of proteins detected by Ribo-seq | The list of re-analyzed Ribo-seq datasets is available in the help section. | |
| Domains | D | Selection of proteins with predicted functional domains | Predictions are done using InterProScan. | |
| AltProts | E | Selection of alternative proteins | This filters the results to only display altProts. | |
| Isoforms | F | Selection of novel isoforms | This filters the results to only display novel isoforms. | |
| Advanced search filters (squares): only appears after clicking on “edit criteria” | Sequence | G | Query box to input an amino acid sequence of interest | This filters the results to only display proteins containing that exact sequence. |
| Type or localization | H | List of supported types of RNA and altORF localizations |
Available options: - 5′UTR: altORF located in the 5′UTR of an mRNA - 3′UTR: altORF located in the 3′UTR of an mRNA - CDS: altORF overlapping a canonical ORF in an mRNA - ncRNA: non-coding RNA - mRNA: messenger RNA |
|
| Reading frame | I | List of possible reading frames |
The +1 frame is assigned to the first nucleotide of the transcript. Available options are 1, 2, or 3. |
|
| Active (triangles) | Order by | a | List of supported sorting rules for the table of results |
The MS, TE, and predicted Domains scores are always sorted in descending order. Available sorting orders: - MS > TE > Domains - Domains > MS > TE - TE > MS > Domains - MW (ascending) > MS > TE > Domains - MW (descending) > MS > TE > Domains - PL (ascending) > MS > TE > Domains - PL (descending) > MS > TE > Domains |
| Column settings | b | Selection of columns to be present in the results table | This opens a pop-up with tick boxes to select the desired columns. | |
| Download TSV | c | Link to download the results of a search | This starts the download of the results as a .tsv file. | |
| Download FASTA | d | Link to download the results of a search | This starts the download of the results as a .FASTA file. |
- a
Symbols and abbreviations: #, flag name in Figure 2; FASTA, text-based format with amino acid sequences of proteins alongside their accession; MS, mass spectrometry; MW, molecular weight; PL, protein length; TSV, tab-separated value; TE, translation event; UTR, untranslated region.
4.Use additional filters to refine the output of the query. Click on “edit search criteria” to open advanced criteria (framed by a gray dashed line in Fig. 2).
5.Use “order by” and “column settings” filters to sort and arrange the table of results.
Exploring the table of results
6.Click on blue box “update search results” (Fig. 2) to view the table of results (Fig. 3).
| Name | Description | Notes |
|---|---|---|
| Protein accession | All proteins annotated in OpenProt have a unique accession number. The unicity is based on the amino acid sequence within a species. Protein accession numbers for refProts are accessions from Ensembl, NCBI RefSeq, and/or UniProtKB. | Accessions start with IP for altProts. Accessions start with II for novel predicted isoforms of refProts. |
| Protein types |
AltProts are predicted from translation of altORFs within an mRNA or ncRNA. RefProts are known from translation of canonical CDSs (mRNA). Isoforms are predicted from translation of altORFs within an mRNA and share clear sequence homology with a RefProt from the same gene. |
Possible entries are RefProt, Isoform, or AltProt. “AltProt” is written in red. |
| Protein length | The length of the protein is reported in amino acids (a.a.). | OpenProt annotates all known proteins and any novel protein longer than 30 amino acids. |
| Experimental evidence: MS | This column reports the mass spectrometry (MS) score for the given protein. | The MS score corresponds to the sum of unique peptides detected per study. |
| Experimental evidence: TE | This column reports the translation event (TE) score for the given protein. | The TE score corresponds to the sum of studies with at least one significant detection of translation. |
| Functional prediction: Domains | This column reports the number of predicted functional domains for the given protein. | Prediction of functional domains is done using InterProScan. |
| Functional prediction: Orthology | This column reports the number of species with at least one ortholog for the given protein, as well as the species concerned. | The species names are abbreviated using the first letters of the species and subspecies, and they are colored based on the identity percentage of the orthologous protein pair (the darker, the higher). |
| Species | This column indicates the species from which the given protein originates. | OpenProt supports 10 species (see Table 3). |
| Gene | This column indicates the gene from which the given protein originates. | The gene name is retrieved from the annotation (Ensembl and/or NCBI RefSeq). |
| Transcript accession | This column indicates the accession number of the transcript from which the given protein originates. | The transcript accession is retrieved from the annotation (Ensembl and/or NCBI RefSeq). |
| Type | This column indicates the type of transcript from which the given protein originates. | Possible entries are ncRNA (non-coding RNA) or mRNA (messenger RNA). |
| Localization | This column indicates the localization of the given altProt on the transcript relative to the canonical protein associated with this transcript. |
Within mRNAs, the localization of altORFs is defined according to the localization of the predicted start codon with respect to that of the refProt. Possible entries are 5′UTR, CDS (overlapping), and 3′UTR. For altORFs within ncRNAs, no localization is inferred. |
| Details | This column contains a link to the page dedicated to the given protein. | This page is detailed in Table 5. |
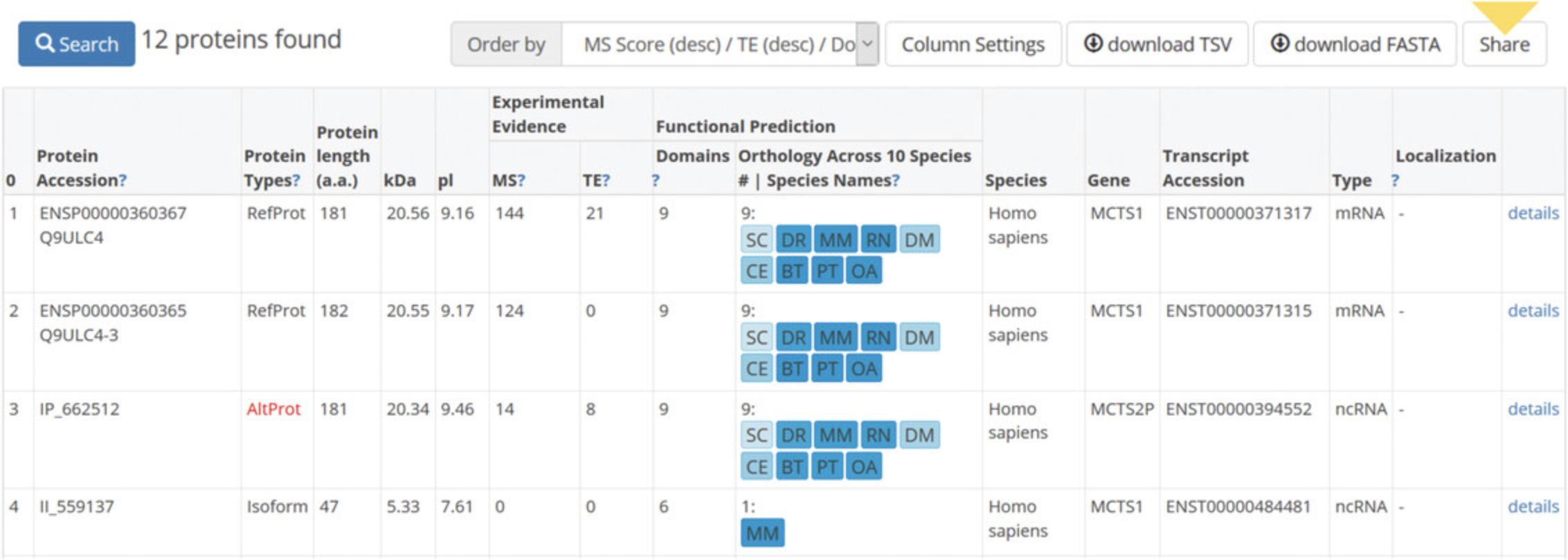
7.Go to bottom of the table to navigate between the different pages of results.
8.Click “share” button, which appears at the top right of the table, next to the sorting and download options, to display a shareable link to this specific search result.
Inspecting a specific protein
9.Click “details” link in the main table of results to navigate to a page dedicated to the queried protein.

| Category | Name | #a | Description | Notes |
|---|---|---|---|---|
| Genome browser | Genome browser | A | The genome browser is centered on the investigated protein. | The browser is controlled by the zoom controls, and keyboard shortcuts can be visualized by clicking on the question mark at the top right corner. |
| Genome | 1 | This track is the genome track. | This track allows you to navigate the genome and visualize up to the nucleotide sequence when sufficiently zoomed. | |
| Transcript | 2 | This track is the transcript track. | This track allows you to visualize the transcript on which the given protein is encoded. | |
| Protein | 3 | This track is the protein track. | This track allows you to visualize the given protein. | |
| Peptide detection | 4 | This track is the mass spectrometry–based peptide detection track. | This track allows you to visualize the peptides that have been identified by mass spectrometry for the given protein. | |
| Browser legend | 5 | At the bottom of the genome browser is the colored legend. |
The legend is as follows: Blue: transcript Green: refProt and identified peptides matching the given refProt Orange: novel isoform and identified peptides matching the given novel isoform Red: altProt and identified peptides matching the given altProt |
|
| General information table | Information table | B | This table regroups general information on the queried protein. | Each line in the table corresponds to the same protein from a different transcript from the same gene. |
| Update browser | - | This tick box controls which transcript is visualized on the genome browser. | Each line corresponds to a different transcript but to the same protein (same amino acid sequence). | |
| Gene | - | This column contains the name of the gene from which the queried protein originates. | In rare exceptions, Ensembl and NCBI RefSeq annotations might not use the same synonym for the gene name. | |
| Annotation | - | This column contains the annotation from which the queried protein is derived. | All supported annotations are listed in Table 3. | |
| Genomic coordinates | - | This column contains the genomic coordinates of the queried protein. | These coordinates do not correspond to the gene or the transcript, but rather to the queried protein mapped back onto the genome. | |
| Strand | - | This column indicates on which genomic strand the queried protein is encoded. | The strand is retrieved from the annotation. | |
| Transcript | - | This column indicates the accession of the transcript from which the queried protein originates. | Transcript accessions are retrieved from the annotation. Click on the accession to navigate to the annotation page of the transcript. | |
| Type | - | This column contains the type of the transcript from which the queried protein originates. | Transcript types are either mRNA (messenger RNA) or ncRNA (non-coding RNA) and are derived from the annotation. | |
| ORF information | Frame | - | This column contains the reading frame of the transcript from which the queried protein originates. | The +1 frame is assigned to the first nucleotide of the transcript. |
| Kozak | - | This column indicates whether the queried ORF is preceded by a Kozak sequence. | The Kozak sequence is derived from the literatureb and is as follows: RNNATGG (where R = A or G and N = any of A, T, C, or G). | |
| High-eff. TIS | - | This column indicates whether the queried ORF is preceded by a high- efficiency TIS motif. | The high-efficiency TIS motif is derived from the literaturec and is as follows: RYMRMVAUGGC (where Y = U or C, M = A or C, R = A or G, and V = A, C, or G). | |
| Localization | - | This column indicates the localization of the queried altProt relative to the canonical protein associated with this transcript. |
Within mRNAs, the localization of altORFs is defined according to the localization of the predicted start codon with respect to that of the refProt. Possible entries are 5′UTR, refpor (overlapping), and 3′UTR. |
|
| - | For altORFs within ncRNAs, no localization is inferred. | |||
| Transcript coordinates | - | This column contains the transcript coordinates of the queried protein. | The transcript coordinates of the queried protein are from the first nucleotide of the start codon to the last nucleotide of the stop codon (1 = first nucleotide of the transcript). | |
| Sequences | Protein | - | This column contains a link to the amino acid sequence of the queried protein. | The link opens a pop-up message with the amino acid sequence of the queried protein. |
| DNA | - | This column contains a link to the nucleotide sequence of the queried protein. | The link opens a pop-up message with the nucleotide sequence of the ORF encoding the queried protein. | |
| Protein page tabs | Mass spectrometry | a | This tab navigates to a page listing mass spectrometry–based evidence for the queried protein. | The mass spectrometry tab is detailed in Figure 5. |
| Translation | b | This tab navigates to a page listing ribosome profiling–based evidence for the queried protein. | The translation tab is detailed in Figure 5. | |
| Domains | c | This tab navigates to a page listing functional domain predictions for the queried protein. | The domains tab is detailed in Figure 5. | |
| Conservation | This tab navigates to a page listing orthologs and paralogs of the queried protein. | The conservation tab is detailed in Figure 5. |
- a
Symbols and abbreviations: #, flag name in Figure 4; TIS, translation initiation sequence.
- b
PMIDs are 7301588 and 12459250.
- c
PMID is 25170020.

10.Use genome browser to visualize the queried protein and the associated transcript.
11.See summary table located below the genome browser to view general information on the queried protein.
12.Click on “mass spectrometry” tab to review experimental detection of the queried protein in mass spectrometry–based proteomic datasets.
13.Click on “translation” tab to review experimental detection of translation of the queried ORF in ribosome profiling datasets.
14.Click on “domains” tab to review prediction of functional domains for the queried protein using multiple domain annotation databases.
15.Click on “conservation” tab to review conservation of the queried protein across species supported by OpenProt.
Basic Protocol 2: USING THE DOWNLOADS INTERFACE
This protocol details the use of the Downloads interface to retrieve a large amount of data stored in the OpenProt resource. The interface is optimized to obtain custom downloads for specific research questions. For example, a researcher may want to download a FASTA file containing only the sequences of altProts and novel isoforms with experimental evidence or a BED file containing the genomic coordinates of all proteins predicted by OpenProt. This protocol will guide novice users in how to exploit the Downloads interface of the OpenProt resource. First, the protocol describes how to navigate to the Downloads interface from the homepage. Then, it details the features available on the interface to tailor results to any query. Finally, the protocol explains the different file formats available.
Necessary Resources
- See Basic Protocol 1.
Navigating from the homepage to the Downloads interface
1.Navigate to Downloads interface according to Basic Protocol 1, steps 1 and 2.
Exploring the Downloads interface
2.Use query filters to define a search.
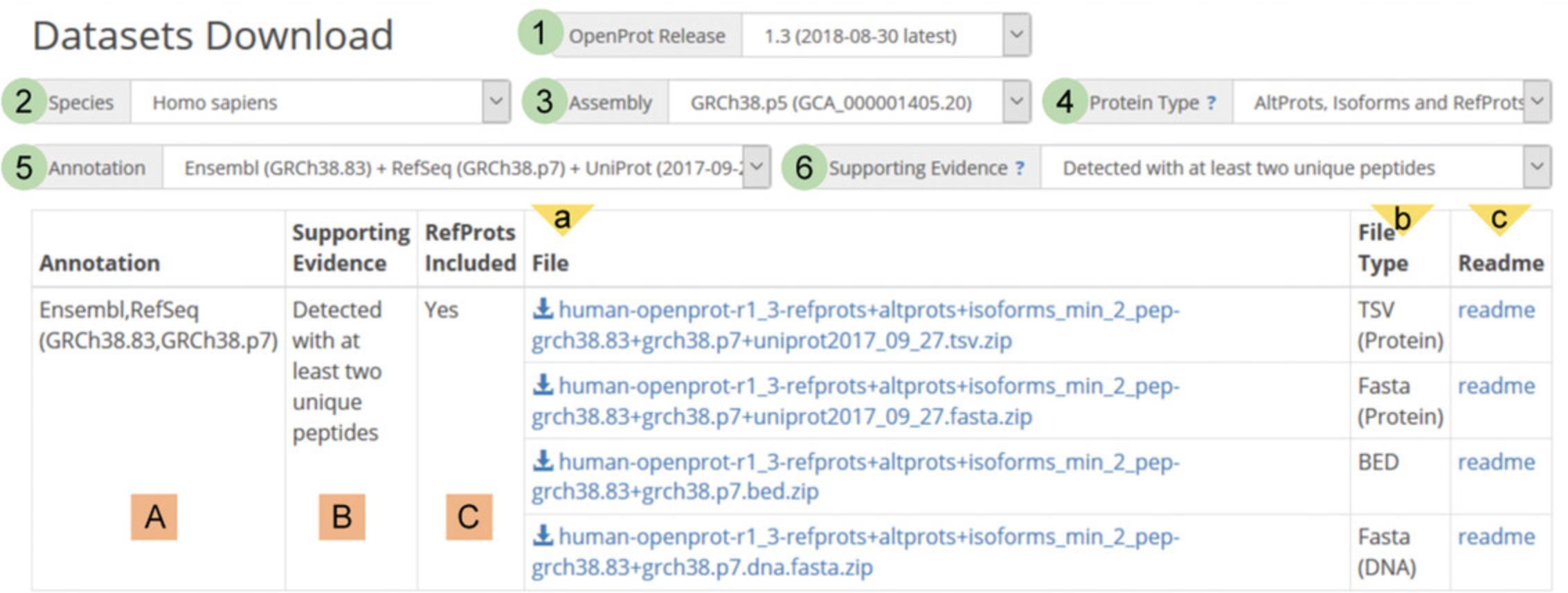
| Category | Name | # | Description | Notes |
|---|---|---|---|---|
| Query interface (circles) | OpenProt release | 1 | List of available OpenProt releases (currently v1.3). | OpenProt is a release-based resource to ensure up-to-date, continuous availability of all the data over time. |
| Species | 2 | List of species supported by OpenProt. | For downloads, all species cannot be selected at once, as this would lead to files of excessive size. | |
| Assembly | 3 | List of genome assemblies supported by OpenProt. |
Supported assemblies: - Hs: GRCh38.p5 - Pt: CHIMP2.1.4 - Mm: GRCm38.p4 - Rn: Rnor_6.0 - Bt: UMD_3.1 - Oa: Oar_v3.1 - Dr: GRCz10 - Dm: Release 6 plus ISO1 MT - Ce: WBcel235 - Sc: R64 |
|
| Protein type | 4 | List of protein types to include in the downloadable files. |
Supported entries: - AltProts and Isoforms = novel proteins predicted by OpenProt - RefProts = known proteins - AltProts, Isoforms, and RefProts = all proteins in OpenProt |
|
| Annotation | 5 | List of supported annotations to include in the downloadable files. | If “AltProts and Isoforms” is chosen as the protein type, the user can choose to include both Ensembl and NCBI RefSeq annotations or either one. | |
| Supporting evidence | 6 | List of available filters on experimental evidence. |
Supported entries: - Detected with at least one unique peptide - Detected with at least two unique peptides - All predicted |
|
| Summary table (squares) | Annotation | A | This column indicates the annotation used in the downloadable files. | This column corresponds to the choice made in the “annotation” box of the query interface (#5 above). |
| Supporting evidence | B | This column indicates the level of supporting evidence used to filter results to include in the downloadable files. | This column corresponds to the choice made in the “protein type” box of the query interface (#4 above). | |
| RefProts included | C | This column indicates whether known proteins are included in the downloadable files. | This column is in line with the choice made on included proteins (#4 above). | |
| File information (triangles) | File | a | This column contains all the downloadable files fitting the search criteria indicated on the query interface. | Click on the file name to start the download. |
| File type | b | TSV (protein): tab-separated values file |
Each TSV file contains the following headers: - Protein accession - Protein type - Protein length - Molecular weight - Isoelectric point - Reading frame - Gene symbol - Chromosome - Genomic coordinates (start) - Genomic coordinates (end) - Strand - Transcript accession - Transcript type - Localization - Transcript coordinates (start) - Transcript coordinates (end) - MS score - TE score - Orthology - Kozak motif - High-efficiency TIS motif - Domains |
|
| FASTA (protein): text-based format with amino acid sequences of proteins alongside their accession |
Each header contains the following: - The protein identifier - Taxonomy (TX) - Organism name (OS) - Gene name (GN) - Transcript accession (TA) The identifier parse rule is >(.*)\| The description parse rule is >(.*) |
|||
| BED: browser extensible data |
Each BED file contains the following information: - Chromosome - chromStart - chromEnd - Protein accession - Score - Strand - thickStart - thickEnd - itemRgb - blockCount - blockSizes - blockStarts |
|||
| FASTA (DNA): text-based format with nucleotide sequences encoding proteins alongside their accession |
Each header contains the following: - The protein identifier - Taxonomy (TX) - Organism name (OS) - Gene name (GN) - Transcript accession (TA) The identifier parse rule is >(.*)\| The description parse rule is >(.*) |
|||
| Readme | c | This column contains a link to open a pop-up Readme file on the corresponding downloadable file. | Files can also be downloaded from the Readme pop-up. |
3.Click on desired file name to start the download.
GUIDELINES FOR UNDERSTANDING RESULTS
OpenProt is a proteogenomic resource that seeks experimental evidence for predicted novel proteins from non-annotated ORFs (Brunet et al., 2019). OpenProt is open source, all methods and codes are published and freely available (Brunet et al., 2019; Samandi et al., 2017), and all supported data are freely accessible and downloadable (Basic Protocols 1 and 2). At OpenProt, we predict all possible ORFs longer than 30 codons throughout the annotated transcriptome for 10 species. This approach was chosen to be as inclusive as possible for predictions and to then retrieve experimental evidence for each prediction. Thus, OpenProt is not dependent on a specific experimental bias, but the user has to be aware that false positives are a reality with such design. Not all predicted proteins in OpenProt are likely expressed. Thus, because noise and nonspecific detections vary across experimental datasets and designs, we encourage users to seek experimental detection across multiple datasets to increase confidence in an altProt and/or the existence of a novel isoform.
Broadly, there are two major usages of the OpenProt resource. First, users may be interested in a specific gene or transcript and wonder whether they are capturing its full coding potential. To that end, users should use the OpenProt Search interface (Basic Protocol 1) and investigate each predicted protein in detail (Fig. 5). Second, users may be interested in analyzing their mass spectrometry–based proteomic data with the OpenProt database. Users should use the OpenProt Downloads interface for such a query (Basic Protocol 2). If users wish to tailor their mass spectrometry database to a specific set of transcripts, we encourage them to download the full database and keep only entries of interest based on the transcript accession (TA field in the FASTA header). Users may also use the OpenProt Search interface to query specific transcripts and download the results as a FASTA file. Please note, however, that for computational reasons, such queries are limited to 2000 genes (or transcripts) at a time.
Crucial OpenProt features and considerations heavily depend on the research question behind the query. For any question or additional information on data analysis and interpretation, contact the OpenProt team via the light blue “contact us” button at the bottom of all OpenProt pages (https://groups.google.com/forum/#!forum/openprot).
COMMENTARY
Background Information
The premises of the OpenProt resource were first published in 2013 (Vanderperre et al., 2013). The former HAltORF database was a mere list of altORFs within the human transcriptome (based on the NCBI RefSeq annotation). Community-driven requests and serendipitous discoveries contributed to the desire and need to develop OpenProt as the first proteogenomic resource to enforce a polycistronic annotation model on both coding RNA (messenger RNA, or mRNA) and non-coding RNA (ncRNA) transcripts (Samandi et al., 2017). The OpenProt resource was first officially released in 2019, contains 10 species, and cumulates experimental evidence using mass spectrometry and ribosome profiling data (Brunet et al., 2019). Using cutting-edge algorithms for ribosome profiling and mass spectrometry data mining (Erhard et al., 2018; Vaudel, Barsnes, Berven, Sickmann, & Martens, 2011, 2015), OpenProt re-analyzed 87 and 114 datasets, respectively. OpenProt not only lists novel proteins with experimental evidence but also allows critical assessment of the evidence by the user. OpenProt is constantly re-analyzing datasets and adding new features, but all data are continuously available thanks to the release-based structure of the resource. Suggestions of new features or additional species or datasets from the community are always welcome and can be submitted via the OpenProt discussion forum (https://groups.google.com/forum/#!forum/openprot).
Critical Parameters and Troubleshooting
We refer the user to the original article for explanation of the mass spectrometry pipeline enforced by OpenProt (Brunet et al., 2019), yet one needs to acknowledge the stringent 0.001% false discovery rate (FDR). Such an FDR balances the use of a large database that can affect the false positive rate in proteomics analyses (Jeong, Kim, & Bandeira, 2012; Nesvizhskii, 2014). Thus, an absence of detection by mass spectrometry in OpenProt does not necessarily mean that the protein does not exist. Such a pipeline will heavily hinder the detection of some proteins. As a guideline, the same standard mass spectrometry analysis filtered at a usual 1% FDR or the stringent 0.001% FDR may only share 40 to 80% of identifications depending on the spectral quality of the dataset (unpub. observ.). Similarly, an absence of detection by ribosome profiling does not mean that there is no evidence of translation. At the moment, OpenProt only incorporates ORFs predicted by the translation analysis pipeline (PRICE) that have a perfect overlap with the ORF predicted by OpenProt. Thus, if a start codon is a non-canonical codon upstream or downstream of the ATG predicted by OpenProt, no translation evidence will be reported by OpenProt. Implementation of such cases will be available in the next OpenProt release. Additionally, one should note that the p-value reported by the PRICE algorithm for each detected ORF is the result of a generalized binomial test (not corrected for multiple comparisons). Hence, the p-value indicates the confidence in the given ORF not being attributable to noise.
Finally, for each protein, a list of paralogs and orthologs is provided in the “conservation” tab (described in Fig. 5). The user should note that this list is restricted to species currently supported by OpenProt (listed in Table 3). For a more exhaustive list, the user may want to use the BLASTp tool (Madden, Tatusov, & Zhang, 1996) to search a specific protein against a reference database such as the non-redundant NCBI or the UniProtKB protein database (Bateman et al., 2017; O'Leary et al., 2016). This analysis may identify proteins with significant sequence similarity in various species.
Acknowledgments
X.R. is a member of the Fonds de Recherche du Québec Santé (FRQS)-supported Centre de Recherche du Centre Hospitalier Universitaire de Sherbrooke. This work was supported by a Canada Research Chair in Functional Proteomics and Discovery of Novel Proteins to X.R. We thank M. Brunelle, J-F. Lucier, M. Levesque, and everybody else involved in the continuous development of the OpenProt resource. We thank the team at Calcul Québec and Compute Canada for their support with the use of the supercomputer mp2 from Université de Sherbrooke. Operation of the mp2 supercomputer is funded by the Canada Foundation of Innovation (CFI), le ministère de l’Économie, de la science et de l'innovation du Québec (MESI), and les Fonds de Recherche du Québec.
Literature Cited
- Andreev, D. E., O'Connor, P. B. F., Fahey, C., Kenny, E. M., Terenin, I. M., Dmitriev, S. E., … Baranov, P. V. (2015a). Translation of 5′ leaders is pervasive in genes resistant to eIF2 repression. eLife , 4, e03971. doi: 10.7554/eLife.03971.
- Andreev, D. E., O'Connor, P. B. F., Zhdanov, A. V., Dmitriev, R. I., Shatsky, I. N., Papkovsky, D. B., & Baranov, P. V. (2015b). Oxygen and glucose deprivation induces widespread alterations in mRNA translation within 20 min. Genome Biology , 16, 90. doi: 10.1186/s13059-015-0651-z.
- Barrett, T., Wilhite, S. E., Ledoux, P., Evangelista, C., Kim, I. F., Tomashevsky, M., … Soboleva, A. (2013). NCBI GEO: Archive for functional genomics data sets—update. Nucleic Acids Research , 41, D991–D995. doi: 10.1093/nar/gks1193.
- Bateman, A., Martin, M. J., O'Donovan, C., Magrane, M., Alpi, E., Antunes, R., … Zhang, J. (2017). UniProt: The universal protein knowledgebase. Nucleic Acids Research , 45, D158–D169. doi: 10.1093/nar/gkw1099.
- Bazzini, A. A., Johnstone, T. G., Christiano, R., Mackowiak, S. D., Obermayer, B., Fleming, E. S., … Giraldez, A. J. (2014). Identification of small ORFs in vertebrates using ribosome footprinting and evolutionary conservation. The EMBO Journal , 33, 981–993. doi: 10.1002/embj.201488411.
- Brar, G. A., & Weissman, J. S. (2015). Ribosome profiling reveals the what, when, where and how of protein synthesis. Nature Reviews Molecular Cell Biology , 16, 651–664. doi: 10.1038/nrm4069.
- Brunet, M. A., Brunelle, M., Lucier, J.-F., Delcourt, V., Levesque, M., Grenier, F., & Roucou, X. (2019). OpenProt: A more comprehensive guide to explore eukaryotic coding potential and proteomes. Nucleic Acids Research , 47, D403–D410. doi: 10.1093/nar/gky936.
- Brunet, M. A., Levesque, S. A., Hunting, D. J., Cohen, A. A., & Roucou, X. (2018). Recognition of the polycistronic nature of human genes is critical to understanding the genotype-phenotype relationship. Genome Research , 28(5), 609–624. doi: 10.1101/gr.230938.117.
- Chen, J., Brunner, A.-D., Cogan, J. Z., Nuñez, J. K., Fields, A. P., Adamson, B., … Weissman, J. S. (2020). Pervasive functional translation of noncanonical human open reading frames. Science , 367, 1140–1146. doi: 10.1126/science.aay0262.
- Cheng, H., Chan, W. S., Li, Z., Wang, D., Liu, S., & Zhou, Y. (2011). Small open reading frames: Current prediction techniques and future prospect. Current Protein & Peptide Science, 12, 503–507. doi: 10.2174/138920311796957667.
- Deutsch, E. W., Bandeira, N., Sharma, V., Perez-Riverol, Y., Carver, J. J., Kundu, D. J., … Vizcaíno, J. A. (2020). The ProteomeXchange consortium in 2020: Enabling ‘big data’ approaches in proteomics. Nucleic Acids Research , 48, D1145–D1152. doi: 10.1093/nar/gkz984.
- Erhard, F., Halenius, A., Zimmermann, C., L'Hernault, A., Kowalewski, D. J., Weekes, M. P., … Dölken, L. (2018). Improved Ribo-seq enables identification of cryptic translation events. Nature Methods , 15, 363–366. doi: 10.1038/nmeth.4631.
- Hao, Y., Zhang, L., Niu, Y., Cai, T., Luo, J., He, S., … Chen, R. (2017). SmProt: A database of small proteins encoded by annotated coding and non-coding RNA loci. Briefings in Bioinformatics , 19(4), 636–643. doi: 10.1093/bib/bbx005.
- Hellens, R. P., Brown, C. M., Chisnall, M. A. W., Waterhouse, P. M., & Macknight, R. C. (2016). The emerging world of small ORFs. Trends in Plant Science , 21, 317–328. doi: 10.1016/j.tplants.2015.11.005.
- Ingolia, N. T. (2014). Ribosome profiling: New views of translation, from single codons to genome scale. Nature Reviews Genetics , 15, 205–213. doi: 10.1038/nrg3645.
- Ingolia, N. T. (2016). Ribosome footprint profiling of translation throughout the genome. Cell , 165, 22. doi: 10.1016/j.cell.2016.02.066.
- Ingolia, N. T., Ghaemmaghami, S., Newman, J. R. S., & Weissman, J. S. (2009). Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science , 324, 218–223. doi: 10.1126/science.1168978.
- Jeong, K., Kim, S., & Bandeira, N. (2012). False discovery rates in spectral identification. BMC Bioinformatics , 13, S2. doi: 10.1186/1471-2105-13-S16-S2.
- Ma, J., Ward, C. C., Jungreis, I., Slavoff, S. A., Schwaid, A. G., Neveu, J., … Saghatelian, A. (2014). Discovery of human sORF-encoded polypeptides (SEPs) in cell lines and tissue. Journal of Proteome Research , 13, 1757–1765. doi: 10.1021/pr401280w.
- Madden, T. L., Tatusov, R. L., & Zhang, J. (1996). Applications of network BLAST server. Methods in Enzymology , 266, 131–141. doi: 10.1016/S0076-6879(96)66011-X.
- Menschaert, G., Criekinge, W. V., Notelaers, T., Koch, A., Crappé, J., Gevaert, K., & Damme, P. V. (2013). Deep proteome coverage based on ribosome profiling aids mass spectrometry-based protein and peptide discovery and provides evidence of alternative translation products and near-cognate translation initiation events. Molecular & Cellular Proteomics, 12, 1780–1790. doi: 10.1074/mcp.M113.027540.
- Nesvizhskii, A. I. (2014). Proteogenomics: Concepts, applications and computational strategies. Nature Methods , 11, 1114–1125. doi: 10.1038/nmeth.3144.
- O'Leary, N. A., Wright, M. W., Brister, J. R., Ciufo, S., Haddad, D., McVeigh, R., … Pruitt, K. D. (2016). Reference sequence (RefSeq) database at NCBI: Current status, taxonomic expansion, and functional annotation. Nucleic Acids Research , 44, D733–D745. doi: 10.1093/nar/gkv1189.
- Olexiouk, V., & Menschaert, G. (2016). Identification of small novel coding sequences, a proteogenomics endeavor. Advances in Experimental Medicine and Biology , 926, 49–64. doi: 10.1007/978-3-319-42316-6_4.
- Olexiouk, V., van Criekinge, W., & Menschaert, G. (2018). An update on sORFs.org: A repository of small ORFs identified by ribosome profiling. Nucleic Acids Research , 46, D497–D502. doi: 10.1093/nar/gkx1130.
- Orr, M. W., Mao, Y., Storz, G., & Qian, S.-B. (2019). Alternative ORFs and small ORFs: Shedding light on the dark proteome. Nucleic Acids Research , 48, 1029–1042. doi: 10.1093/nar/gkz734.
- Perez-Riverol, Y., Csordas, A., Bai, J., Bernal-Llinares, M., Hewapathirana, S., Kundu, D. J., … Vizcaíno, J. A. (2019). The PRIDE database and related tools and resources in 2019: Improving support for quantification data. Nucleic Acids Research , 47, D442–D450. doi: 10.1093/nar/gky1106.
- Raj, A., Wang, S. H., Shim, H., Harpak, A., Li, Y. I., Engelmann, B., … Pritchard, J. K. (2016). Thousands of novel translated open reading frames in humans inferred by ribosome footprint profiling. eLife , 5, e13328. doi: 10.7554/eLife.13328.
- Samandi, S., Roy, A. V., Delcourt, V., Lucier, J.-F., Gagnon, J., Beaudoin, M. C., … Roucou, X. (2017). Deep transcriptome annotation enables the discovery and functional characterization of cryptic small proteins. eLife , 6, e27860. doi: 10.7554/eLife.27860.
- Vanderperre, B., Lucier, J.-F., Bissonnette, C., Motard, J., Tremblay, G., Vanderperre, S., … Roucou, X. (2013). Direct detection of alternative open reading frames translation products in human significantly expands the proteome. PLOS One , 8, e70698. doi: 10.1371/journal.pone.0070698.
- Vaudel, M., Barsnes, H., Berven, F. S., Sickmann, A., & Martens, L. (2011). SearchGUI: An open-source graphical user interface for simultaneous OMSSA and X!Tandem searches. Proteomics , 11, 996–999. doi: 10.1002/pmic.201000595.
- Vaudel, M., Burkhart, J. M., Zahedi, R. P., Oveland, E., Berven, F. S., Sickmann, A., … Barsnes, H. (2015). PeptideShaker enables reanalysis of MS-derived proteomics data sets. Nature Biotechnology , 33, 22–24. doi: 10.1038/nbt.3109.
- Wilkinson, M. D., Dumontier, M., Aalbersberg, I. J. J., Appleton, G., Axton, M., Baak, A., … Mons, B. (2016). The FAIR Guiding Principles for scientific data management and stewardship. Scientific Data , 3, 160018. doi: 10.1038/sdata.2016.18.
- Wu, P.-Y., Phan, J. H., & Wang, M. D. (2013). Assessing the impact of human genome annotation choice on RNA-seq expression estimates. BMC Bioinformatics , 14, S8. doi: 10.1186/1471-2105-14-S11-S8.
- Xie, S.-Q., Nie, P., Wang, Y., Wang, H., Li, H., Yang, Z., … Xie, Z. (2016). RPFdb: A database for genome wide information of translated mRNA generated from ribosome profiling. Nucleic Acids Research , 44, D254–D258. doi: 10.1093/nar/gkv972.
- Yates, A. D., Achuthan, P., Akanni, W., Allen, J., Allen, J., Alvarez-Jarreta, J., … Flicek, P. (2020). Ensembl 2020. Nucleic Acids Research , 48, D682–D688. doi: 10.1093/nar/gkz966.
Key References
- Brunet et al. (2019). See above.
Corresponds to the official release of OpenProt and explains all the underlying methods in detail.
- Orr et al. (2019). See above.
Recent review painting an exhaustive picture of altORFs and small ORFs.
- Brunet et al. (2018). See above.
Complete review of the impact of altORF omission in experimental design and investigation of pathologies.
- Nesvizhskii (2014). See above.
Reviews the concepts of proteogenomics methods to better explore the proteome.
- Ingolia (2016). See above.
Exhaustive presentation of the ribosome profiling technique.
Internet Resources
OpenProt.
Citing Literature
Number of times cited according to CrossRef: 3
- Jessica J. Mohsen, Alina A. Martel, Sarah A. Slavoff, Microproteins—Discovery, structure, and function, PROTEOMICS, 10.1002/pmic.202100211, 23 , 23-24, (2023).
- Sébastien Leblanc, Marie A. Brunet, Jean-François Jacques, Amina M. Lekehal, Andréa Duclos, Alexia Tremblay, Alexis Bruggeman-Gascon, Sondos Samandi, Mylène Brunelle, Alan A. Cohen, Michelle S. Scott, Xavier Roucou, Newfound Coding Potential of Transcripts Unveils Missing Members of Human Protein Communities, Genomics, Proteomics & Bioinformatics, 10.1016/j.gpb.2022.09.008, (2022).
- Marie A Brunet, Jean-François Lucier, Maxime Levesque, Sébastien Leblanc, Jean-Francois Jacques, Hassan R H Al-Saedi, Noé Guilloy, Frederic Grenier, Mariano Avino, Isabelle Fournier, Michel Salzet, Aïda Ouangraoua, Michelle S Scott, François-Michel Boisvert, Xavier Roucou, OpenProt 2021: deeper functional annotation of the coding potential of eukaryotic genomes, Nucleic Acids Research, 10.1093/nar/gkaa1036, 49 , D1, (D380-D388), (2020).

