Generating Nuclei Segmentations for vEM using Cellpose
Alyson Petruncio, Aubrey Weigel, CellMap Project Team, Emma Avetissian, Jeff L Rhoades
Abstract
This protocol details the process of generating nucleus segmentations in FIB-SEM (Focused Ion Beam Scanning Electron Microscopy) image stacks of cells and tissues using Cellpose. These segmentations are crucial for gaining biological insights and training deep learning models on cell and organelle segmentation. The protocol covers the application of Cellpose for fine-tuning 2D models specifically for nuclei segmentation. Once fine-tuned, the model is used to generate 3D predictions for the entire dataset. These segmentations can be further refined through manual approaches, utilized for visualization, or employed in training other machine learning models.
Steps
Introduction:
The purpose of this protocol is to provide comprehensive instructions on how to create nuclei segmentations in FIB-SEM datasets on OpenOrganelle.org using Cellpose.
Cellpose is a generalist algorithm developed by Stringer et al., (2020) to segment cellular structures. Cellpose also provides software for manual segmentation and model training. In this protocol we discuss the use of Cellpose v3.0.9.
In this protocol you can expect to find:
- OpenOrganelle Overview
- Using Fiji to save files in the cloud locally as 2D tiffs
- Cellpose in 2D and Model Training
- Cellpose in 3D and Predictions
- Final Touches
OpenOrganelle Overview
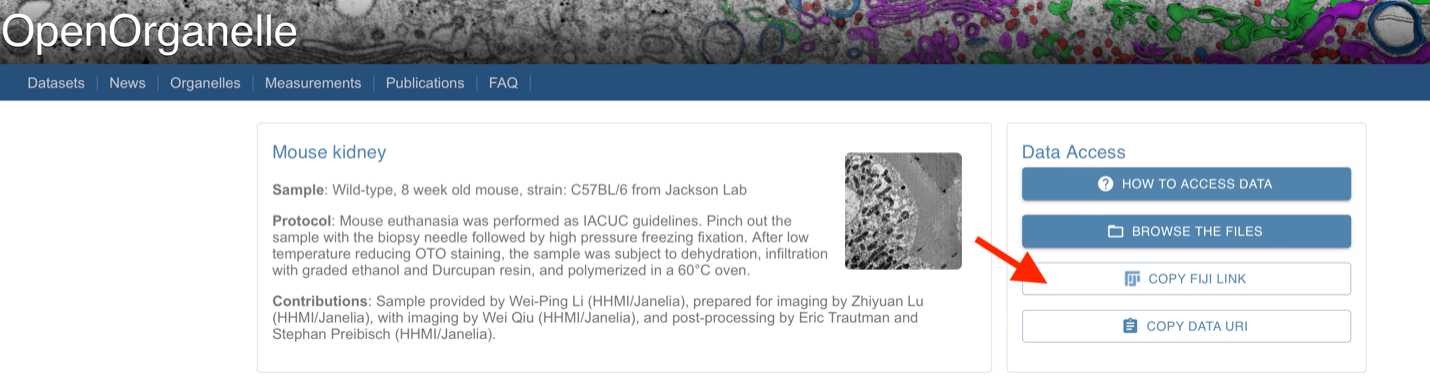
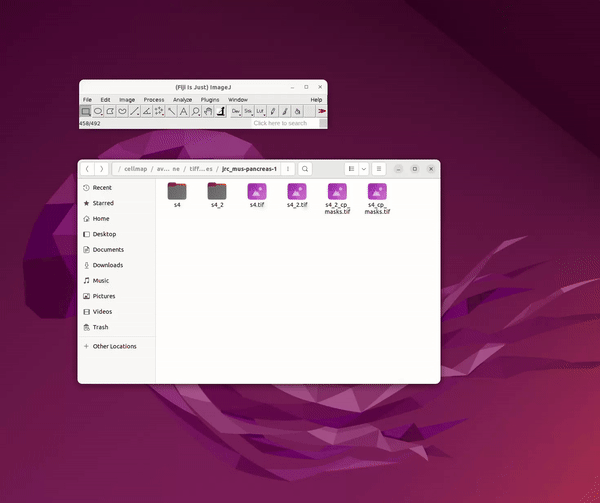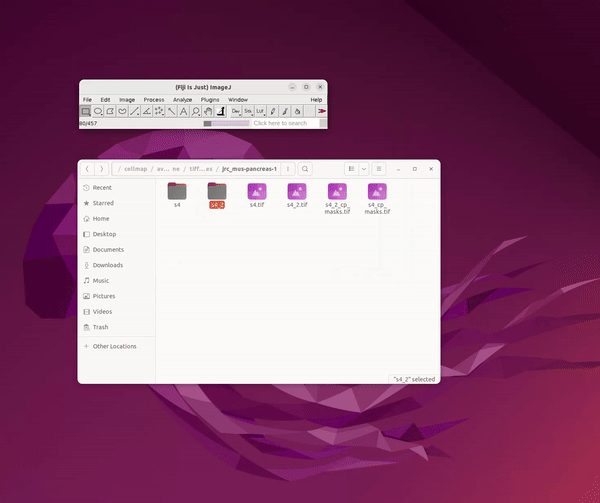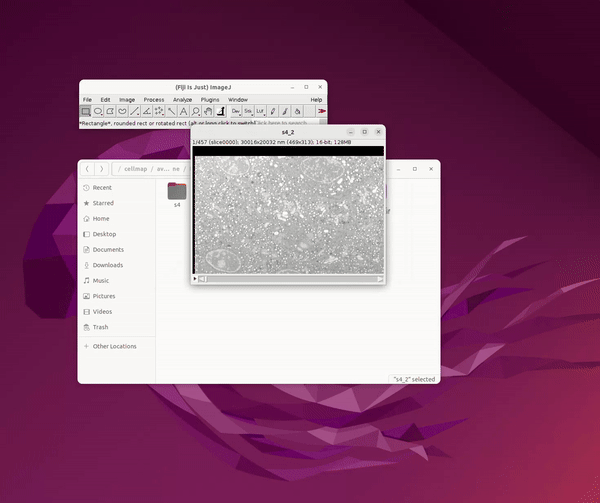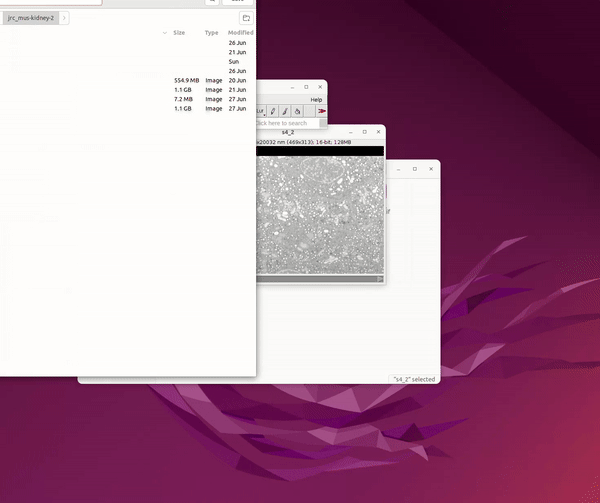
OpenOrganelle (Heinrich et al, 2021) is a collection of focused ion beam scanning electron microscopy (FIB-SEM) datasets curated by the Janelia CellMap Project Team. Each dataset on OpenOrganelle includes important background information about the sample, protocol, contributors, and more. The “VIEW” button opens a new tab to view the chosen dataset in Neuroglancer. Each dataset is also accompanied by a Fiji link which copies the location of the dataset in the S3 bucket (see Figure 1). This link was used to read data into Fiji and save as 2D slices for Cellpose.
Using Fiji to save files in the cloud locally as 2D tiffs
Once the Fiji link from OpenOrganelle has been copied, it can be pasted into Fiji.
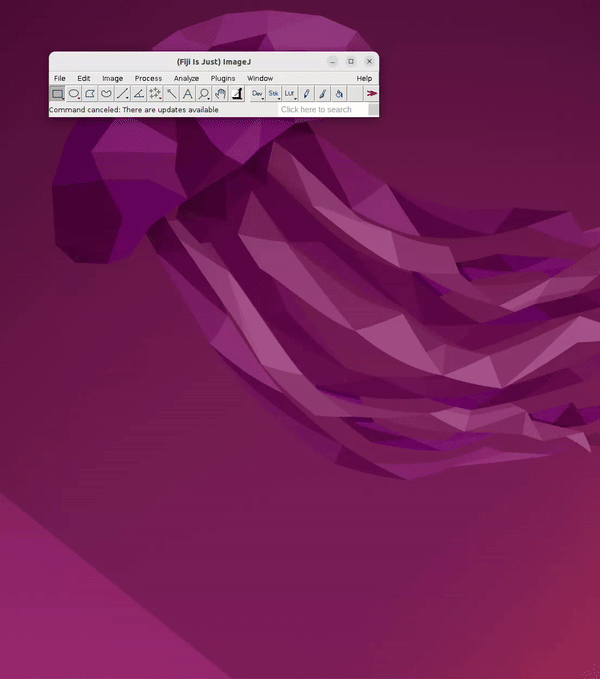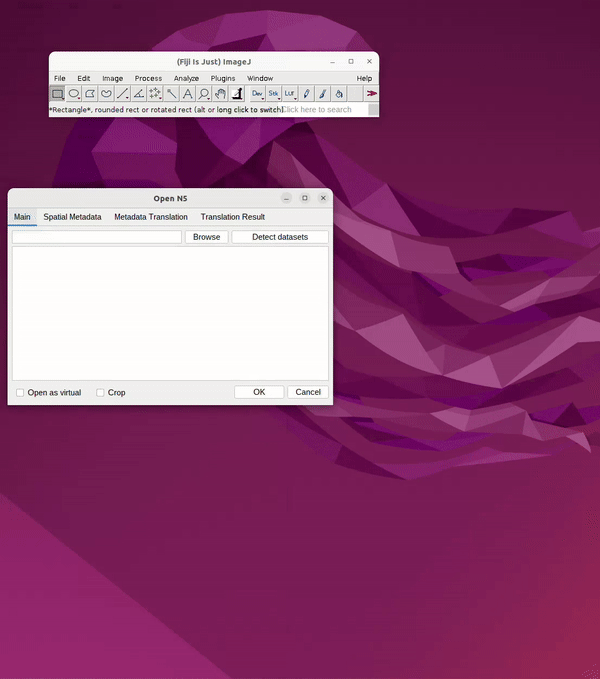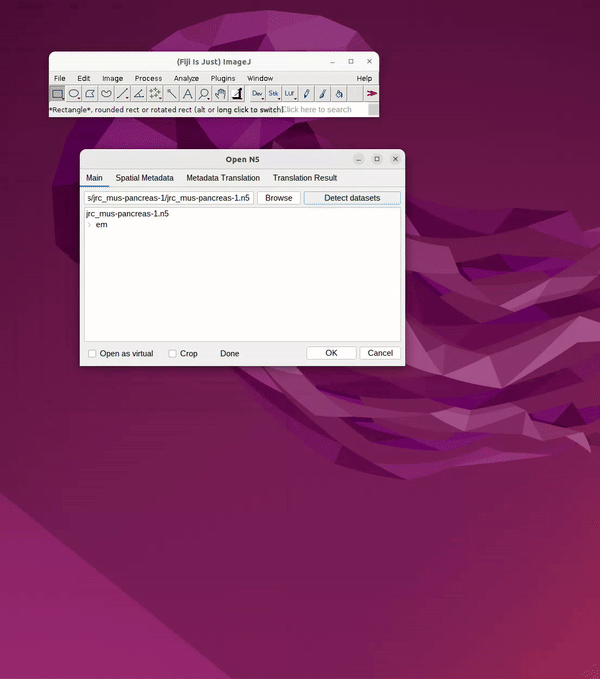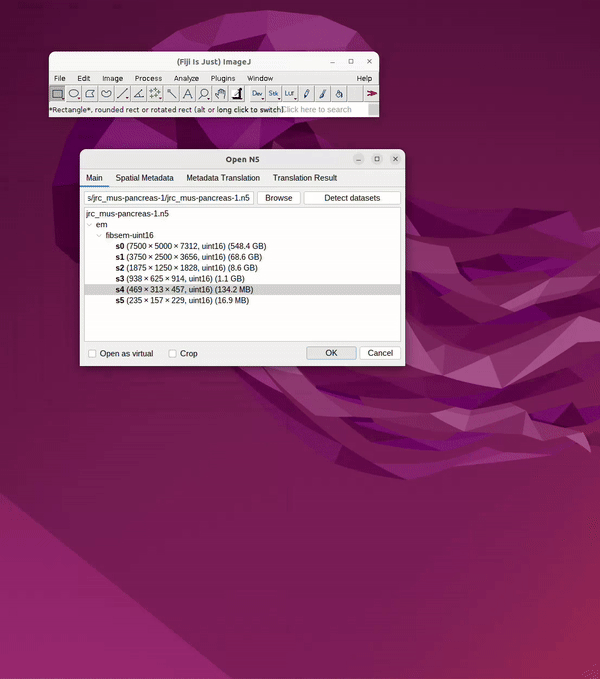
Head to: File → Import → HDF5/N5/Zarr/...
Paste the link into the search bar.
Click Detect Datasets
Navigate through the file container by clicking the dropdown, e.g. “em”.
Choose resolution, e.g., s0 (original), s1 (2x downsample), s3 (4x downsample), etc.
Click OK. See Figure 2.
Once the dataset is open in Fiji, it can be saved as a sequence of 2D slices for reading into Cellpose.
From the open dataset window go to: File Save As Image Sequence → Save into a new folder r
See Figure 3.
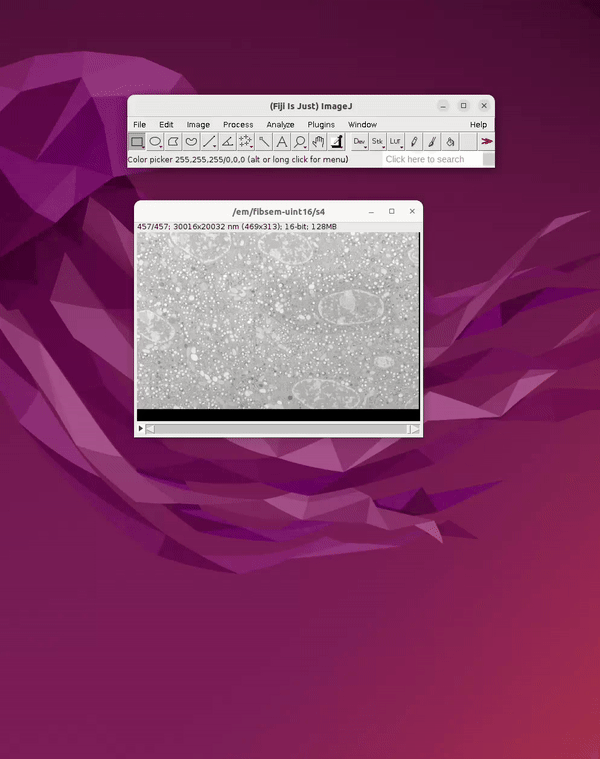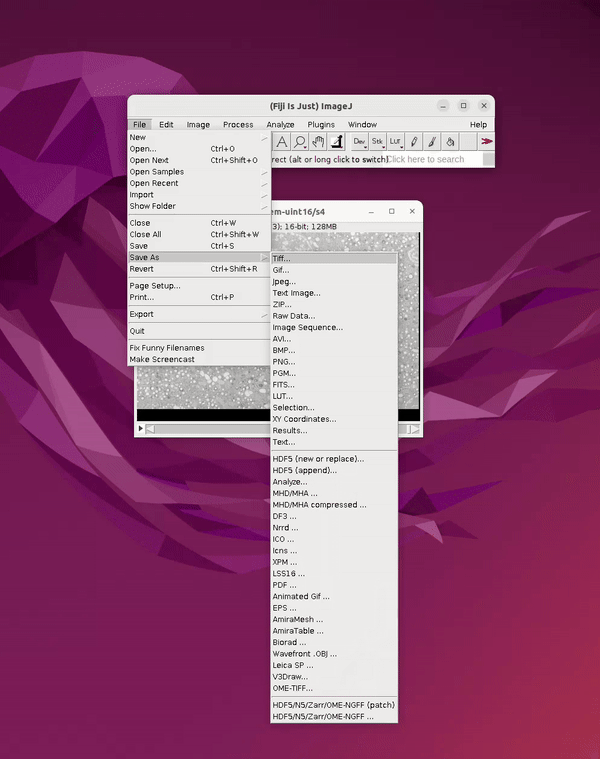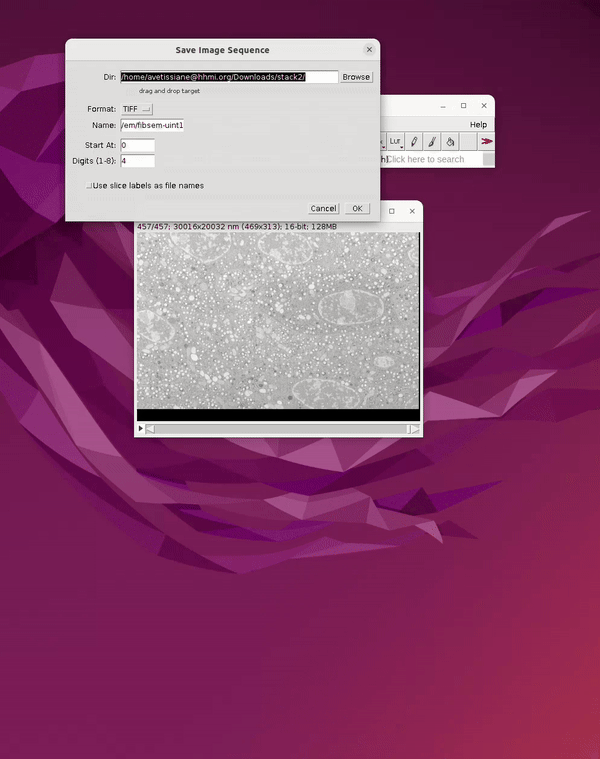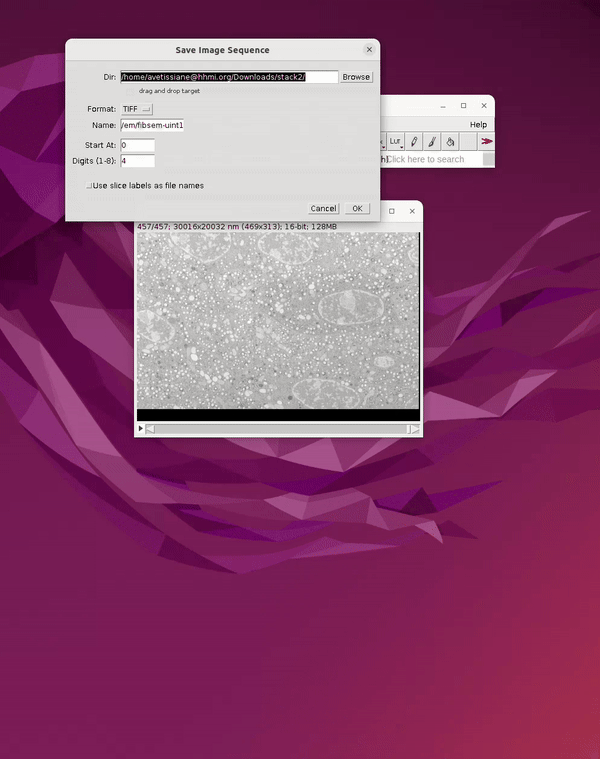
Before closing the image in Fiji, save a second copy of the dataset as a 3D tiff. This will be used for generating 3D predictions in a later step in Cellpose. If the image is closed before creating the 3D tiff, you can simply drag the folder containing the image sequence into the main Fiji bar to open it. To save it as a 3D tiff:
File → Save As → Tiff
See Figure 4.
Cellpose in 2D and Model Training
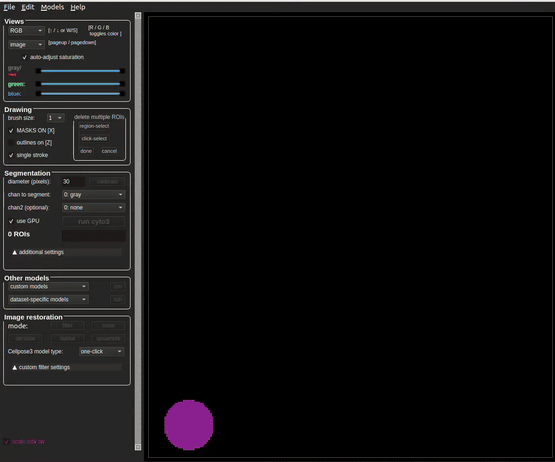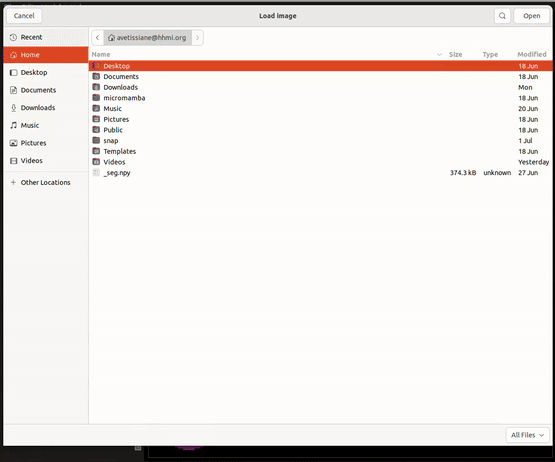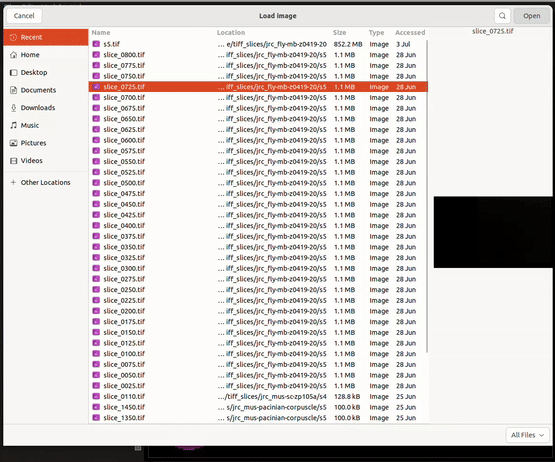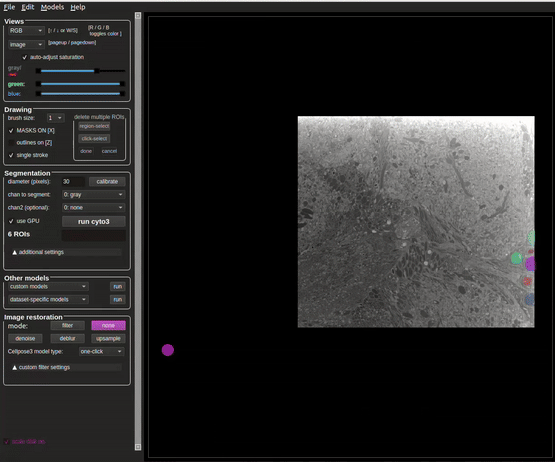
To begin using Cellpose on the dataset, open the program (see Appendix). More detailed instructions for general Cellpose use can be found on https://cellpose.readthedocs.io/en/latest/gui.html. Within Cellpose go to: File → Load Image → Select a slice for segmentation
See Figure 5.
| A | B |
|---|---|
| Zoom | Hold right click and drag |
| Zoom | Scroll wheel |
| Pan | Hold left click and drag |
| Create mask | Tap right click and drag |
| Remove mask | Ctrl left click |
| Undo remove mask | Ctrl Y |
| Toggle mask visibility | X |
| Toggle mask outlines | Z |
Table 1: Basic key binds for annotation in Cellpose
Annotate the nuclei as accurately as possible.
At first, it may be beneficial to use an existing model to make annotating easier. Out of the box, the cyto2_cp3 model tends to give promising results for nuclei in FIB-SEM volumes of tissue. If the model does not catch anything, try annotating a couple slices manually and training a model. Continue to use and improve the model by periodically adding more training slices.
Refer to Figure 6 for what a completed slice might look like.
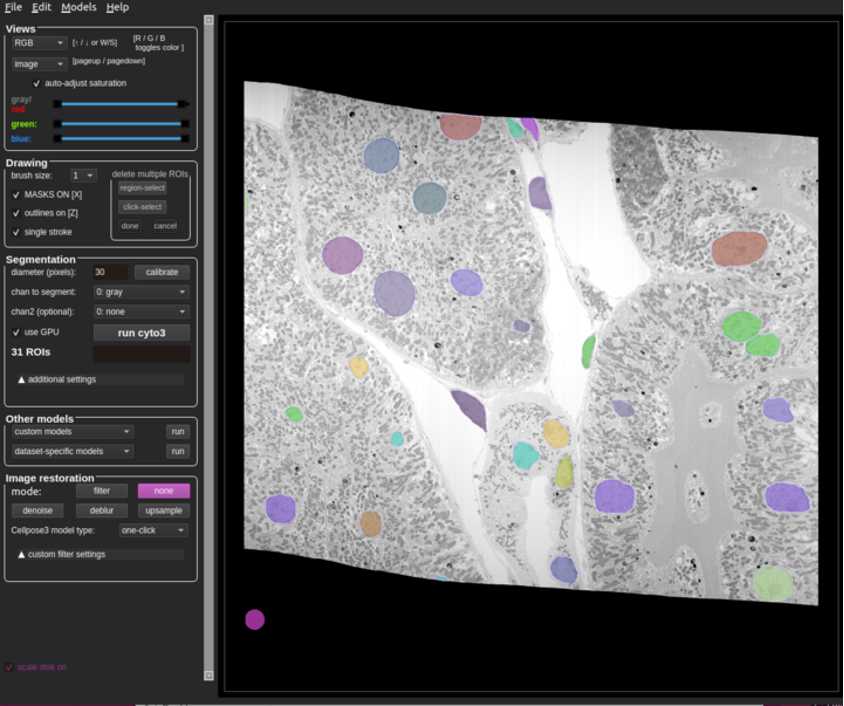
Once the annotation is complete, save the slice along with the masks.
File → *Save masks and image (as _seg.npy)
This file will automatically be saved into the folder where the rest of the 2D slices can be found.
To train a model on annotated slices, go to:
Models → Train new model with image+masks in folder
Set the initial model to cyto2_cp3
Adjust n_epochs to change the number of epochs (we generally went with 1000)
Name the model for organizational purposes. Click “OK” and wait for the training to finish.
![Figure 7. Screenshot of the terminal while training is in progress. The number after “[INFO]” displays the current epoch. Figure 7. Screenshot of the terminal while training is in progress. The number after “[INFO]” displays the current epoch.](https://static.yanyin.tech/literature_test/protocol_io_true/protocols.io.n2bvjnq8pgk5/fig3.3.png)
To run this model on a new 2D slice
Open the blank 2D slice and locate the section labeled “Other models” on the left-hand side of the program.
Open the dropdown for custom models, select the new model, and click run.
Repeat this process, making use of previous models, annotating additional slices until the model performs satisfactorily on 2D slices.
Cellpose in 3D and Predictions
Cellpose can extend models that were trained exclusively on 2D data to perform predictions in 3D. Once the model demonstrates a satisfactory performance in 2D, we attempt to run a prediction in 3D.
Close Cellpose
Into the terminal enter:
python -m cellpose --Zstack
This will alert Cellpose to load data as 3D
Now load in the previously saved 3D volume by:
File → Load Image → Select the tiff file
Once the tiff has loaded, run the custom model as described for 2D in Step 15.
This may take a while (on the order of tens of minutes) and progress can be tracked in the terminal.
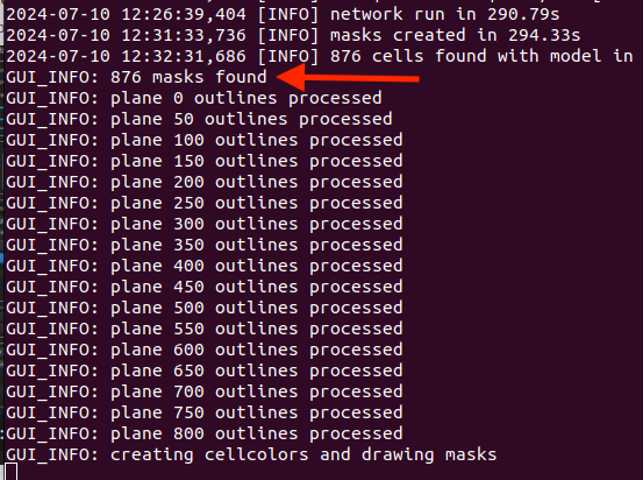
The process will be complete when the masks are visible (see Figure 8).
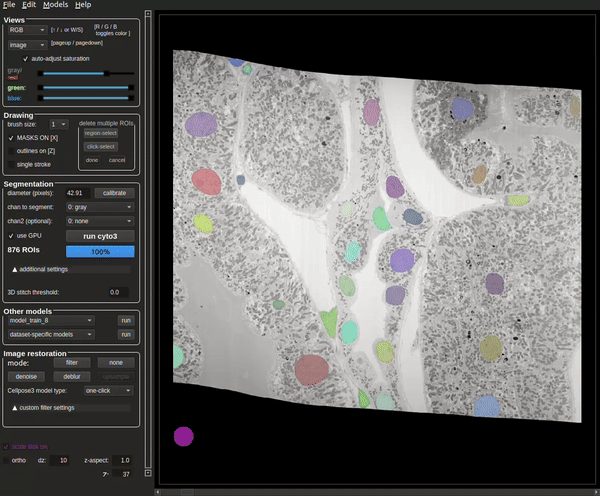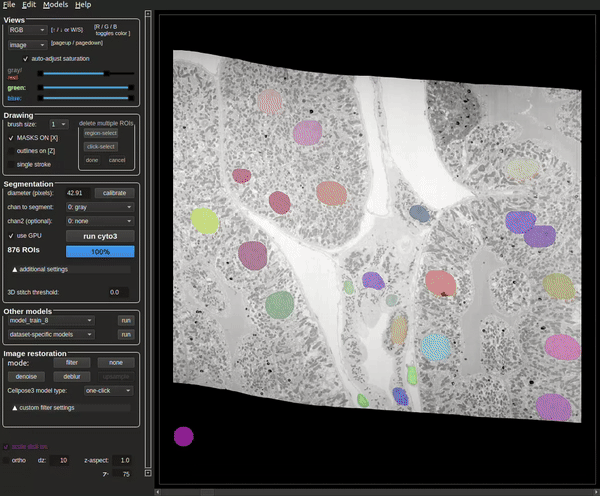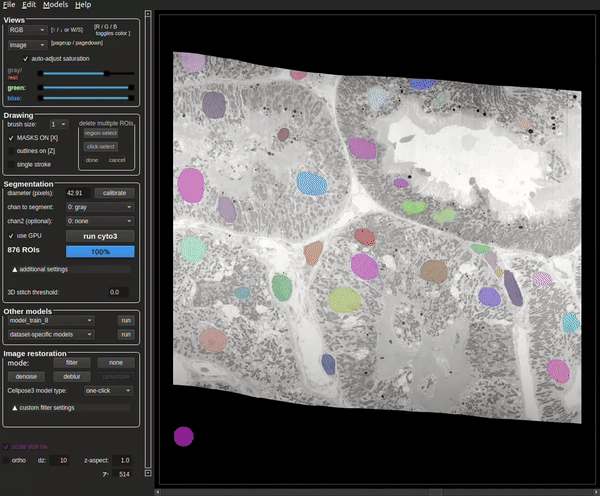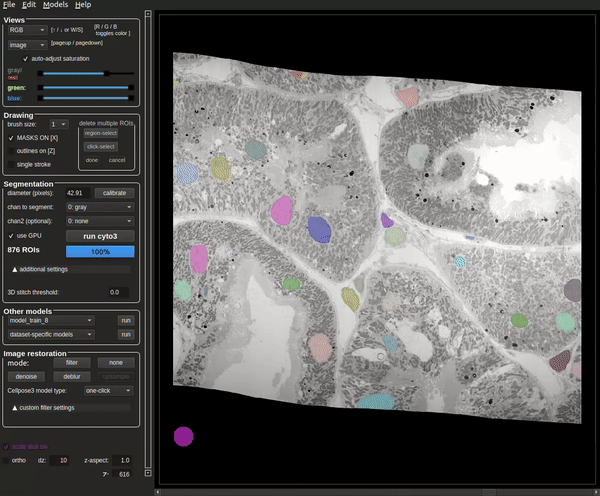
The terminal will also contain how many masks were found at the end of the run (see Figure 9).