Fixation and Laser Capture Microdissection of Plant Tissue for RNA Extraction and RNASeq Library Preparation
Christopher Saski, Christopher Saski, Suchitra Chavan, Suchitra Chavan, Elise Schnabel, Elise Schnabel, Julia Frugoli, Julia Frugoli
Abstract
To study the transcriptome of individual plant cells at specific points in time, we developed protocols for fixation, embedding, and sectioning of plant tissue followed by laser capture microdissection (LCM) and processing for RNA recovery. LCM allows the isolation of individual cell types from heterogeneous tissue sections and is particularly suited to plant processing because it does not require the breakdown of cell walls. This approach allows accurate separation of a small volume of cells that can be used to study gene expression profiles in different tissues or cell layers. The technique requires neither separation of cells by enzymatic digestion of any kind nor cell-specific reporter genes, and it allows storage of fixed and embedded tissue for months before capture. The methods for fixation, embedding, sectioning, and capturing of plant cells that we describe yield high-quality RNA suitable for making libraries for RNASeq. © 2023 The Authors. Current Protocols published by Wiley Periodicals LLC.
Basic Protocol 1 : Tissue Preparation for Laser Capture Microdissection
Basic Protocol 2 : Tissue Sectioning
Basic Protocol 3 : Laser Capture Microdissection of Embedded Tissue
Basic Protocol 4 : RNA Extraction from Laser Capture Microdissection Samples
INTRODUCTION
Laser capture microdissection (LCM) is a powerful and useful technique for isolating cells of interest from a heterogeneous cell population. Using a laser unit and a microscope, it is easy and fast to procure individual cells of interest from tissue sections under direct microscopic visualization (Curran et al., 2000). Direct visualization of tissue sections with intact histology is possible and allows reliable procurement of specific cells. The precision and speed of the laser beam used to cut the tissue help to capture a large number of pure cells from a heterogeneous tissue sample in a very short period (Simone et al., 1998). The method is practical for the rapid and efficient isolation of specific populations of cells that are free of contaminants and amenable to a variety of molecular analyses, including as a source of confirming information for computational analysis in single-cell RNASeq experiments (Schnabel et al., 2023). In the following protocols, we describe the preparation of tissue for LCM (Basic Protocols 1 and 2) and the use of LCM for individual cell types (Basic Protocol 3) to generate RNASeq libraries (Basic Protocol 4) at defined time points for the cells involved in nodulation.
One of the most important aspects of the procedures is maintaining an RNase-free workspace throughout each step of each protocol. This requires the use of gloves, sterile filter tips, and RNase-free glassware and water always. It is strongly recommended to dedicate a laboratory bench to RNA experiments and to cover it with paper that is changed frequently to minimize contamination. All consumables should be immersed in diethyl pyrocarbonate (DEPC)-treated water and autoclaved. Note that DEPC-treated water is RNase free after autoclaving but can no longer destroy additional RNase because autoclaving destroys DEPC. When using equipment such as centrifuges, incubators, and other items that cannot be autoclaved (for example, cooling plates), clean surfaces thoroughly with RNase AWAY or a similar product. Always use autoclaved DEPC-treated water whenever water is needed. Specific details regarding treating consumables with DEPC are mentioned in the Materials sections below when applicable; treatment with UV light can also destroy RNase, depending on the shape of the object. It is assumed that molecular biology consumables (such as tubes) that are wrapped in the factory are RNase free, but if this is a concern, treat them as described for plastic cassettes.
Basic Protocol 1: TISSUE PREPARATION FOR LASER CAPTURE MICRODISSECTION
Tissue preparation for LCM involves infiltration, fixation, and embedding of tissue. Preparation of harvested tissue is performed in a cold, RNase-free environment to maintain stability of the RNA. Then, the tissue is dehydrated using a series of fixative solutions to simultaneously preserve enough visual details to identify specific cells for harvesting and allow maximum recovery of DNA, RNA, or protein from the harvested cells (in this case, RNA). Although there are several histological methods available for infiltration and fixation, it is important to understand the differences in structure between plant and animal cells to select the appropriate technique. Cryosectioning, commonly used for animal tissue, is undesirable for plant tissue due to the difficulty in stabilizing the vacuolated cells and the loss of tissue integrity caused by freezing and thawing (Kerk et al., 2003). Plant tissues lose their histological integrity in cryosections even with prior fixation and treatment with cryoprotectants, which makes the identification of target cells difficult (Nelson et al., 2006). The most commonly used fixatives are acetic acid, ethanol (EtOH), and xylene, but methanol and increasing concentrations of EtOH are an alternative; removing water from the plant cell and infusing the cell with methanol and EtOH will stabilize and maintain the integrity of the cellular contents. We use these fixative reagents for infiltration to increase the recovery of RNA because the cross-linking fixative formaldehyde–acetic acid–ethanol (FAA) reduces this recovery. The choice of infiltration and fixation methods should be a balance between the preservation of histological detail and maximum recovery of cellular contents, whether DNA, RNA, or protein (Kerk et al., 2003).
After infiltration and fixation, the tissue is embedded in either paraffin or polyester wax. Embedding of tissue enables better visual observation during cell harvesting and maintains the stability of tissue. The embedded block serves as an archive that can be resampled or re-sectioned even after an extended period (Kerk et al., 2003). Paraffin embedding is commonly used, but in the current protocol, polyester wax is used. Paraffin melts at 55° to 65°C, whereas polyester wax melts at 38° to 40°C. The modifications to the embedding protocol avoid degradation of RNA at high temperatures, thus maximizing recovery of RNA.
Materials
-
Diethyl pyrocarbonate (DEPC)-treated water (see recipe)
-
RNase AWAY (Thermo Fisher Scientific) and spray bottle if purchased in bulk
-
Seedlings of the plant of interest—e.g., Medicago truncatula
-
Fixative solution (see recipe), 4°C
-
EtOH/polyester wax solution (see recipe), 37°C
-
100% methanol (anhydrous, ultra-pure grade), 4°C
-
70% (v/v) EtOH solution (see recipe), 4°C
-
96% (v/v) EtOH solution (see recipe), 4°C
-
100% EtOH (anhydrous, molecular biology grade), 4°C
-
Polyester wax (e.g., Steedman's wax, Electron Microscopy Sciences, cat. no. 19312)
-
Drierite
-
Glass slide-staining box
-
Autoclave
-
Slimsette plastic biopsy cassettes with recessed cover (Thermo Scientific)
-
Aluminum foil
-
Tissue forceps (4.5 in.)
-
Cooling plate that can maintain −20°C (e.g., Tissue-Tek Cold Plate)
-
No. 2 pencil
-
Single-edge razor blades, individually wrapped
-
Sealed chamber and vacuum pump capable of pulling and holding 20-kPa vacuum
-
RNase-free pipet tips (filter tips recommended)
-
RNase-free beaker
-
37°C incubator
-
Individually wrapped plastic drinking straws
-
Parafilm
-
Disposable transfer pipet
-
Airtight container (e.g., plastic food storage box with airtight lid) or sealable plastic bag
Preparation for infiltration and fixation before tissue harvest
1.Submerge a clean glass slide-staining box in DEPC-treated water overnight. The following day, remove all water from the box, close lid, wrap with aluminum foil, and autoclave. Cool to room temperature and store at 4°C until ready for use.
2.Submerge Slimsette plastic biopsy cassettes with a recessed cover in DEPC-treated water overnight. The following day, remove excess water from cassettes, wrap each cassette individually with aluminum foil, and autoclave. Cool to room temperature and store at 4°C until ready for use.
3.Submerge tissue forceps in DEPC-treated water overnight. The following day, remove forceps from the solution, wrap with aluminum foil, and autoclave. Cool to room temperature and store at 4°C until ready for use.
4.Prepare a cooling plate for infiltration by storing overnight at −20°C.
Infiltration and fixation of tissue
5.Cover cooling plate from step 1 with aluminum foil, spray with RNase AWAY, and clean according to directions on the RNase AWAY bottle. Then, pour approximately 5 to 10 ml DEPC-treated water onto cooling plate.
6.Label plastic biopsy cassette from step 2 with a No. 2 pencil.
7.Collect clean plant tissue of choice from Medicago truncatula seedlings and place into cold DEPC-treated water on the RNase-free cooling plate. Align tissue as desired for sectioning (such as alignment of root tips from seedlings; Fig. 1A).

8.Cut portions of tissue intended for fixation (e.g., the maturation zones from each root) with a single-edge razor blade and place them in the labeled biopsy cassette (Fig. 1B). Immediately place the cassette in the cold fixative solution inside the slide-staining box, kept at 4°C by placing on ice (Fig. 1C), making sure that the cassette is completely immersed in the fixative solution. Keep slide-staining box on ice throughout infiltration to maintain the temperature at 4°C.
9.Apply a 20-kPa vacuum. Quickly release and reapply vacuum every 10 min for a total of 30 min to force the solution into the tissue.
10.Prepare EtOH/polyester wax solution 2 to 4 hr prior to step 16 (Fig. 2A).

11.Replace fixative solution in the box with cold 100% methanol. Place the box in a cold room or refrigerator and incubate for 2 hr at 4°C.
12.Pour off methanol, removing the last drops with an RNase-free pipet tip. Replace with cold 70% EtOH solution and incubate for 30 min at 4°C.
13.Pour off 70% EtOH, removing the last drops with a pipet. Replace with cold 96% EtOH solution and incubate for 30 min at 4°C.
14.Replace 96% EtOH with cold 100% EtOH and incubate for 30 min at 4°C. Repeat this step twice, for a total of three changes of EtOH.
15.Replace 100% EtOH with the EtOH/polyester wax solution from step 10 and incubate overnight at 37°C.
16.The following day, melt pure polyester wax at 37°C for 2 to 4 hr before replacing the EtOH/polyester wax solution in the box with the melted wax. Cover box with aluminum foil and store at 37°C for 2 hr. Repeat this step twice for a total of three changes of wax.
17.Proceed immediately to embedding.
Embedding of tissue
18.Melt polyester wax in an RNase-free beaker at 37°C 2 to 4 hr prior to embedding (Fig. 2A).
19.Cover the −20°C cooling plate with aluminum foil and clean it with RNase AWAY but do not add DEPC-treated water.
20.Using a razor blade and gloved hands, cut a plastic drinking straw to ∼4 cm in length and then cut it horizontally and wrap it with Parafilm to seal the ends of the straw, resulting in a boat-like structure (Fig. 2C). Place this boat on the −20°C cooling plate.
21.Heat the forceps from step 3 by placing them inside a 37°C incubator for 2 to 5 min. Remove the cassette from the wax (step 16) and open it inside the 37°C incubator, using the glass lid of the box as an RNase-free platform. Orient roots or other tissue in a single bundle using the RNase-free forceps (Fig. 2D).
22.Pour a few drops of melted wax into the cut straw boat using a disposable transfer pipet and let the boat and wax cool for a few minutes by placing it on the −20°C cooling plate. Then, carefully place the bundle of roots in the boat, immediately pouring enough wax to cover the roots and fill the straw. Place the boat in the labeled cassette that originally contained the tissue on top of the −20°C cooling plate (Fig. 2E). Allow embedded samples to solidify on the −20°C cooling plate for 2 to 4 hr.
23.After the embedded samples are hardened, cover them with Parafilm, label them, and store them in an airtight container or sealable plastic bag at 4°C or −20°C until ready for sectioning (see Basic Protocol 2). Add Drierite to the container or bag to combat moisture and mold (Fig. 2F).
Basic Protocol 2: TISSUE SECTIONING
Sectioning of tissue involves cutting embedded tissue into ribbons of sections and arranging the ribbons on polyethylene naphthalate (PEN) slides. Traditionally, during sectioning, the cut ribbons are floated in a warm-water bath and then loaded onto PEN membrane slides, which are then kept on a slide warmer at 32°C overnight (Gomez et al., 2009). However, we have made modifications to the regular sectioning technique to maximize RNA recovery. The major change is loading the wax ribbons containing tissue directly onto pre-wetted PEN slides and then drying the slides for 2 to 5 min at 28°C. The lower temperature and reduced time used avoid exposure of the RNA to heat but still allow affixing of the polyester wax–embedded sections to the membranes while reducing the risk of RNA contamination and degradation posed by the water bath and overnight incubation of the slides at 32°C. The procedure still allows maximum coverage of the PEN slides by permitting loading of multiple ribbons onto a single slide.
Materials
-
Polyester wax (e.g., Steedman's wax, Electron Microscopy Sciences, cat. no. 19312)
-
RNase AWAY (Thermo Fisher Scientific)
-
DEPC-treated water (see recipe)
-
Embedded root tissue (see Basic Protocol 1)
-
Drierite
-
37°C incubator
-
PEN membrane slides appropriate to LCM microscope (see Basic Protocol 3), either regular or RNase free
-
No. 2 pencil
-
Tissue forceps (4.5 in.)
-
Camel hairbrush (size 2)
-
Microslide box(es)
-
Sterile hood with UV lamp (only required if PEN slides are not purchased RNase free)
-
Microtome and appropriate PTFE-coated high-profile microtome blades
-
Slide warmer with lid and ability to hold 28°C
-
Single-edge razor blades, individually wrapped
-
RNase-free plastic biopsy cassette with recessed cover (see Basic Protocol 1, step 2)
Preparation for sectioning and laser capture
1.Melt polyester wax at 37°C 2 to 4 hr prior to sectioning.
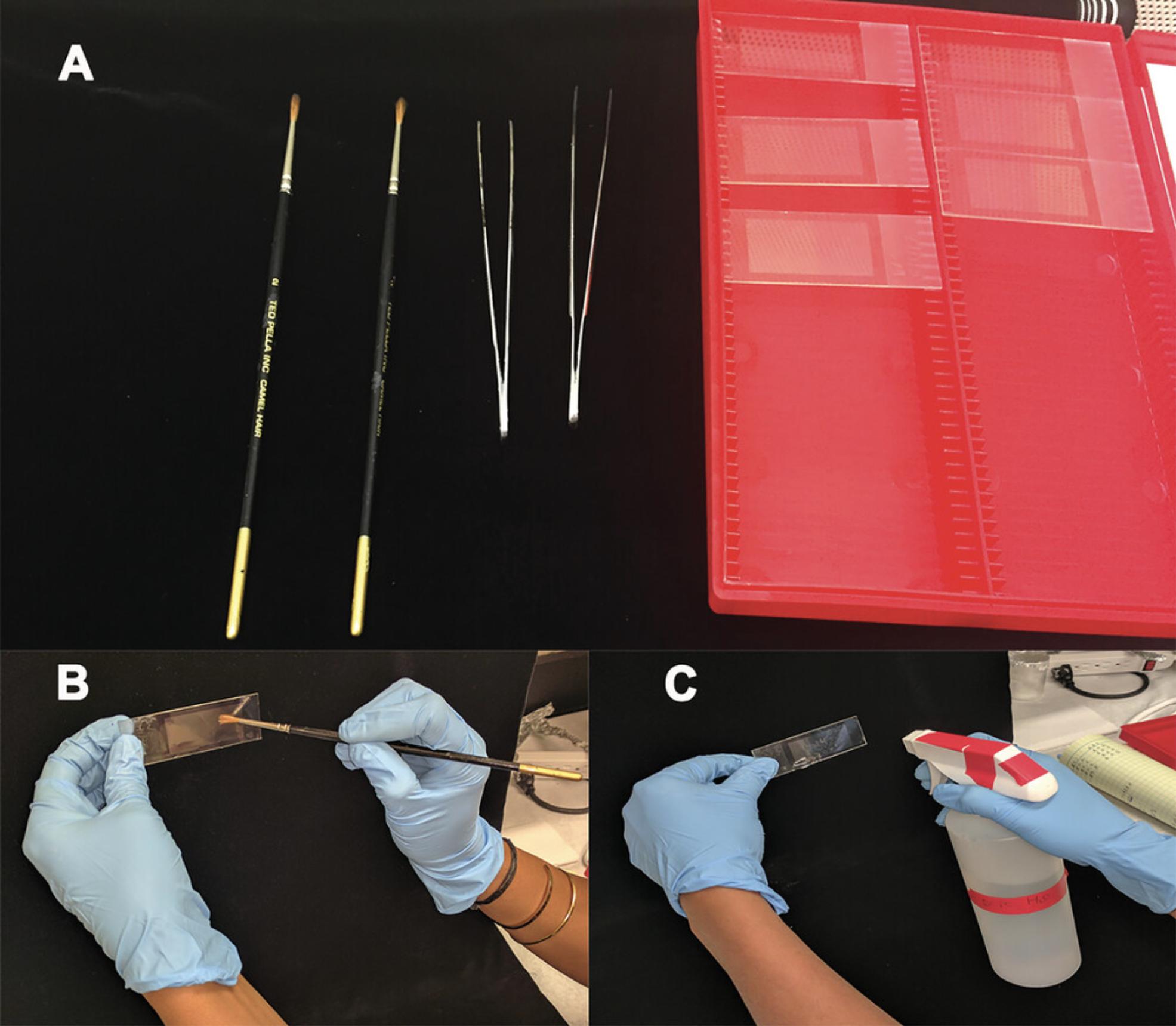
2.Label 5 PEN membrane slides with a No. 2 pencil. On the day of use, treat PEN slides, tissue forceps, a camel hair brush, and an open microslide box with UV light in a sterile hood for 30 min to reduce RNase contamination and then store everything in microslide box (Fig. 3A).
3.Clean the microtome and slide warmer with RNase AWAY. Wipe any surface that will come in contact with gloves, wax, slides, or tissue with RNase AWAY as well, focusing on the microtome's universal cassette clamp, hand rest, handwheel with a centering handle, standard specimen clamp, and knife holder for disposable blades. Set the slide warmer to 28°C.
Sectioning
4.Pre-wet membranes on the PEN slides with approximately 10 to 12 µl DEPC-treated water using the camel hairbrush (Fig. 3B). Alternatively, if preferred, spray DEPC-treated water on the slides so that the water covers the entire slide and forms a uniform coating (Fig. 3C).
5.Using a single-edge razor blade, cut ∼1 cm off the block of embedded root tissue, remove the Parafilm and plastic straw coating from the embedded sample, and affix the sample in the center of an RNase-free plastic biopsy cassette with a recessed cover using melted polyester wax from step 1 (Fig. 4A). Place a new PTFE-coated high-profile microtome blade on the microtome.

6.Place the cassette on the microtome and begin sectioning the embedded sample at the desired thickness, using the RNase-free forceps to manipulate the ribbons as they are cut (Fig. 4B). Load the cut ribbons containing tissue sections onto the wetted PEN slides (step 4) using forceps and/or the brush.
7.Once a slide is covered with ribbons, place it on the slide warmer for approximately 3 to 5 min or until the slide is dry (Fig. 4C).
8.Once the ribbons have adhered to the PEN slides, place the slides in the microslide box along with Drierite and store at −20°C (Fig. 4D).
Basic Protocol 3: LASER CAPTURE MICRODISSECTION OF EMBEDDED TISSUE
After sectioning, the PEN slides are de-waxed for LCM. Paraffin-embedded samples are treated with xylene prior to LCM to remove the paraffin, which allows the laser to cut cells of interest. However, there are reports of paraffin embedding and xylene causing significant RNA damage (Evers et al., 2011). Employing polyester wax instead allows the use of 100% EtOH to remove the wax and perform LCM. After LCM, RNA can be extracted from the harvested cells using commercially available kits specific to LCM tissue.
Materials
-
PEN slides carrying sectioned root tissue (see Basic Protocol 2)
-
100% EtOH (anhydrous, molecular biology grade)
-
RNase AWAY (Thermo Fisher Scientific)
-
Distilled water
-
Ambion RNAqueous-Micro Kit (Life Technologies, cat. no. AM1931)
- 50 micro filter cartridge assembly
- 50 micro elution tubes
- 15-ml wash solution 1 concentrate
- 28-ml wash solution 2/3 concentrate
- 25 lysis solution
- 200-μl LCM additive
- 55-μl DNase 2U/μl
- 600-μl 10X DNase I buffer
- 150-μl DNase inactivation reagent
- 5-ml elution solution
-
RNase-free petri dish
-
RNase-free microscope slide box
-
Sealable plastic food container
-
LCM microscope (e.g., Leica LMD6500 microscope)
-
0.2-ml thin-walled 8-tube strip with domed caps (handled to keep RNase free)
-
Permanent marker
-
RNase-free pipet tips (filter tips recommended)
-
Standard tabletop centrifuge with tube-strip adaptor
De-waxing prior to LCM
1.Remove a PEN slide carrying sectioned root tissue from −20°C. Place slide in ∼10 ml of 100% EtOH in an RNase-free petri dish) and gently swirl for ≤3 min to dissolve the wax on the slide (Fig. 5A), removing the slide as soon as it is clear of wax (Fig. 5B).
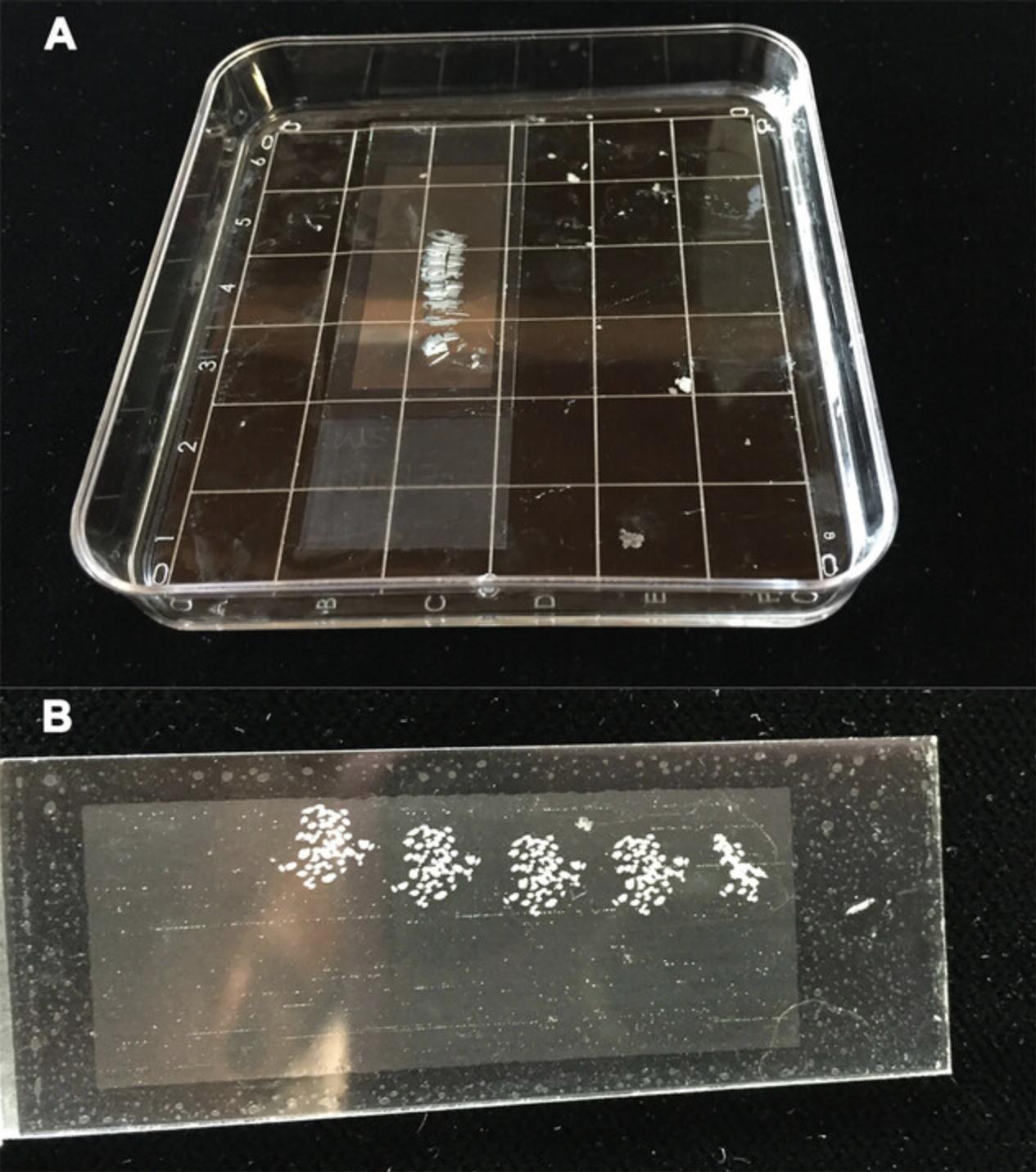
2.Evaporate any excess EtOH from the PEN slide at room temperature in an RNase-free space. Use the PEN slide immediately for LCM or store it in an RNase-free microscope slide box inside an airtight container (a sealable plastic food container) for ≤5 days at room temperature.
LCM
3.Clean the LCM microscope and adjacent workspace with RNase AWAY, including the universal collector for 8-tube strip caps, the holder for slides, the motorized stage of the microscope, and any other parts of the microscope that will come in contact with the slides or tissue.
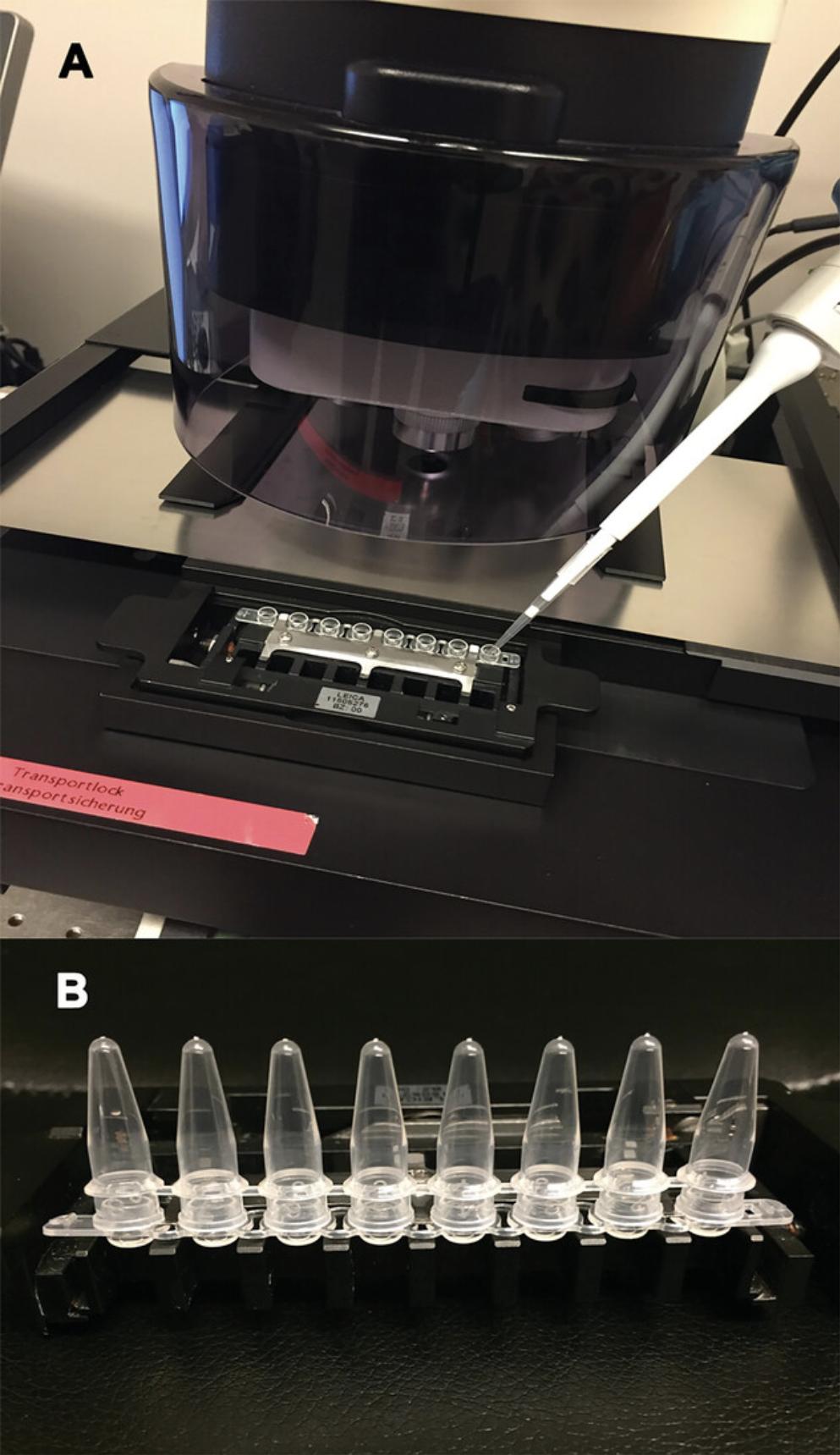
4.Label a 0.2-ml thin-walled 8-tube strip with domed caps with a permanent marker and load it onto the tissue collector of the microscope. Using an RNase-free pipet tip, load 1 cap with 40 µl distilled water (Fig. 6A). Following the microscope manufacturer's instructions, calibrate the collector tube to make sure that cut samples fall inside the tube cap.
5.Load a PEN slide onto the microscope and calibrate the laser beam on a blank section of the slide before cutting tissue. In the case of a Leica microscope, adjust for power, magnification, aperture, speed, specimen balance, and light intensity to ensure sharp cuts and rapid descent of the sample into the collector tube cap. Once you have determined the settings to be used, record them for future work. Then, load 40 µl lysis solution from the Ambion RNAqueous-Micro Kit in each collector tube cap.
6.Begin cutting desired tissue from the PEN slide into the caps. When all tissue on the slide has been cut or 100 to 120 min have passed (whichever comes first), remove the tube holder and place 0.2-ml thin-walled 8-strip tubes on the caps (Fig. 6B). Fix caps tightly on the tubes and centrifuge for 30 s at 5724 × g to force the lysis solution containing the cut tissue to fall into the tube. Immediately label tubes and store them at −80°C until RNA extraction (see Basic Protocol 4).
Basic Protocol 4: RNA EXTRACTION FROM LASER CAPTURE MICRODISSECTION SAMPLES
Typically, RNA yields will be ultra-low (picogram amounts) and will require the use of high-sensitivity, low-abundance assays. The purity of nucleic acids can be determined by UV spectroscopy using a NanoDrop 8000 (Thermo Fisher Scientific). Purity is considered acceptable when both the 260/280 and the 260/230 values are >2.0. RNA integrity can be determined using an Agilent 2100 Bioanalyzer system along with the Agilent RNA 6000 Pico assay. RNA quantity is best determined with a dsRNA dye-binding assay to avoid quantification of trace amounts of residual DNA. The preferred technique is to use a Qubit fluorometer and a high-sensitivity assay kit (Qubit® RNA HS Assay Kit, Thermo Fisher Scientific, cat. no. Q32852). If the RNA integrity number (RIN) is above 6 but the quantity is too low for downstream library preparation, the original RNA can be stored at −80°C until you have amassed sufficient quantity and quality for your application, and all the RNA can then be combined during library preparation. There is some debate about how long low-concentration samples can be stored, but ≤6 months is recommended. Olivieri et al. (2014) reported storing a 25-ng/µl human RNA sample at −80°C for 8 months without loss.
The quantity and quality of RNA required for library preparation for RNASeq will vary depending on the processing methodology used to generate the library. RINs are an unambiguous assessment of RNA integrity, and RIN values between 6 and 10 (as determined on a bioanalyzer) are appropriate for most RNASeq applications; however, if the quantity is low, higher quality (RIN ≥8.0) is better. It is important to note here that if RNAs with RIN values <8.0 are used, the results can be significantly impacted by 3′ biases because of general 5′ degradation of the transcripts. Caution should thus be taken regarding splice-form and isoform analyses, and this issue should also be considered prior to expression profiling analyses.
Using the TruSeq Stranded mRNA Library Prep Kit (Illumina, cat. no. RS-122-2103), the manufacturer's suggested minimum total amount of RNA to generate a library with sufficient complexity to include rare transcriptional events (∼25 M read pairs) without amplification is 100 ng. In our experience, libraries can be successfully generated with as little as 25 ng RNA, and analysis of libraries generated from even lower amounts of input RNA (Schuierer et al., 2017) suggest that 10 ng of high-quality RNA is sufficient to produce high-quality results. For situations where inputs are <50 ng, ribosomal depletion can be used, or RNA can be amplified to generate cDNA in sufficient amounts for sequencing using the SMART-Seq v4 Ultra Low Input RNA Kit (Clontech, cat. no. 634888); the lower limit of input suggested by the manufacturer is 60 pg. Thus, amplification of RNA can be used to produce a sequencing library from lower-abundance samples but requires an RIN value in the range of 7 to 10. To compare libraries, the same protocol must be used for all libraries, and yield varies by tissue; in our case, one tissue drove the need to amplify all libraries.
Irrespective of whether the RNA is amplified before library construction or used directly, commercial spike-in standards should be used to enable an objective, quantitative assessment of the sequencing results, including determining amplification biases, if appropriate. The exogenous spike-ins used in our experimentation are the Ambion ERCC RNA Spike-In Control Mixes (Life Technologies, cat. no. 4456740 and 4456739) (Devonshire et al., 2010; Jiang et al., 2011; Reid, 2005).
Materials
- RNA collector tubes containing LCM sample, stored at −80°C (see Basic Protocol 3, step 4)
- Ambion RNAqueous-Micro Kit (Life Technologies, cat. no. AM1931)
- 50 micro filter cartridge assembly
- 50 micro elution tubes
- 15-ml wash solution 1 concentrate
- 28-ml wash solution 2/3 concentrate
- 25 lysis solution
- 200-μl LCM additive
- 55-μl DNase 2U/μl
- 600-μl 10X DNase I buffer
- 150-μl DNase inactivation reagent
- 5-ml elution solution
1.Remove a collector tube kept at −80°C and let it thaw on ice.
2.Perform RNA extraction using the RNAqueous-Micro Kit according to the manufacturer's instructions.
3.Remove a 5-µl aliquot of the extracted RNA for quantification and integrity analysis and store it and the remaining extracted RNA at −80°C until RNASeq library preparation.
REAGENTS AND SOLUTIONS
Use distilled, deionized water in all recipes and protocol steps.
Diethyl pyrocarbonate (DEPC)-treated water
Mix 800 µl DEPC with 1 L water. Stir overnight and autoclave before using to get rid of DEPC. Use during entire experiment whenever water is required. Store at room temperature for ≤1 month.
Ethanol (EtOH)/polyester wax solution
Melt polyester wax (e.g., Steedman's wax, Electron Microscopy Sciences, cat. no. 19312) in an RNase-free beaker at 37°C, resulting in a milky white liquid. Mix equal volumes of 100% EtOH (anhydrous) and melted polyester wax. Store at 37°C for ≤1 day.
EtOH solution, 70%
Mix 70 ml 100% EtOH (anhydrous, molecular biology grade) with 30 ml autoclaved DEPC-treated water (see recipe), being sure to use RNase-free glassware (cylinders/beakers/bottles). Store at 4°C for ≤1 month.
EtOH solution, 96%
Mix 96 ml 100% EtOH (anhydrous, molecular biology grade) with 4 ml autoclaved DEPC-treated water (see recipe), being sure to use RNase-free glassware (cylinders/beakers/bottles). Store at 4°C for ≤1 month.
Fixative solution
Add Triton X-100 to 100% methanol (anhydrous, ultra-pure grade) to a final concentration of 0.1% (v/v) and mix by pipetting up and down. Store at 4°C for ≤1 month.
COMMENTARY
Background Information
A laser was first used for cell separation in the 1920s (Gilbrich-Wille, 2013), and this approach has been evolving ever since. Various methods are used for microdissection of samples to obtain a single cell type from a heterogeneous sample. Because of the presence of cell walls in plants, isolation of individual cells for RNA recovery has relied on the formation of protoplasts, which have lost their tissue identity (Efroni & Birnbaum, 2016), or on the enzymatic degradation of cell walls in intact roots, for example, followed by sorting of cells based on their expression of a fluorescent reporter that marks certain cell types (Geng et al., 2013). The latter approach is usually limited to use in Arabidopsis and rice and maize plants, which are amenable to transformation and are easily handled in large quantities (Evrard et al., 2012; Li & Arumuganathan, 2003; Li et al., 2004). Some techniques applied to animal tissue, including sectioning of frozen tissue blocks to select for specific cell populations, irradiation of stained sections to destroy unwanted cells, and microdissection with manual tools such as sharp needles (Emmert-Buck et al., 1996; Outlaw & Zhang, 2001; Schrader et al., 2004), are not applicable to plant tissue. Further, plant tissue and cells present specific challenges in comparison to animal tissue due to the presence of plant cell walls, vacuoles, chloroplasts, and secondary metabolites, making LCM a better choice (Cosgrove, 2005; Giannella et al., 1997). In addition, the methods applied to animal tissue fail to offer the precision, ease, and efficiency required to generate RNA at a quality and quantity essential for sequencing. For example, manual microdissection is precise, but it is also time consuming and allows degradation of RNA to occur.
To address the objectives of resolving the progression of events at the tissue/cellular level in Medicago truncatula /rhizobia symbiosis and understanding the signaling occurring between cells in the maturation zone in space and time, we were unable to use most of the above-mentioned approaches when we began the experiments. Our goal of measuring the transcriptome for each maturation zone cell type involved in nodule formation in space and time led us to LCM in 2017. Multiple protocols using plant cells have since been developed for doing single-cell RNASeq (Ryu et al., 2019) or single-nuclei RNASeq (Conde et al., 2021; Thibivilliers et al., 2020). These techniques require a sequencing platform that is quite expensive for a single lab. In addition, because of the plant cell wall, the protocols require optimized digestion of the tissue of choice and rapid processing of the protoplasts or nuclei in cold rooms near the point of sequencing (less than an hour from harvest to loading the sequencer), making scheduling on a common instrument difficult. We discovered that we could use the information from an LCM experiment (Schnabel et al., 2023) to inform single-nuclei RNASeq done in the same system and have combined the data for more information (Pereira et al. in review).
In animal systems, tissue sections can be stained prior to laser capture to differentiate between cell types (for example, tumor and nontumor cells); this aids visualization of the cells and does not alter the cellular constituents. In addition, immunohistochemical staining and soluble immunoassays conducted on LCM samples have confirmed that cellular proteins are preserved. The addition of a tissue fixation step prior to sectioning allows capture of different cell types by permitting examination of their morphology without staining (Jensen, 2013). The technique uses sterile disposable films on microscope slides, which minimizes contamination and allows one-step transfer of cells that is far faster than manual microdissection. Another advantage of LCM is that it does not require cell-specific markers or tags, genetically altered cell lines, special tissue or cell properties (e.g., epidermal peels), or differential cell size or density achieved through fractionation (Nelson et al., 2006). Depending on how they are handled during fixation and cutting, the harvested cells of interest can provide DNA, RNA, and protein for gene expression analysis.
The advantages of using LCM are numerous, but it does have disadvantages, which vary based on the target cells and tissue used for the procedure. For example, if the chosen cell type is quite rare in the tissue of interest, LCM can require a large amount of tissue and time. In this case, thousands or millions of cells may need to be dissected to obtain sufficient material for further analysis (Jensen, 2013) and single-cell RNASeq might be better. Although the laser beam cuts cells accurately, it also tends to damage adjacent cells while cutting, and although the damaged cells might be lost during downstream processing, they also might contaminate the cut cells. Nevertheless, the enrichment of cells up to 100-fold can clearly differentiate the signal of any underrepresented cell types in any tissue (Jensen, 2013). The main limitation of LCM is the requirement of identifying cells of interest based on morphological characteristics, which usually requires skilled personnel. In contrast, single-cell RNASeq can identify many more cell types using computational clustering, but the computation requires some knowledge of tissue-specific expression to confirm assignments, making the two techniques complementary in a system.
Prior to LCM, plant tissue requires chemical fixation; this fixation is essential to stabilize the cell contents (in this case, RNA) and to preserve the histological integrity of the cells. The fixation procedure depends on the tissue type and on the downstream analysis to be performed. For example, animal tissues are mostly cryosectioned, and the amount of RNA harvested from tissue prepared with methanol-EtOH/acetic acid as the fixative is higher than that from tissue treated with cross-linking fixatives such as formaldehyde (Kerk et al., 2003). The fixation methods used also depend on whether the intent is to extract DNA, RNA, or protein; a single fixation procedure cannot be used to extract all of the above. Any single fixation protocol is also unlikely to be optimal for all plant tissue sources, and the user should optimize fixation and processing protocols on a case-by-case basis (Nelson et al., 2006).
To extract RNA, one needs not only to fix the RNA in place but also to be very careful to avoid contamination, and many of the fixation and embedding protocols for preparing plant tissue for sectioning do not protect RNA. Additionally, the microscope itself can be an issue. The LCM microscope is expensive for a single laboratory or even as a shared piece of equipment used exclusively for RNA extraction (Curran et al., 2000). Because it is often shared by multiple users, extra steps are also required to prevent sample contamination with RNase. In situations where a microscope is reserved on a time basis, it is important to add the extra time required for these steps to the reservation.
Plant tissue is usually fixed and embedded before sectioning (Weigel & Glazebrook, 2008), though there are some tissues, such as woody plant xylem tissues, for which cryosectioning is preferred (Blokhina et al., 2017; Knapp et al., 2012). Fixatives used in the preparation of tissue for LCM with the intent of downstream RNA analysis depend on the resin to be used for embedding. Examples in the literature include xylene embedded in paraffin for seeds (Belmonte et al., 2013), but any method using xylene can be problematic from a safety standpoint. Polyester wax, which is compatible with methanol fixation, has been successfully used in M. truncatula (Gomez & Harrison, 2009; Hogekamp et al., 2011; Roux et al., 2014); methanol is a better fixative for RNA (Srinivasan et al., 2002) and is easier to discard. It has been reported that non-cross-linking alcoholic reagents always yielded superior results as nucleic-acid fixatives compared to aldehydes (Giannella et al., 1997; Noguchi et al., 1997; Srinivasan et al., 2002). In particular, EtOH and methanol preserve nucleic acids better because they cause little chemical change. Further, absolute (100%) EtOH and methanol are known to be excellent fixatives for preserving both high-molecular-weight DNA and RNA. The low molecular weight and rapid tissue penetration of these alcohols are thought to provide uniform tissue fixation and minimal loss of cellular content (Giannella et al., 1997; Noguchi et al., 1997; Srinivasan et al., 2002).
The evolving advances in RNASeq techniques that can be used with low-abundance samples, e.g., input amounts in the picogram range with minimized endpoint biases, have expanded the use of LCM from quantitative PCR to RNASeq library preparation, allowing the capture of much more information. The general workflow for preparing RNASeq libraries is RNA isolation for reverse transcription into cDNA; low-abundance samples (<100 ng total RNA) are pre-amplified to generate sufficient amounts for input into library preparation. Because very small amounts of RNA are used for amplification, certain nonlinear coverage biases can arise from these procedures, which can impact quantitative accuracy during data analysis. There are now several commercially available kits for amplification of cDNA from very-low-input samples (5 to 50 pg), such as the Ovation RNA amplification kit (NuGEN), SMARTer amplification kit (Clontech), TransPlex WTA2 kit (Sigma-Aldrich), and SuperAmp kit (Miltenyi Biotech); the various approaches and bias comparisons are detailed elsewhere (Shanker et al., 2015). To account for biases, exogenous RNA spike-ins, such as ERCC standards, can be included prior to amplification to determine relative and absolute transcript counts (Jiang et al., 2011). However, current methods are well suited to investigate the polyadenylated (poly(A)+) transcript class of RNA molecules. To account for poly(A)- transcript classes, such as microRNAs, lncRNAs, circular RNAs, or even prokaryotic RNAs, ribosomal depletion combined with random hexamer priming can be used to capture the poly(A)+ and poly(A)- transcript species.
Critical Parameters
As mentioned in the Introduction, one of the most important aspects of the procedures is maintaining an RNase-free workspace throughout all steps, followed by keeping temperatures as low as is practical. Specifically, the following are critical parameters:
- 1.The use of gloves as well as sterile filter tips and RNase-free glassware and water is always required.
- 2.Dedicate a laboratory bench for RNA experiments and cover it with paper that is changed frequently to minimize contamination.
- 3.All consumables should be immersed in DEPC-treated water and autoclaved. Note that DEPC-treated water is RNase free after autoclaving but can no longer destroy additional RNase because autoclaving destroys the DEPC. Therefore, when using equipment such as centrifuges, incubators, and other items that cannot be autoclaved (for example, cooling plates), clean surfaces thoroughly with RNase AWAY.
- 4.Always use autoclaved DEPC-treated water whenever water is required. Specific details regarding treating consumables with DEPC are mentioned in the Materials section when applicable; treatment with UV light can also destroy RNase depending on the shape of the object.
- 5.It is assumed molecular biology consumables (such as tubes) that are wrapped in the factory are RNase free as they come out of the wrapping, but if this is a concern, treat them as described for plastic cassettes.
Troubleshooting
- 1.If the resulting RNA is degraded, the most likely reason is contamination by exogenous RNase or improper infiltration before embedding (due to endogenous RNase). To avoid contamination, always maintain an RNase-free environment (see Critical Parameters) and keep the temperature low whenever possible. To ensure proper infiltration, make sure that the tissue is free to float in the cassettes and that the cassettes are submerged in the infiltration solutions (see Basic Protocol 1). Follow the directions regarding the length of time for holding materials at different time points in the protocols and the length of time for capture; in each of these cases, time is an issue for RNA integrity. Remember that once the PEN slide is de-waxed, all the tissue should be cut within 5 days, or the RNA will start degrading (see Basic Protocol 3). Therefore, the number of slides de-waxed at any one time should reflect the amount of tissue that you can cut using LCM in that time.
- 2.If the RNA quality is satisfactory but the quantity of RNA is low, more tissue area needs to be cut using the LCM microscope (see Basic Protocol 3) and/or a thicker section needs to be produced (see Basic Protocol 2). Each tissue type will require some preliminary experiments to determine the amount of tissue necessary for a given application.
Understanding Results
The RNA isolated using the above procedures should have an RIN value in the range of 7 to 10. In our experience, simply cutting whole root sections instead of collecting individual tissue types, i.e., 71 mm2 of M. truncatula roots cut in sections of 12-µm thickness, yielded 135 ng RNA with an RIN of 6.9, as determined on a bioanalyzer. In another experiment, half of that amount, or 35 mm2 of M. truncatula roots cut in sections of 12-µm thickness, yielded 70 ng RNA, suggesting that the yield is proportional to the area being cut. However, yield varies by individual tissue and must be determined on a case-by-case basis.
Time Considerations
- 1.Infiltration of tissue samples will take 1.5 days (see Basic Protocol 1).
- 2.Embedding of infiltrated tissue samples will take several hours (see Basic Protocol 1). This should be done on the same day as when the last wash with polyester wax is completed.
- 3.Sectioning of tissue samples should be done within 1 hr of placing the embedded tissue on the microtome because polyester wax melts at 37°C and can be soft at room temperature (see note to Basic Protocol 2).
- 4.LCM should be done in 2-hr increments based on the start time of setting up the tubes because the lysis solution will crystallize and no longer protect the RNA otherwise (see Basic Protocol 3).
- 5.RNA isolation with the kit mentioned takes hours, and depending on whether amplification is required, the creation of RNA libraries can take 3 to 4 days (see Basic Protocol 4).
Acknowledgments
We would like to acknowledge John Harada and Julie Marie Pelletier of UC Davis for initial introduction to an LCM protocol in their laboratory and Nathalie Rodde of INRA for sharing tips on the fixation protocol used in the laboratory of Pascal Gamas as well as tips for using polyester wax. We would also like to acknowledge the Clemson Light Imaging Facility and Dr. Terri Bruce and Dr. Rhonda Powell for use of and training on the laser capture microscope and the Clemson Computational Genomics Laboratory for RNA assessment and RNASeq library preparation. Finally, we thank the 2017 cohort of Camp Lasernode, an NSF-sponsored workshop, for using and commenting on an initial draft of this protocol; members of the Frugoli laboratory for reading and commenting on the manuscript; and the NSF IOS#1444461 for supporting this work. As this work was funded by the NSF, NSF-PAR deposit is required.
Author Contributions
Suchitra Chavan : investigation, methodology, validation, visualization, writing original draft; Elise Schnabel : data curation, investigation, methodology, supervision, writing original draft; Christopher Saski : methodology, resources, supervision, writing original draft; Julia Frugoli : conceptualization, formal analysis, funding acquisition, methodology, project administration, resources, visualization, writing original draft, writing review and editing
Conflicts of Interest
The authors state that they are not aware of any conflicts of interest.
Open Research
Data Availability Statement
Data sharing is not applicable to this article as no datasets were generated or analyzed during the current study.
Literature Cited
- Belmonte, M. F., Kirkbride, R. C., Stone, S. L., Pelletier, J. M., Bui, A. Q., Yeung, E. C., Hashimoto, M., Fei, J., Harada, C. M., Munoz, M. D., Le, B. H., Drews, G. N., Brady, S. M., Goldberg, R. B., & Harada, J. J. (2013). Comprehensive developmental profiles of gene activity in regions and subregions of the Arabidopsis seed. Proceedings of the National Academy of Sciences of the United States of America , 110, E435–E444. https://doi.org/10.1073/pnas.1222061110
- Blokhina, O., Valerio, C., Sokołowska, K., Zhao, L., Kärkönen, A., Niittylä, T., & Fagerstedt, K. (2017). Laser capture microdissection protocol for xylem tissues of woody plants. Frontiers in Plant Science , 7, 1965. https://doi.org/10.3389/fpls.2016.01965
- Conde, D., Triozzi, P. M., Balmant, K. M., Doty, A. L., Miranda, M., Boullosa, A., Schmidt, H., Pereira, W., Dervinis, C., & Kirst, M. (2021). A robust method of nuclei isolation for single-cell RNA sequencing of solid tissues from the plant genus Populus, PLOS ONE , 16, e0251149. https://doi.org/10.1371/journal.pone.0251149
- Cosgrove, D. J. (2005). Growth of the plant cell wall. Nature Reviews Molecular Cell Biology , 6, 850–861. https://doi.org/10.1038/nrm1746
- Curran, S., McKay, J. A., McLeod, H. L., & Murray, G. I. (2000). Laser capture microscopy. Journal of Clinical Pathology: Molecular Pathology , 53, 64–68. https://doi.org/10.1136/mp.53.2.64
- Devonshire, A. S., Elaswarapu, R., & Foy, C. A. (2010). Evaluation of external RNA controls for the standardisation of gene expression biomarker measurements. BMC Genomics [Electronic Resource] , 11, 662. https://doi.org/10.1186/1471-2164-11-662
- Efroni, I., & Birnbaum, K. D. (2016). The potential of single-cell profiling in plants. Genome Biology , 17, 65. https://doi.org/10.1186/s13059-016-0931-2
- Emmert-Buck, M. R., Bonner, R. F., Smith, P. D., Chuaqui, R. F., Zhuang, Z., Goldstein, S. R., Weiss, R. A., & Liotta, L. A. (1996). Laser capture microdissection. Science , 274, 998–1001. https://doi.org/10.1126/science.274.5289.998
- Evers, D. L., He, J., Kim, Y. H., Mason, J. T., & O'Leary, T. J. (2011). Paraffin embedding contributes to RNA aggregation, reduced RNA yield, and low RNA quality. The Journal of Molecular Diagnostics , 13, 687–694. https://doi.org/10.1016/j.jmoldx.2011.06.007
- Evrard, A., Bargmann, B. O., Birnbaum, K. D., Tester, M., Baumann, U., & Johnson, A. A. (2012). Fluorescence-activated cell sorting for analysis of cell type-specific responses to salinity stress in Arabidopsis and rice. Methods in Molecular Biology , 913, 265–276. https://doi.org/10.1007/978-1-61779-986-0_18
- Geng, Y., Wu, R., Wee, C., Xie, F., Wei, X., Chan, P., Tham, C., Duan, L., & Dinneny, J. (2013). A spatiotemporal understanding of growth regulation during the salt stress response in Arabidopsis. The Plant Cell , 25, 2132–2154. https://doi.org/10.1105/tpc.113.112896
- Giannella, C., Zito, F. A., Colonna, F., Paradiso, A., Marzullo, F., Alaibac, M., & Schittulli, F. (1997). Comparison of formalin, ethanol, and Histochoice fixation on the PCR amplification from paraffin-embedded breast cancer tissue. European Journal of Clinical Chemistry and Clinical Biochemistry , 35, 633–635.
- Gilbrich-Wille, C. (2013). Leica Microsystems. History of Laser Microdissection. (Retrieved from https://www.leica-microsystems.com/science-lab/history/history-of-laser-microdissection/) last accessed on 6 May 2013
- Gomez, S. K., & Harrison, M. J. (2009). Laser microdissection and its application to analyze gene expression in arbuscular mycorrhizal symbiosis. Pest Management Science , 65, 504–511. https://doi.org/10.1002/ps.1715
- Gomez, S. K., Javot, H., Deewatthanawong, P., Torres-Jerez, I., Tang, Y., Blancaflor, E. B., Udvardi, M. K., & Harrison, M. J. (2009). Medicago truncatula and Glomus intraradices gene expression in cortical cells harboring arbuscules in the arbuscular mycorrhizal symbiosis. BMC Plant Biology [Electronic Resource] , 9, 10. https://doi.org/10.1186/1471-2229-9-10
- Hogekamp, C., Arndt, D., Pereira, P. A., Becker, J. D., Hohnjec, N., & Küster, H. (2011). Laser microdissection unravels cell-type-specific transcription in arbuscular mycorrhizal roots, including CAAT-box transcription factor gene expression correlating with fungal contact and spread. Plant Physiology , 157, 2023–2043. https://doi.org/10.1104/pp.111.186635
- Jensen, E. (2013). Laser-capture microdissection. The Anatomical Record , 296, 1683–1687. https://doi.org/10.1002/ar.22791
- Jiang, L., Schlesinger, F., Davis, C. A., Zhang, Y., Li, R., Salit, M., Gingeras, T. R., & Oliver, B. (2011). Synthetic spike-in standards for RNA-seq experiments. Genome Research , 21, 1543–1551. https://doi.org/10.1101/gr.121095.111
- Kerk, N. M., Ceserani, T., Tausta, S. L., Sussex, I. M., & Nelson, T. M. (2003). Laser capture microdissection of cells from plant tissues. Plant Physiology , 132, 27–35. https://doi.org/10.1104/pp.102.018127
- Kleber, R., & Kehr, J. (2006). Preparation and quality assessment of RNA from cell-specific samples obtained by laser microdissection. In J. Salinas & J. J. Sanchez-Serrano (Eds.), Methods in molecular biology, Arabidopsis protocols , Vol. 323 (pp. 367–377). New Jersey: Humana Press.
- Knapp, E., Flores, R., Scheiblin, D., Modla, S., Czymmek, K., & Yusibov, V. (2012). A cryohistological protocol for preparation of large plant tissue sections for screening intracellular fluorescent protein expression. Biotechniques , 52, 31–37. https://doi.org/10.2144/000113778
- Li, L., & Arumuganathan, K. (2003). Microcloning of maize chromosome 9 by using a flow-sorting technique. Plant Molecular Biology Reporter , 21, 359–364. https://doi.org/10.1007/BF02772585
- Li, L., Arumuganathan, K., Gill, K. S., & Song, Y. (2004). Flow sorting and microcloning of maize chromosome 1. Hereditas , 141, 55–60. https://doi.org/10.1111/j.1601-5223.2004.01847.x
- Nelson, T., Tausta, S. L., Gandotra, N., & Liu, T. (2006). Laser microdissection of plant tissue: What you see is what you get. Annual Review of Plant Biology , 57, 181–201. https://doi.org/10.1146/annurev.arplant.56.032604.144138
- Noguchi, M., Furuya, S., Takeuchi, T., & Hirohashi, S. (1997). Modified formalin and methanol fixation methods for molecular biological and morphological analyses. Pathology International , 47, 685–691. https://doi.org/10.1111/j.1440-1827.1997.tb04442.x
- Olivieri, E. H., Franco, Lde. A., Pereira, R. G., Mota, L. D., Campos, A. H., & Carraro, D. M. (2014). Biobanking practice: RNA storage at low concentration affects integrity. Biopreservation and Biobanking , 12, 46–52. https://doi.org/10.1089/bio.2013.0056
- Outlaw, W. H. Jr., & Zhang, S. (2001). Single-cell dissection and microdroplet chemistry. Journal of Experimental Botany , 52, 605–614. https://doi.org/10.1093/jexbot/52.356.605
- Reid, L. H. (2005). Proposed methods for testing and selecting the ERCC external RNA controls. BMC Genomics [Electronic Resource] , 6, 1–18. https://doi.org/10.1186/1471-2164-6-150
- Roux, B., Rodde, N., Jardinaud, M. F., Timmers, T., Sauviac, L., Cottret, L., Carrère, S., Sallet, E., Courcelle, E., Moreau, S., Debellé, F., Capela, D., de Carvalho-Niebel, F., Gouzy, J., Bruand, C., & Gamas, P. (2014). An integrated analysis of plant and bacterial gene expression in symbiotic root nodules using laser-capture microdissection coupled to RNA sequencing. Plant Journal , 77, 817–837. https://doi.org/10.1111/tpj.12442
- Ryu, K. H., Huang, L., Kang, H. M., & Schiefelbein, J. (2019). Single-cell RNA sequencing resolves molecular relationships among individual plant cells. Plant Physiology , 179, 1444–1456. https://doi.org/10.1104/pp.18.01482
- Schnabel, E., Thomas, J., El-Hawaz, R., Gao, Y., Chavan, S., Feltus, F. A., & Frugoli, J. A. (2023). Laser capture microdissection transcriptome of roots responding to rhizobia reveals spatiotemporal signaling between tissues. bioRxiv , 2023.03.14.532650 , https://doi.org/10.1101/2023.03.14.532650
- Schrader, J., Nilsson, J., Mellerowicz, E., Berglund, A., Nilsson, P., Hertzberg, M., & Sandberg, G. (2004). A high-resolution transcript profile across the wood-forming meristem of poplar identifies potential regulators of cambial stem cell identity. Plant Cell , 16, 2278–2292. https://doi.org/10.1105/tpc.104.024190
- Schuierer, S., Carbone, W., Knehr, J., Petitjean, V., Fernandez, A., Sultan, M., & Roma, G. (2017). A comprehensive assessment of RNAseq protocols for degraded and low quantity samples. BMC Genomics , 18, 442. https://doi.org/10.1186/s12864-017-3827-y
- Shanker, S., Paulson, A., Edenberg, H. J., Peak, A., Perera, A., Alekseyev, Y. O., Beckloff, N., Bivens, N. J., Donnelly, R., Gillaspy, A. F., Grove, D., Gu, W., Jafari, N., Kerley-Hamilton, J. S., Lyons, R. H., Tepper, C., & Nicolet, C. M. (2015). Evaluation of commercially available RNA amplification kits for RNA sequencing using very low input amounts of total RNA. Journal of Biomolecular Techniques , 26, 4–18. https://doi.org/10.7171/jbt.15-2601-001
- Simone, N. L., Bonner, R. F., Gillespie, J. W., Emmert-buck, M. R., & Liotta, L. A. (1998). Laser-capture microdissection: Opening the microscopic frontier to molecular analysis. Trends in Genetics , 14, 272–276. https://doi.org/10.1016/S0168-9525(98)01489-9
- Srinivasan, M., Sedmak, D., & Jewell, S. (2002). Effect of fixatives and tissue processing on the content and integrity of nucleic acids. American Journal of Pathology , 161, 1961–1971. https://doi.org/10.1016/S0002-9440(10)64472-0
- Thibivilliers, S., Anderson, D., & Libault, M. (2020). Isolation of plant root nuclei for single cell RNA sequencing. Current Protocols in Plant Biology , 5, e20120. https://doi.org/10.1002/cppb.20120
- Weigel, D., & Glazebrook, J. (2008). Fixation, embedding, and sectioning of plant tissues. Cold Spring Harbor Protocols , 2008, pdb.prot4941. https://doi.org/10.1101/pdb.prot4941
Citing Literature
Number of times cited according to CrossRef: 1
- Sareena Sahab, Matthew J Hayden, John Mason, German Spangenberg, An Efficient Fluorescence‐Activated Protoplast Sorting (FAPS) and Regeneration Protocol for Canola (Brassica napus), Current Protocols, 10.1002/cpz1.70008, 4 , 9, (2024).

