Creating Planet Microbe Data Packages
Kai Blumberg, Alise J Ponsero, Bonnie Hurwitz
Abstract
Marine microbial ecology requires the systematic comparison of biogeochemical and sequence data to analyze environmental influences on the distribution and variability of microbial communities. With ever-increasing quantities of metagenomic data, there is a growing need to make datasets Findable, Accessible, Interoperable, and Reusable (FAIR) across diverse ecosystems. FAIR data is essential to developing analytical frameworks that integrate microbiological, genomic, ecological, oceanographic, and computational methods. Although community standards defining the minimal metadata required to accompany sequence data exist, they haven't been consistently used across projects, precluding interoperability. Moreover, these data are not machine-actionable or discoverable by cyberinfrastructure systems. By making ‘omic and physicochemical datasets FAIR to machine systems, we can enable sequence data discovery and reuse based on machine-readable descriptions of environments or physicochemical gradients.
In this work, we developed a novel technical specification for dataset encapsulation for the FAIR reuse of marine metagenomic and physicochemical datasets within cyberinfrastructure systems. This includes using Frictionless Data Packages enriched with terminology from environmental and life-science ontologies to annotate measured variables, their units, and the measurement devices used. This approach was implemented in Planet Microbe, a cyberinfrastructure platform and marine metagenomic web-portal. Here, we discuss the data properties built into the specification to make global ocean datasets FAIR within the Planet Microbe portal. We additionally discuss the selection of, and contributions to marine-science ontologies used within the specification. Finally, we use the system to discover data by which to answer various biological questions about environments, physicochemical gradients, and microbial communities in meta-analyses. This work represents a future direction in marine metagenomic research by proposing a specification for FAIR dataset encapsulation that, if adopted within cyberinfrastructure systems, would automate the discovery, exchange, and re-use of data needed to answer broader reaching questions than originally intended.
Steps
Setup
Step one including downloading the relevant repositories. You'll want to download the relevant repositories to the same directory.
Open a terminal or shell program and navigate to a base working directory.
Clone the planet-microbe-datapackages repository:
git clone git@github.com:hurwitzlab/planet-microbe-datapackages.git
```Note you could also use the https version
Clone the planet-microbe-scripts repository
git clone git@github.com:hurwitzlab/planet-microbe-scripts.git
Prepare datapackage.json
Prepare data for creation of Planet Microbe Frictionless Datapackage
How to create the datapackage.json wrapper for your tabular dataset(s).
Prepare ONTOLOGY_MAPPING.tsv file(s)
Make sure to do this for every individual TSV file that is part of one datapackage product
This can be done by transposing the column headers of your tabular dataset to be the items in the first row of a new csv file. Simply copy and paste the column headers of the original dataset using excel (or similar program) and in a new sheet past them (transposed).
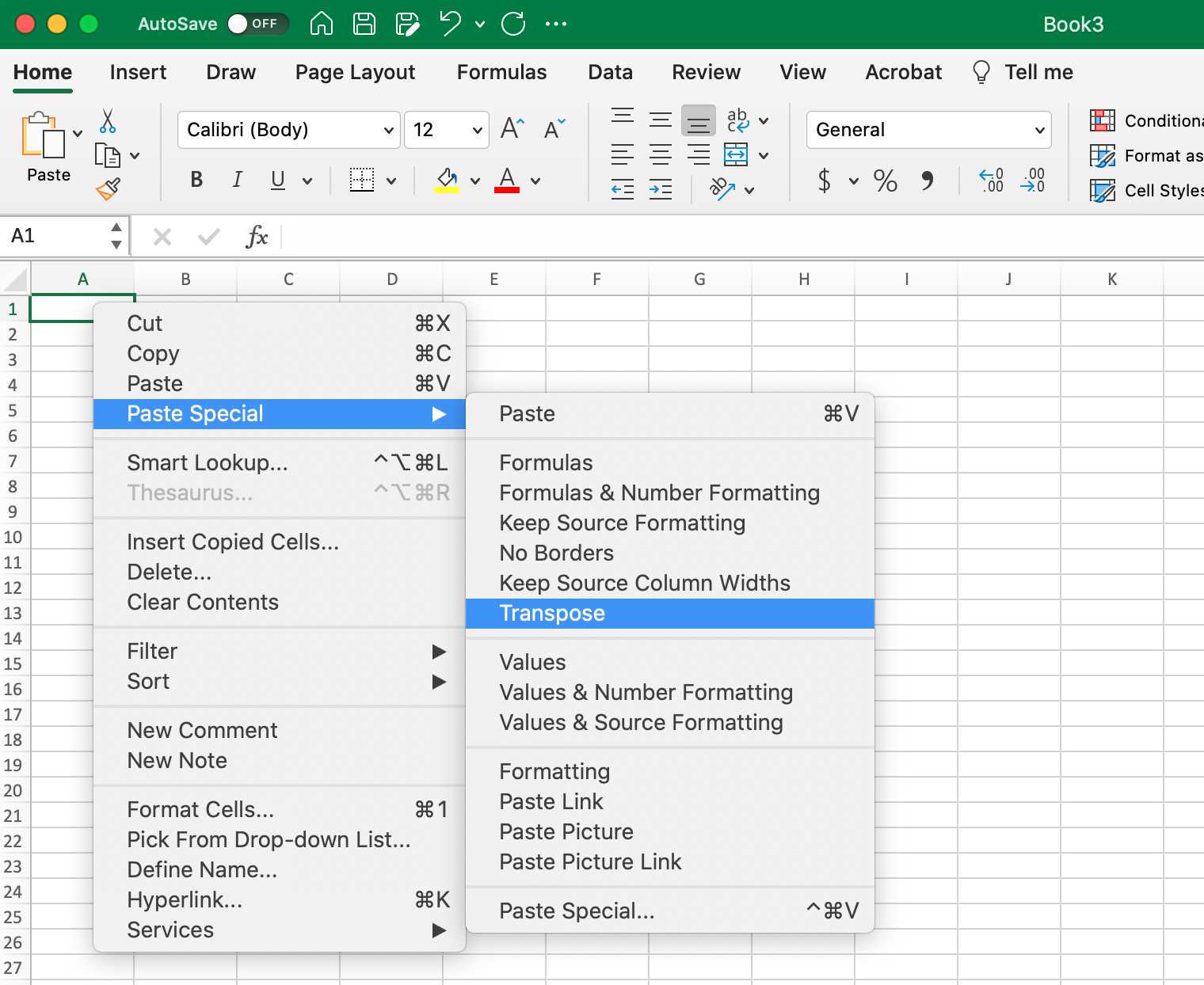
Next add the following as the new column headers:
| A | B | C | D | E | F | G | H | I | J | K | L |
|---|---|---|---|---|---|---|---|---|---|---|---|
| parameter | rdf type purl label | rdf type purl | pm:searchable | units label | units purl | measurement source purl label | measurement source purl | pm:measurement source protocol | pm:source url | frictionless type | frictionless format |
Populate the new ONTOLOGY_MAPPING.tsv one column at a time.
The "pm:searchable" column is a boolean flag (TRUE or FALSE) for columns that have a corresponding "rdf type purl" annotation columns. If the column from the dataset is about a physicochemical variable from the pmo_searchable_terms.tsv that you want to made searchable make sure to fill it out completely and set the "pm:searchable" to "TRUE". Otherwise the for a given column it can be set to false and everything else left blank.
- Fill in the "rdf type purl" column with IRIs from of terms from this pmo_searchable_terms.tsv.
- Fill in the "units purl" column with IRIs from of terms from the UO (Units Ontology) which you can view and browser from here.
- Fill in the "measurement source purl" columns with term from the OBI (Ontology for Biomedical Investigations) device hierarchy, which you can view and browser from here.
- Fill in the "pm:source url" column with any link relevant to the dataset columns collection or source e.g., "https://www.ncbi.nlm.nih.gov/bioproject/213098"
- Fill in the "frictionless type" column with one of: "string", "number", "date", or "datetime".
- If necessary, fill in the "frictionless format" column for dates datetypes with custom formatting string patterns, e.g., "%Y-%m-%dT%H:%M" or default for ISO standard datetimes.
- If uploading INSDC submited data (from EBI or NCBI) make sure to fill out one column with a "Biosample identifier assigned by the National Center for Biotechnology Information" aka "http://purl.obolibrary.org/obo/PMO_00000122"
- If uploading MIxS compliant data make sure to annotated the appropriate columns for following ENVO MIxS triad:
biome http://purl.obolibrary.org/obo/ENVO_00000428
environmental feature http://purl.obolibrary.org/obo/ENVO_00002297
environmental material http://purl.obolibrary.org/obo/ENVO_00010483
Note that the columns "rdf type purl label', "units label" and "measurement source purl label" do not need to be filled in those are just for your convenience as you fill out the accompanying "purl" columns.
An example filled out ONTOLOGY_MAPPING.tsv file might look like the following:
Creating Data Package Templates
This example command is how one can generate a tabular data package JSON template for the OSD data set:
cd planet-microbe-scripts
cat example_ontology_mappings/OSD.tsv | ./scripts/schema_tsv_to_json.py > example_data_packages/osd/datapackage.json
```The JSON was then hand-edited to add missing information and correct names, types, and units.
For more information on FD Table Schemas see [http://frictionlessdata.io/specs/table-schema/](http://frictionlessdata.io/specs/table-schema)
This can be replicate for a new dataset by running the similar command with the newly created ONTOLOGY_MAPPING.tsv file(s). Recreate this command in the new directory with the new project files in the [planet-microbe-datapackages repository](https://github.com/hurwitzlab/planet-microbe-datapackages). E.g.,
cd planet-microbe-datapackages/NEW_DATASET/
cat ONTOLOGY_MAPPING.tsv | #PATH_TO_planet-microbe-scripts_REPO#./scripts/schema_tsv_to_json.py > datapackage_component1.json
Finalize the datapackage_component.json file
Open the file in a text editor such as atom or sublime text and modify the following information specific to the resource in question.
{
"name": "#ADD NAME e.g., sample",
"title": "#ADD tile",
"profile": "#tabular-data-resource",
"pm:resourceType": "#ADD TYPE",
"path": "#ADD FILEPATH e.g., FILEPATH.tsv",
"dialect": {
"delimiter": "#Add delimiter e.g., \t",
"header": true,
"caseSensitiveHeader": true
},
"format": "csv",
"mediatype": "text/tab-separated-values",
"encoding": "UTF-8",
"hash": "OPTIONALLY ADD file e.g., hasheac36d6747691e1061718509828598b1",
"schema": {
"fields": [ ...
}
Note the following "pm:resourceTypes" are accepted:
"pm:resourceType": "niskin",
"pm:resourceType": "campaign",
"pm:resourceType": "sample",
"pm:resourceType": "sampling_event",
"pm:resourceType": "ctd",
The following is an skeleton of the Planet Microbe Datapackage main json file for a new project
{
"@context": {
"pm": "http://purl.obolibrary.org/obo/PMO_00000000"
},
"profile": "tabular-data-package",
"name": "#ADD NAME",
"title": "#ADD TITLE",
"description": "#ADD DESCRIPTION",
"pm:selfUrl": "https://github.com/hurwitzlab/planet-microbe-datapackages/tree/master/#ADD_DIR_NAME",
"homepage": "#ADD HOMEPAGE",
"keywords": [
"#ADD KEYWORDS",
"#ADD MORE KEYWORDS"
],
"sources": [
{
"title": "#ADD SOURCE(s)",
"path": "ADD URL FOR SOURCE"
}
],
"licenses": [
{
"name": "CC-BY-3.0",
"title": "Creative Commons Attribution 3.0 Unported",
"path": "https://creativecommons.org/licenses/by/3.0/"
}
],
"resources": [
{#ADD datapackage_component1.json CODE HERE},
{... #Add more datapackage components if needed}
]
}
```Save this as a new (main) **datapackage.json** file for the project.
For each tabular dataset file that was prepared in the above steps open up the "datapackage_componentX.json" files one at a time and paste the entirety of the json file into the {}'s inside the "resources" block. This way each file will be annotated in a FAIR way that can be uploaded into the Planet Microbe Database.
Make sure to also fill in the other information about the datapackage for example the "name" "description" "keywords" etc.
Optional Step, add constraints for select fields
Optionally for fields which should follow a numeric constraint such as such as Latitude and Longitude, add and or modify the json file for each column header for each resource to change them from something like the following:
{
"name": "Latitude",
"type": "number",
"format": "default",
"rdfType": "http://purl.obolibrary.org/obo/OBI_0001620",
},
into something like this with a constraint block specified with numerical values e.g., -90 and 90 for Latitude.
{
"name": "Latitude",
"type": "number",
"format": "default",
"rdfType": "http://purl.obolibrary.org/obo/OBI_0001620",
"constraints": {
"required": false,
"minimum": -90,
"maximum": 90
},
Validate datapackage.json
Validating Data Packages
First, make sure you have a Python 3 virtual environment setup:
Alternatively create a conda environment:
Run the validation script:
Example command to validate a Datapackage:
scripts/validate_datapackage.py ../planet-microbe-datapackages/OSD/datapackage.json
Optional Step, run GoodTables validation for constraints
This is based on the original goodtables script avaiable from: https://goodtables.readthedocs.io/en/latest/
First install the goodtables library using pip:
Example call using the script

