Concentration, extraction and quantification of SARS-CoV-2 in wastewater via droplet digital PCR
Jim Huang, August Luc, Sarah Kane, Rose Nash, Susan De Long, Carol Wilusz
Disclaimer
DISCLAIMER – FOR INFORMATIONAL PURPOSES ONLY; USE AT YOUR OWN RISK
The protocol content here is for informational purposes only and does not constitute legal, medical, clinical, or safety advice, or otherwise; content added to protocols.io is not peer reviewed and may not have undergone a formal approval of any kind. Information presented in this protocol should not substitute for independent professional judgment, advice, diagnosis, or treatment. Any action you take or refrain from taking using or relying upon the information presented here is strictly at your own risk. You agree that neither the Company nor any of the authors, contributors, administrators, or anyone else associated with protocols.io, can be held responsible for your use of the information contained in or linked to this protocol or any of our Sites/Apps and Services.
Abstract
This protocol describes the method used at Colorado State University to extract and quantify SARS-CoV-2 in wastewater collected from sewers and at wastewater treatment plants. The method relies on concentration via ultrafiltration, silica column based purification, and quantification via digital droplet reverse transcription PCR. Endogenous and exogenous controls are described.
Steps
Preparation
Sample Description and Treatment
This protocol is designed for use with untreated influent from wastewater treatment plants. Samples should be provided in 50ml conical tubes with ~40ml of influent in each tube. Optimally, the samples should be flow proportional, collected over a 24 hour time period, and stored at ~4°C for transport to the laboratory for processing. The protocol is also appropriate for grab samples and for flow/time proportional samples collected from sewers.
Work Area Set Up
- Wipe down Biosafety Cabinet (BSC) with 70% ethanol
- Thaw sufficient BCoV spike for the number of samples [(N+2)x 14.5μl] - store on ice
- Perform Start Up Procedure on Innovaprep and insert effluent tube into beaker containing 10% bleach
- Ensure sufficient Elution Fluid, Ultrafiltration Tips etc are available to complete the run
Safety information
If processing 10 or more samples, the BSC will be crowded. Be careful to ensure the air flow is not blocked and that you remove any unnecessary items.
Bovine Coronavirus Spike-In
- Lyophilized BCoV (10 dose) is reconstituted in 5 ml PBS, 0.01% Tween 20 and stored in aliquots at -80°C
- A working stock at 1:500 dilution in PBS, 0.01% Tween 20 is prepared and stored in 500 μl aliquots at -80°C
- The concentration of the working stock should be in the range of 3000 genome copies per μl
Clarification and Concentration of Wastewater
Sample Pre-Treatment
Addition of BCoV allows virus recovery and inhibition of PCR due to the wastewater matrix to be detected. Tween-20 is added as recommended by Innovaprep. Perform this step on ice.
- Thaw the BCoV aliquot on ice
- Vortex and centrifuge at 16,000g for 5 minutes
- Transfer the supernatant to a fresh tube. This is to ensure the viral particles are evenly distributed.
- Adjust the volume of each wastewater sample to 40 ml
- Add 400 μl of 10% Tween 20
- Add 14.3 μl of BCoV spike (~30-50,000 genome copies)
- Mix well by inverting
- Store remaining BCoV spike on ice until Step 6 is complete
Filtration to remove particulates
Clarification of the sample is necessary to avoid clogging at the ultrafiltration step. This protocol uses filtration but centrifugation can also be employed and is cheaper.
- Pass each sample through a 0.22μM PES filter (diameter >8 cm)
Innovaprep Ultrafiltration
- Insert a fresh ultrafiltration tip into the device
- Insert the tip into the sample
- Start Run (see note for program details)
- If run stops before all liquid is aspirated, Press Return and Start Run again. This is more likely to happen with turbid samples, or if the tip is faulty.
- Remove the empty sample bottle and replace with a 15 ml conical tube
- Press Elute
- Store the eluate on ice until all samples have been concentrated
- Transfer 140 μl of eluate to a fresh tube containing 560 μl of Qiagen AVL buffer and mix well before proceeding to Step 8
- The remaining eluate can be stored at -80°C
Spike Lysis
Spike Lysis
- Add 5μl Quick Extract DNA Extraction Solution to 10μl BCoV Spike from Step 5
- Heat in thermocycler at 65°C for 15 min then 98°C for 2 min, then 4°C forever
- Add 85 μl H2O and store on ice for use in ddPCR
- Can store at -80°C for reference
Note
Altenative Lysis Protocol using GT Molecular Lysis Buffer
Extraction of RNA
QiaAMP Viral RNA Extraction Protocol
- Add 560 μl 100% ethanol
- Vortex for 15 sec
- Apply 630 μl to Epoch column in a 2ml collection tube
- Centrifuge 6,000xg for 1 minute.
- Decant the filtrate
- Load the remaining sample onto the column
- Centrifuge 6,000xg for 1 minute
- Add 500μl of AW1 and spin 1 minute at 6,000xg for 1 minute
- Add 500μl of AW2 and spin at max speed for 3 minutes
- Place the column in a fresh 2mL collection tube and spin again for 1 minute at max speed
- Place the column in a fresh, labeled 1.5ml tube and add 60μl of RNase free water to each column
- Incubate for 1 minute
- Spin at 6,000xg for 3 minutes
- Store samples on ice (if using immediately) or at -80°C for long term storage
Citation
By this stage there is generally very little RNA, so quality control via nanodrop is not informative. If samples are frozen before processing then more RNA is recovered, presumably because freezing lyses cells and microbes and releases nucleic acids.
Preparation for ddPCR
SARS-CoV-2 Positive Control
This reagent is used as a positive control for ddPCR detection of SARS-CoV-2. The variant listed below works well with the primer set described here, other variants may or may not be detected.
- Add 5μl Quick Extract DNA Extraction Solution to 10μl heat-inactivated virus
- Heat in thermocycler at 65°C for 15 min then 98°C for 2 min, then 4°C forever
- Add 85 μl H2O
- Dilute to desired concentration, aliquot and store at -80°C
Citation
The resulting solution will be at around 105 copies per μl. We recommend further dilution to between 10 and 100 copies per μl for use as a positive control in RT-ddPCR.
Primer / Probe Mixes
See Materials for Sequences.
Primer/probe mixes are made as described below, aliquoted, and stored at -20°C in the dark. After thawing on ice, aliquots are stored at 4°C.
100 μM Forward Primer 100 μl
100 μM Reverse Primer 100 μl
100 μM Probe 25 μl
H2O 1275 μl
One-Step Reverse Transcription Droplet Digital PCR
Reaction Set Up
- Thaw all required reagents on ice:
Reverse Transcriptase
300mM DTT
Primer Probe Mixes
BioRad 1-step RT-ddPCR supermix for probes
Wastewater RNA
Positive controls
SARS-CoV-2 /BCoV Reactions
- Create a mastermix sufficient for all reactions adding reagents in the order shown
- Include 1-2 safety reactions to ensure there is enough mastermix i.e. if you have 10 reactions make a mastermix for 11-12 reactions
| A | B |
|---|---|
| Reagent | Volume per reaction (µl) |
| Nuclease free H2O | 0.9 |
| SARS-CoV-2 N1 primer/probe mix | 1.65 |
| BCoV primer/probe mix | 1.65 |
| 1-step supermix | 5.5 |
| 300 mM DTT | 1.1 |
| Reverse Transcriptase | 2.2 |
| Template RNA | 9.0 |
| Final Volume | 22.0 |
Table: Reaction mixture for RT-ddPCR.
- Pipette each template RNA into one well of an 8-tube strip.
- Add 13µl mastermix to each well
- Vortex briefly and spin to the bottom of the tube
F+ / PMMoV Reactions
- Dilute template RNA 1:100 for F+ and PMMoV reactions - add 3 μl RNA to 273 μl H2O
- Create a mastermix sufficient for all reactions adding reagents in the order shown
- Include 1-2 safety reactions to ensure there is enough mastermix i.e. if you have 10 reactions make a mastermix for 11-12 reactions
| A | B |
|---|---|
| Reagent | Volume per reaction (µl) |
| Nuclease free H2O | 0.9 |
| F+ primer/probe mix | 1.65 |
| PMMoV primer/probe mix | 1.65 |
| 1-step supermix | 5.5 |
| 300 mM DTT | 1.1 |
| Reverse Transcriptase | 2.2 |
| Template RNA (1:100 diluted) | 9.0 |
| Final Volume | 22.0 |
Table: Reaction mixture for RT-ddPCR.
- Pipette each diluted template RNA into one well of an 8-tube strip.
- Add 13µl mastermix to each well
- Add ddPCR buffer control for probes to any empty wells in the 8 tube strip
- Mix and spin to the bottom of the tube
It is important to include both positive and negative controls. The negative control should contain H2O instead of RNA template.
Droplet Generation
Place a DG8 cartridge in the holder and click to close
- Using a multi-channel pipette, transfer 20 μl of each reaction to the "Sample" row
- Dispense 70 μl AutoDG* (droplet generator) oil for probes in the "Oil" row
- Seal with a clean gasket, insert into droplet generator and press Start
- Using a p200 gently transfer the droplets to a 96-well plate
- Repeat until all samples are in the 96-well plate
- Turn on the plate sealer.
- Place a foil seal on top of the plate. Seal at 180 °C for 5 sec
- We find that Auto-DG oil reproducibly gives a higher droplet count than Manual-DG oil, even when using a manual droplet generator.
Thermocycling
Using a BioRad C1000 Touch deep-well device:
- 50°C for 1 hr (reverse transcription step)
- 95°C for 10 min
- [94°C for 30 sec; 55°C for 1 min] x 44;
- 98°C for 10 min
- 4°C forever
Droplet Reading
Follow the manufacturer's instructions for using the QX200 reader
Allow 2.5 min per sample plus 10-15 minutes for set up
- Open the reader, insert the plate, lock in the metal grid on top and close the device
- Open the Quantasoft program
- For each well, select ABS (for Absolute Quantification), Ch1 FAM (SARS-CoV-2 or F+), Ch2 HEX (BCoV or PMMoV) and 1-step RT-ddPCR kit for probes.
- Click RUN
Data Analysis
Setting Thresholds in Quantasoft Analysis Pro
Follow the manufacturer's instructions for using the software
-
On the Droplets tab, select all wells. Verify that the droplet count is over 10,000 in all wells
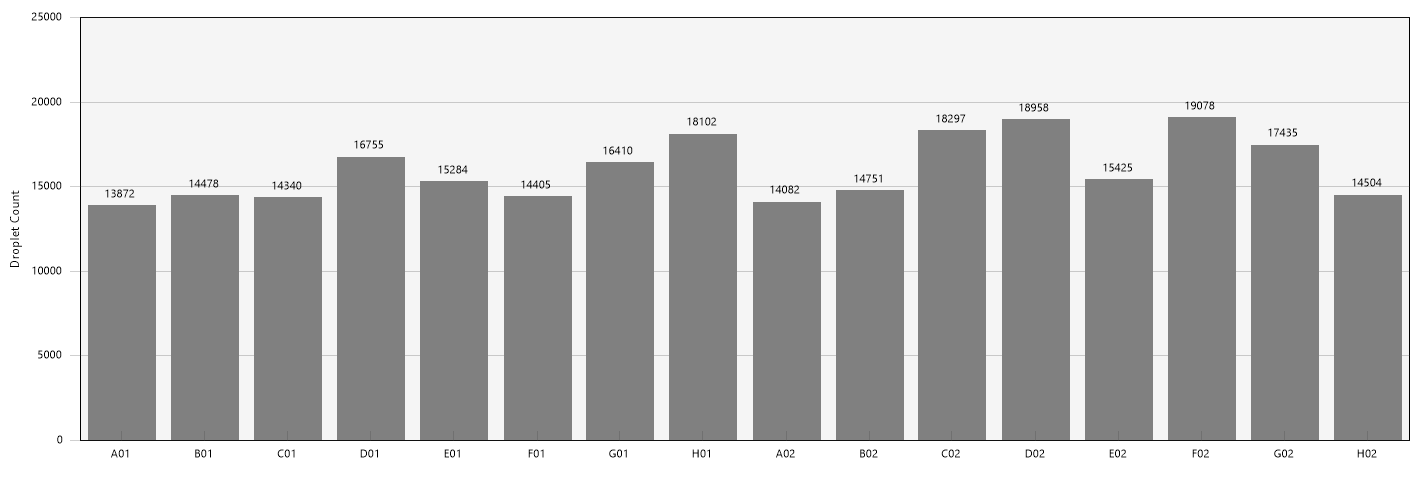
Figure: Droplet counts should be over 10,000. They generally fall within the 12,000-19,000 range. -
In the 2D Amplitude view, select all wells containing the SARS/BCoV probes
-
Position the thresholds (pink +) such that they fall between positive and negative droplets
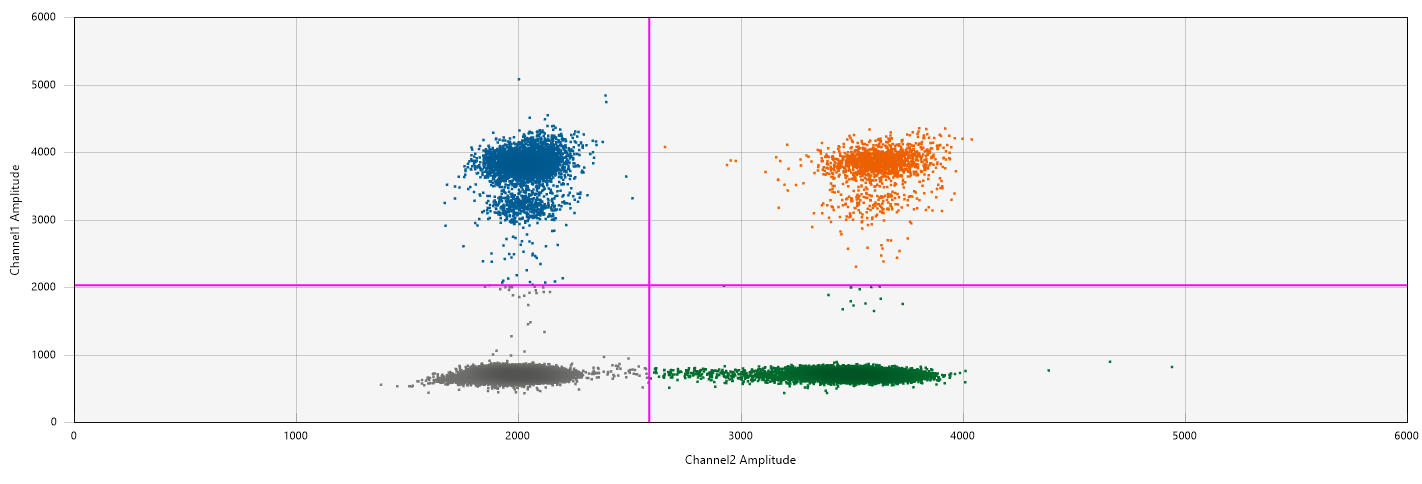
Figure : F+ or SARS-N1 positive droplets are blue, PMMoV or BCoV positive droplets are green, negative droplets are grey. Droplets that are positive for both F+ and PMMoV, or BCoV and SARS-N1 are orange. -
Check on the 1D Amplitude view that the threshold in both channels lies above all the droplets in the no template control.
-
Set the thresholds for the F+/PMMoV wells in the same way (but not at the same time because the correct position will be different).
Exporting Data from Quantasoft Manager Pro to MS Excel
- On 1D or 2D Amplitude view, select all the wells
- On the Well Data panel menu select "Export to Excel"
- While you can select all the data, the values required for downstream calculations or reporting to the NWSS/CDC are:
Well
Sample
Target
Copies/20 ul
Accepted [# droplets]
PoissonConfMax
PoissonConfMin
Calculating SARS-CoV-2 Genome Copies per L
The following values are required to convert the copies per 20µl [ C ] into copies per L:
Volume of wastewater sample [ W =40ml]
Total volume of eluate from innovaprep [ IP P = ranges from 200-900 µl]
Volume of eluate used in RNA extraction [ IE E = 140µl]
Total volume of RNA eluted after extraction [ RT T = 60 µl]
Volume of RNA used in reaction [ RU U = 8.2 µl]
Using the protocol described above:
Calculating Genome Copies per L for Endogenous Controls
The same calculation is used as above, but should be multiplied by 100 to take into account that the RNA template for the PMMoV and F+ reaction is diluted 100 fold.
Using the protocol described above:
Determining BCoV Recovery
Calculate the copies of BCoV that were added at the start of the protocol:
- Use the copies per 20µl in the BCoV positive control reaction to determine how many copies of BCoV were added to each sample at the beginning.
- Then calculate the number of copies of BCoV that would have been recovered if you had processed the entire 40ml of wastewater and express as a percentage of the copies that were added.
Quality Controls and Troubleshooting
One possible use of the controls is for normalization. In our experience with Colorado WWTPs normalization to either exogenous (BCoV) or endogenous controls (PMMoV, F+) did not improve correlations with community infection rate or reduce variability.
The controls can also be used to detect failure of reactions due to inhibitors and to distinguish between inhibition and degradation:
Both the reverse transcription step and the PCR step can be inhibited by compounds that carry through the extraction step (either reagents such as phenol or guanidine, or contaminants such as humic acids). Inhibition of the RT step will generally result in fewer positive droplets (because there will be less cDNA produced) and confounds accurate quantification. Inhibition of the PCR step can result in reduced fluorescence amplitude i.e. poor separation of the positive and negative droplets, because each cycle will produce fewer copies of the cDNA template). This may not affect quantification if the separation remains adequate for threshold setting. Inhibition is not template specific - all targets in a reaction mix will be affected. Low BCoV recovery (<5%) can indicate inhibition of the RT step. Importantly, inhibition can often be abrogated by using less of the RNA containing the inhibitor. Therefore the PMMoV/F+ reaction will generally give good results even when inhibitors are present because it uses a 100 fold diluted RNA template. If BCoV recovery fails QC, it is helpful to rerun the reaction with half or quarter the amount of RNA template (assuming the SARS-N1 levels are high enough to still be detected).
In contrast, degradation of the RNA sample will result in fewer positive droplets for all four targets (i.e. Low BCOV recovery and low PMMoV/F+). This problem can only be remedied by repeating the extraction to make fresh RNA.
RNA quality cannot be easily evaluated prior to the ddPCR assay because even good quality RNA is generally very low abundance (too low for nanodrop), partially degraded and represents a mixture of nucleic acids from a variety of microbes, viruses and human cells (confounds Tapestation/Bioanalyzer). Fluorescent measurements (such as Qubit) may be informative but add time and expense.


