CUTAC for FFPEs
Steven Henikoff
Abstract
For more than a century, Formalin Fixed Paraffin Embedded (FFPE) sample preparation has been the preferred method for long-term preservation of biological material. However, the use of FFPE samples for epigenomic studies has been difficult because of chromatin damage from long exposure to high concentrations of formaldehyde. Previously, we introduced Cleavage Under Targeted Accessible Chromatin (CUTAC), an antibody-targeted chromatin accessibility mapping protocol based on CUT&Tag. Recently, we showed that simple modifications of our single-tube CUTAC protocol are sufficient to produce high-resolution maps of paused RNA Polymerase II (RNAPII) at enhancers and promoters using FFPE samples. We found that transcriptional regulatory element differences produced by FFPE-CUTAC distinguish between tumor specimens and identify regulatory element markers with high confidence and precision. Our simple step-by-step workflow makes possible affordable epigenomic profiling of archived biological samples for biomarker identification, clinical applications and retrospective studies.
Before start
Steps
REAGENT SETUP (for up to 16 samples)
Cross-link reversal buffer Mix 800 µL Tris-HCl pH8.0, 195 µL dH2O and 5 µL Triton-X100.
Binding buffer Mix 200 μl 1M HEPES-KOH pH 7.9*, 100 μl 1M KCl, 10 μl 1M CaCl2 and 10 μl 1M MnCl2, and bring the final volume to 10 mL with dH2O. Store the buffer at 4 °C for up to several months. *HEPES-NaOH pH 7.5 is OK.
Triton-Wash buffer Mix 1 mL 1 M HEPES pH 7.5, 1.5 mL 5 M NaCl, 250 µl Triton-X100 and 12.5 μl 2 M spermidine, bring the final volume to 50 mL with dH2O, and add 1 Roche Complete Protease Inhibitor EDTA-Free tablet. Store the buffer at 4 °C for up to 2 days.
Antibody buffer Mix 5 µl 200X BSA with 1 ml Triton-Wash buffer and chill on ice.
CUTAC-DMF Tagmentation buffer Mix 780 µl dH2O, 200 µl N,N-dimethylformamide, 10 µl 1 M TAPS pH 8.5, 5 µl Triton-X100 and 5 µl 1 M MgCl2 (10 mM TAPS, 5 mM MgCl2, 20% DMF, 0.05% Triton-X100). Store the buffer at 4 °C for up to 1 week.
TAPS wash buffer Mix 1 mL dH2O, 10 µl 1 M TAPS pH 8.5, 0.4 µl 0.5 M EDTA (10 mM TAPS, 0.2 mM EDTA). Store at room temperature.
1% SDS/ProtK Release solution (For 32 samples) Mix 20 µl 10% SDS and 2 µl 1 M TAPS pH 8.5 in 158 µl dH2O. Just before use add 20 µL Thermolabile Proteinase K (NEB cat. no. P8111S).
6% Triton Mix 600 µl 10% Triton-X100 + 400 µl dH2O. Store at room temperature.
Option 1: Deparaffinize FFPE section affixed to slide using xylene (1 hr)
In a fume hood, immerse slide(s) in xylene for 10 min, then transfer to fresh xylene for 5 min.
Transfer slide(s) to a 50:50 mixture of xylene and 100% ethanol for 3 min. This can be reused or discarded in toxic waste container.
Transfer slide(s) to 100% ethanol for 3 min. Repeat once.
Immerse slide(s) 95% ethanol for 3 min.
Immerse slide(s) in 70% ethanol for 3 min.
Immerse slide(s) in 50% ethanol for 3 min.
Rinse slide(s) with tap water or tap-distilled water with change(s).
Option 1: Process deparaffinized FFPE sample for CUT&Tag ( 1.5 hr).
Dice with a razor blade, scrape and pick up the 10 µm section and deposit it into a 1.5 mL tube containing 400 µL Cross-link reversal buffer (800 mM Tris-HCl pH8.0, 0.05% Triton-X100).
Incubate 8-16 hours at 85 °C in a heating block.
Resuspend and withdraw enough of the ConA bead slurry, ensuring that there will be ~5 μl for each final sample. For example, we add 160 µl ConA bead slurry to 1.5 mL of Binding buffer for 32 samples.
Mix by pipetting. Place the tube on a magnet stand to clear (~1 min).
Withdraw the supernatant completely, and remove the tube from the magnet stand. Add 2 mL Binding buffer (for 32 samples) and mix by vortexing.
Add 240 µl to each tube while vortexing, where one section will be used for 4 replicate samples (60 µL per sample). Place on Rotator 10-20 min.
Pass through a 22 gauge 1" needle using a Luer-lock glass syringe 20 times to break up tissue.
Option 2: Deparaffinize FFPE section affixed to slide with mineral oil
Scrape off excess paraffin and continuously scrape all of part of the paraffin-embedded 10-µm section. Using tweezers ( e.g. #3 watchmaker's forceps) carefully lift off the curls and plunge into mineral oil in a 2 ml tube using 100 µl per final sample.
Heat 10 min 80oC to dissolve the curls. Add 1 volume Cross-link reversal buffer (800 mM Tris-HCl pH8.0, 0.05% Triton-X100. Vortex 10 sec, centrifuge briefly, and pass 20 times through a 22 gauge needle on a 1 ml Luer Lock syringe. Centrifuge at 3000xg 1 min. The paraffin will partially solidify in the upper layer while the tissue partitions to the lower (aqueous) layer.
Heat briefly to melt the upper layer and remove without disturbing the lower layer using a wide-bore or cut-off 200 µl low-bind pipette tip. Add 1 volume mineral oil, vortex, centrifuge, heat and decant the upper layer.
Repeat Step 18 until the interface is clear or nearly so. Using a wide-bore 200 µl pipette tip transfer 100 µl @ to PCR tubes.
Place tubes in a thermocycler and incubate at 85 °C for at least 2 hours. We have found that mappability improves with incubations at 80-90 °C overnight (8-16 hr).
Resuspend and withdraw enough of the ConA bead slurry, ensuring that there will be ~5 μl for each final sample. For example, we add 160 µl ConA bead slurry to 1.5 mL of Binding buffer for 32 samples. Place the pipette tip below the meniscus to avoid coating the beads with oil and discharge the beads while mixing by pipetting.
Mix by pipetting. Place the tube on a magnet stand to clear (~1 min).
Withdraw the supernatant completely, and remove the tube from the magnet stand. Add 2 mL Binding buffer (for 32 samples) and mix by vortexing.
Resuspend in 160 µL Binding buffer. Add 5 µl to each sample while vortexing. Place on Rotator 10-20 min.
Bind primary antibody (2 hr)
After a quick spin, place the tubes on the magnet stand to clear and withdraw the liquid.
For each CUT&Tag and CUTAC sample, mix the primary antibody 1:25 with Antibody buffer. Resuspend beads in 25 µl per sample followed by vortexing.
Place on a rotator at room temperature and incubate at least 1 hr on Rotator at room temperature.
1h 0m 0s
Bind secondary antibody (1.5 hr)
After a quick spin, place the tubes on the magnet stand to clear and withdraw the liquid..
Mix the secondary antibody 1:100 in Wash buffer and squirt in 25 µl per sample followed by vortexing.
Place the tubes on a rotator or nutator and rotate or nutate at room temperature for 1 hr.1h 0m 0s
After a quick spin (<500 x g or just enough to remove the liquid from the sides of the tube), place the tubes on the magnet stand to clear and remove and discard the supernatant with two successive draws, using a 20 µl tip with the pipettor set for maximum volume.
With the tubes still on the magnet stand, carefully add 500 µl of Wash buffer. The surface tension will cause the beads to slide up along the side of the tube closest to the magnet.
Slowly withdraw 460 µl of supernatant with a 1 mL pipette tip without disturbing the beads.
After a quick spin (< 500 x g or just enough to remove the liquid from the sides of the tube), place the tubes back into the magnet stand and remove the remaining supernatant with a 20 µl pipettor multiple times if necessary, to remove the entire supernatant without disturbing the beads. Proceed immediately to the next step.
Bind pA-Tn5 adapter complex (1.5 hr)
Mix pAG-Tn5 pre-loaded adapter complex in Triton-Wash buffer following the manufacturer's instructions ( e.g. 1:20 for EpiCypher pAG-Tn5).
Pipette in 25 µl per sample of the pA-Tn5 mix followed by vortexing.
After a quick spin, place the tubes on a rotator at room temperature for 1 hr or 4 °C overnight.1h 0m 0s
After incubating in the rotator, perform a quick spin and place the tubes in the magnet stand.
Carefully remove the supernatant using a 20 µl pipettor as in Step 31.
With the tubes still on the magnet stand, add 500 µl of the Triton-Wash buffer.
Slowly withdraw 460 µl with a 1 ml pipette tip without disturbing the beads as in Step 33.
After a quick spin, place the tubes back on the magnet stand and remove and discard the supernatant with a 20 µL pipettor using multiple draws. Proceed immediately to Step 43.
Tagment (1.5 hr, performed in parallel with standard CUT&Tag and CUTAC samples)
Resuspend the bead/FFPE pellet in 50 µl CUTAC-DMF tagmentation solution (5 mM MgCl2, 10 mM TAPS, 20% DMF, 0.05% Triton-X) while vortexing. Incubate 1 hr 55 °C in thermocycler.
Place tubes on a magnet stand and remove and discard the supernatant with a 20 µL pipettor using multiple draws then resuspend the beads in 50 µL TAPS wash and mix by vortexing.
Fragment Release (2.5 hr)
After a quick spin, place tubes on the magnet stand, and withdraw the liquid with a 20 µL pipettor using multiple draws.
Resuspend the beads by squirting in 5 µL 1% SDS/ProtK Release solution followed by vortexing.
After a quick spin, incubate at 37 ºC for 1 hr and 58 ºC for 1 hr (programmed in succession in a PCR cycler with a heated lid) to release pA-Tn5 from the tagmented DNA. 2h 0m 0s
PCR (1 hr)
To the PCR tube containing the bead slurry add 15 µl of Triton neutralization solution + 2 µl of 10 µM Universal or barcoded i5 primer + 2 µl of 10 µM uniquely barcoded i7 primers, using a different barcode for each sample. Vortex on full speed and place tubes in the metal tube holder on ice.
Add 25 µl NEBnext (non-hot-start), vortex to mix, and perform a quick spin. Place the tubes immediately in the thermocycler and proceed immediately with the PCR.
Begin the cycling program with a heated lid on the thermocycler:
Cycle 1: 58 °C for 5 min (gap filling)
Cycle 2: 72 °C for 5 min (gap filling)
Cycle 3: 98 °C for 5 min
Cycle 4: 98 °C for 10 sec
Cycle 5: 63 °C for 30 sec
Cycle 6: 72 °C for 1 min
Repeat Cycles 4-6 11 times
Hold at 8 °C
Post-PCR Clean-up (30 min)
After the PCR program ends, remove tubes from the thermocycler and add 65 µL of SPRI beads (ratio of 1.3 µL of SPRI beads to 1 µL of PCR product). Mix by pipetting up and down.
Let sit at room temperature 5-10 min.0h 5m 0s
Place on the magnet stand for a few minutes to allow the solution to clear.
Remove and discard the supernatant.
Keeping the tubes in the magnet stand, add 200 µL of 80% ethanol.
Completely remove and discard the supernatant.
Repeat Steps 55 and 56.
Perform a quick spin and remove the remaining supernatant with a 20 µl pipette, avoiding air drying the beads by proceeding immediately to the next step.
Remove from the magnet stand, add 22 µl 10 mM Tris-HCl pH 8 and vortex at full speed. Let sit for 5 min to 1 hr. 0h 5m 0s
Place on the magnet stand and allow to clear.
Remove the liquid to a fresh 1.5 mL tube with a pipette, avoiding transfer of beads.
Tapestation analysis and DNA sequencing
Determine the size distribution and concentration of libraries by capillary electrophoresis using an Agilent 4200 TapeStation with D1000 reagents or equivalent.
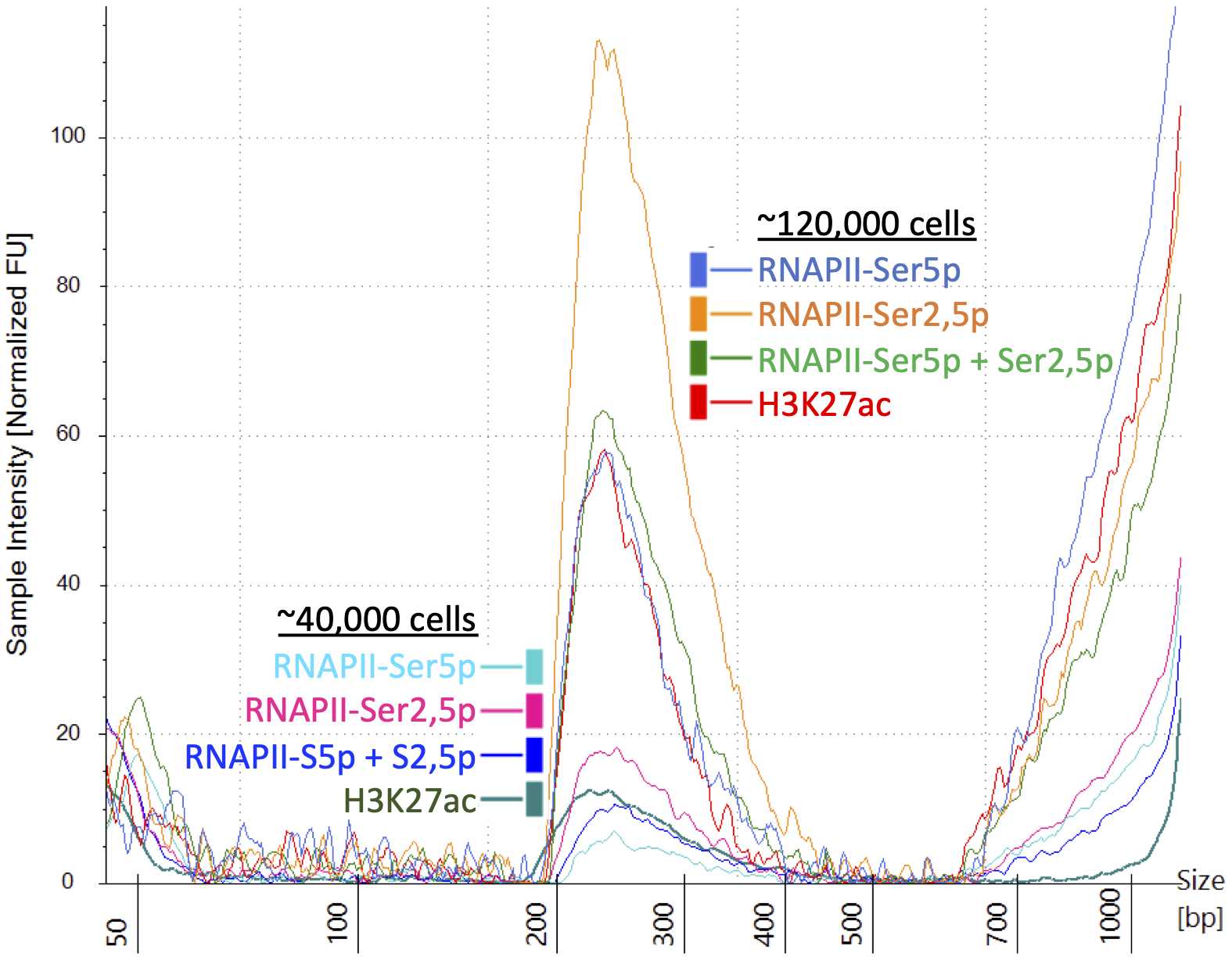
Mix barcoded libraries to achieve equal representation as desired aiming for a final concentration as recommended by the manufacturer. After mixing, perform an SPRI bead cleanup if needed to remove any residual PCR primers.
Perform paired-end Illumina sequencing on the barcoded libraries following the manufacturer’s instructions.
Data processing and analysis
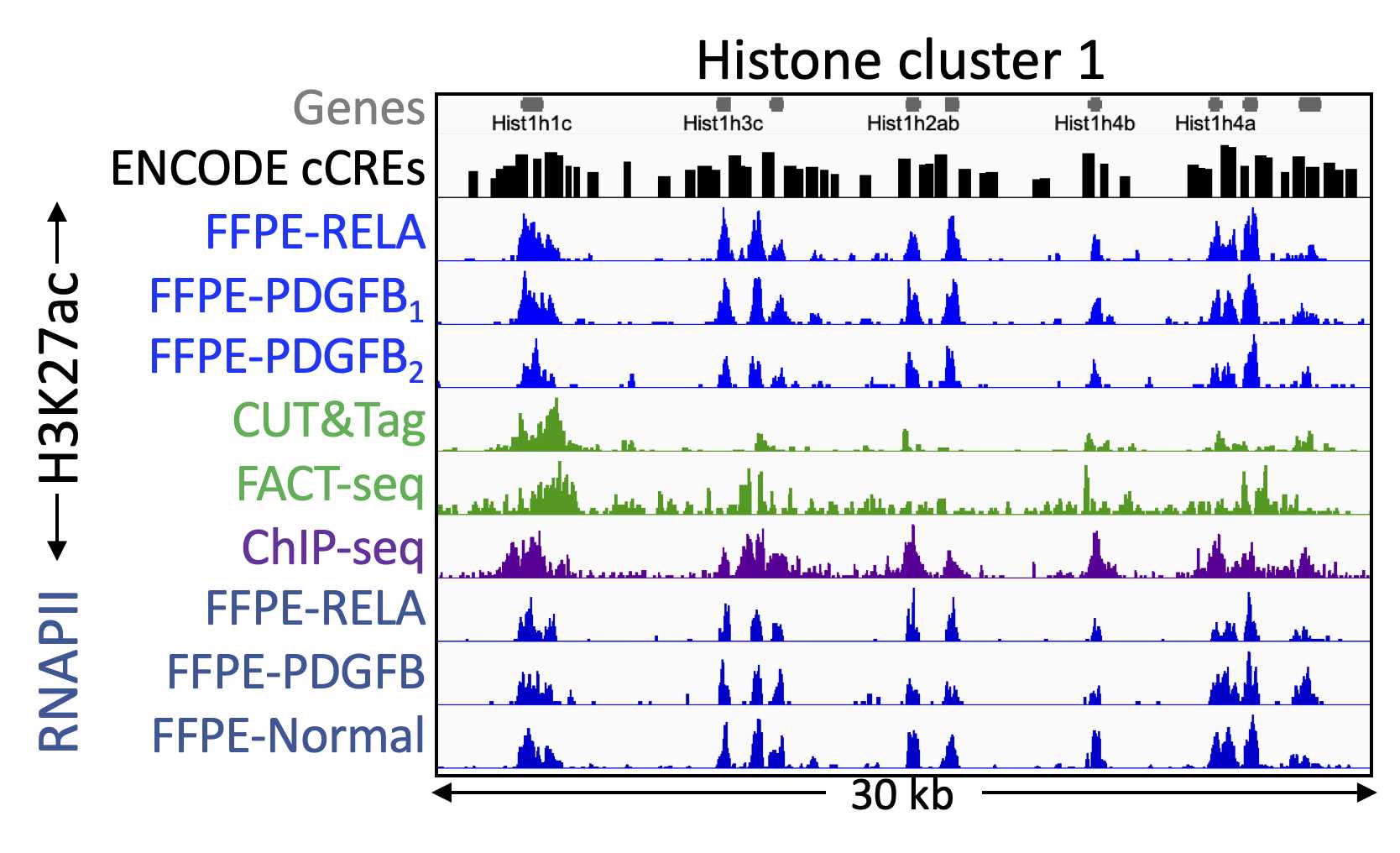
Align paired-end reads to hg19 using Bowtie2 version 2.3.4.3 with options: --end-to-end --very-sensitive --no-unal --no-mixed --no-discordant --phred33 -I 10 -X 700. For mapping E. coli carry-over fragments, we also use the --no-overlap --no-dovetail options to avoid possible cross-mapping of the experimental genome to that of the carry-over E. coli DNA that is used for calibration. Tracks are made as bedgraph files of normalized counts, which are the fraction of total counts at each basepair scaled by the size of the hg19 genome.
Our CUT&Tag Data Processing and Analysis Tutorial on Protocols.io provides step-by-step guidance for mapping and analysis of CUT&Tag sequencing data. Most data analysis tools used for ChIP-seq data, such as bedtools, Picard and deepTools, can be used on CUT&Tag data. Analysis tools designed specifically for CUT&RUN/Tag data include the SEACR peak caller also available as a public web server and CUT&RUNTools.