CRISPR Genome Editing Made Easy Through the CHOPCHOP Website
Kornel Labun, Kornel Labun, Maximilian Krause, Maximilian Krause, Yamila Torres Cleuren, Yamila Torres Cleuren, Eivind Valen, Eivind Valen
Cas9
CRISPR
CRISPRi/a
gene knock-out
genome editing
guide design
knock-down
knock-in
nickase
RNA targeting
Abstract
The design of optimal guide RNA (gRNA) sequences for CRISPR systems is challenged by the need to achieve highly efficient editing at the desired location (on-target editing) with minimal editing at unintended locations (off-target editing). Although laboratory validation should ideally be used to detect off-target activity, computational predictions are almost always preferred in practice due to their speed and low cost. Several studies have therefore explored gRNA-DNA interactions in order to understand how CRISPR complexes select their genomic targets. CHOPCHOP (https://chopchop.cbu.uib.no/) leverages these developments to build a user-friendly web interface that helps users design optimal gRNAs. CHOPCHOP supports a wide range of CRISPR applications, including gene knock-out, sequence knock-in, and RNA knock-down. Furthermore, CHOPCHOP offers visualization that enables an informed choice of gRNAs and supports experimental validation. In these protocols, we describe the best practices for gRNA design using CHOPCHOP. © 2021 The Authors. Current Protocols published by Wiley Periodicals LLC.
Basic Protocol 1 : Design of gRNAs for gene knock-out
Alternate Protocol 1 : Design of gRNAs for dCas9 fusion/effector targeting
Support Protocol : Design of gRNAs for targeting transgenic or plasmid sequences
Basic Protocol 2 : Design of gRNAs for RNA targeting
Basic Protocol 3 : Design of gRNAs for sequence knock-in
Alternate Protocol 2 : Design of gRNAs for knock-in using non-homologous end joining
Basic Protocol 4 : Design of gRNAs for knock-in using Cas9 nickases
INTRODUCTION
The CRISPR system allows for programmable targeting of genomes and transcriptomes, and has thus been repurposed for a range of applications in biological systems. At its core, CRISPR is a modular two-part system requiring a guide RNA (gRNA) and a Cas protein. The gRNA is a sequence of ∼100-nt that consists of two parts: one that interfaces with the Cas protein and one that guides the complex to its target. The latter is a recognition site of ∼20 bp that contains the complementary sequence of its genomic or transcriptomic target. In addition, most Cas proteins require a specific sequence adjacent to the target. This is referred to as a protospacer adjacent motif (PAM) for DNA or a protospacer flanking sequence (PFS) for RNA. It is typically specific for each Cas protein. For example, the CRISPR system harvested from Streptococcus pyogenes uses a Cas9 protein requiring an NGG PAM motif. When the Cas protein recognizes its preferred PAM in the genome, the adjacent sequence can base-pair with the 20-bp recognition site of the gRNA. If base-pairing succeeds, the Cas protein will create a double-strand break (DSB) in the genomic DNA. Even though the complementary target of the gRNA's recognition site may be unique in the genome, base-pairing and PAM recognition are not perfect. Thus, there is a possibility that a gRNA will guide the Cas protein to off-target locations, and that DSBs will occur even when bulges and mismatches are present in these sites. Achieving efficient target recruitment and DSB generation at the intended locus is very important, but higher on-target efficiency often results in higher off-target frequency. Thus, computational tools are needed that can assess and balance high on-target efficiency and low off-target frequency.
CHOPCHOP (https://chopchop.cbu.uib.no/) was one of the first tools created to design and visualize gRNAs (Montague, Cruz, Gagnon, Church, & Valen, 2014). It was designed to be simple to operate without the need for expert knowledge but to be highly customizable for expert users. To this end, it provides preset options optimized for specific CRISPR applications and updates them regularly to include the most recent scientific advances. For expert users, it also provides a long and growing list of options for customizing how gRNAs are selected and ranked. Here, we explain the basic use of CHOPCHOP step by step, focusing on the most common pitfalls and errors users may experience. All protocols will result in a ranked list of gRNAs for a target of interest, visualized in a user-friendly interface. The results can be downloaded in multiple formats.
Strategic Planning explains how to make the best use of the CHOPCHOP output. It outlines general considerations for selecting an experimental mode that tailors CHOPCHOP towards your needs. Specifically, we highlight the need for your own evaluation of CHOPCHOP ranking and the need for validation. Basic Protocol 1 introduces the most basic use of CHOPCHOP to design a gRNA to create a DSB at a specific genomic location for a knock-out. Alternate Protocol 1 applies the same targeting principles for recruiting Cas9 effectors. A Support Protocol explains how to target transgenic or plasmid sequences. Basic Protocol 2 describes the use of the relatively recently discovered Cas13 to target RNA for knock-down. Basic Protocol 3 outlines a protocol for knock-in of specific sequences with homology or microhomology arms using homology-directed repair (HDR). Alternative Protocol 2 shows how to perform knock-in using non-homologous end joining (NHEJ) when this is preferred over HDR. Finally, Basic Protocol 4 describes knock-in using the nickase system, where two Cas proteins are recruited to the target site, thus minimizing off-target events at the expense of efficiency.
STRATEGIC PLANNING
CHOPCHOP aims to simplify gRNA design for CRISPR. It runs on a web browser, requires very few input parameters, provides genomes and annotations for hundreds of organisms, and provides an output with visualization of PCR primers and restriction enzymes sites. To use CHOPCHOP efficiently and correctly, some strategic planning of the experiment and expected outcomes is necessary. Here, we highlight the most important considerations for basic gRNA design and give a brief overview of advanced options.
Before using CHOPCHOP, it is important to define your aim (e.g., knock-out of a specific gene) and the required experimental procedure. Depending on these choices, you will select the correct Cas protein and the application. These selections will load default presets that tailor the gRNA design and results output towards your specific needs. Experienced users can adjust these presets under “Options” (see Critical Parameters and the various protocols for specific examples).
It is important to familiarize yourself with your target of interest. Although CHOPCHOP provides an output that takes gene structure into account, prior knowledge ensures that you make the best use of the presented results. Before using CHOPCHOP, it is advised to study the gene for features important for the experimental outcome. Some considerations include:
What is more important in my experiment: achieving the best on-target efficiency or minimizing off-targets? CHOPCHOP prioritizes off-targets first and then on-target efficiency. Is it important to target one specific genetic locus or can a wider region be targeted?
Can multiple gRNAs be used in parallel? One must remember that multiple gRNAs can cause very large deletions and chromosomal aberrations. Is it likely that SNPs are present in the targeted region? SNP databases can be checked if this is of concern. Do I want to target one isoform of a gene or all possible isoforms?
For a knock-out, where are the functionally relevant domains of the protein?
For effector targeting, which regions would result in the largest effect (e.g., on expression)?
Could certain features of the target reduce the chances of a positive outcome? For example, CpG islands can reduce on-target efficiency. It is possible to import results from CHOPCHOP to the UCSC Genome Browser to review such features. For a knock-in, which repair strategy should be used (HDR vs. NHEJ)? HDR is precise and efficient in cell cultures, but very limited in vivo and in non-reproducing cells. NHEJ can be more error-prone, but can be used in vivo and in difficult cell types such as neurons. Would a nickase approach be a valuable alternative to a single-gRNA knock-in? Nickases are thought to produce less off-target mutations, as single-strand breaks (SSBs) are efficiently repaired and the chance of having two SSBs through the two chosen gRNAs in close proximity is rather low. For more information, see Basic Protocol 4.
CHOPCHOP is designed to make CRISPR editing easy, but cannot replace proper experimental design or ensure that gRNAs are perfectly matched to the needs of the specific experiment. It is a good idea to familiarize yourself with your target and any potential overlapping genomic features. CHOPCHOP enables off-target searching, but does not scan for features that could affect neighboring genes or elements (e.g., overlapping genes, insulator elements, or enhancers). It is in the best interest of the experimenter to choose from the ranked results to find one that best fits the needs of the experiments.
Considerations for Gene Knock-outs
Important questions that should be considered are:
When targeting a region close to the start codon, are there in-frame start codons downstream (highlighted in green in CHOPCHOP) that could initiate and translate a truncated protein? Are the most important functional domains downstream of the gRNA target site? Could alternative splicing compensate for an introduced mutation? Could the target site disrupt splicing? Are any SNPs present in or close to the edit site?
Considerations for dCas9 Fusion/Effector Targeting
Targeting of Cas9 effectors follows the same principles as Cas9 targeting for genome editing, but the choice of target is typically different. In these experiments, it is often necessary to target a very specific region (e.g., a promoter, enhancer, or splice site), and screening with restriction enzymes and primers is not applicable. Instead, the location and efficiency of gRNA binding are the only considerations of importance. It is therefore advised to define the location and the acceptable window of recruitment around the desired locus before starting. To ensure efficient recruitment to the desired locus, one can consider using multiple gRNAs spaced a minimum a of ∼50 bp apart (Ran et al., 2013).
Considerations for Knock-ins
Knock-in strategies are summarized in Figure 1 and can be achieved by (1) using Cas9 to create DSBs (Fig. 1A) that are repaired by HDR or NHEJ, or (2) using Cas9 nickase to create pairs of SSBs (nicks; Fig. 1B), thereby increasing chances for HDR. Both options are presented in the protocols below.

Knock-ins using HDR
Knock-in with Cas9 and gRNAs follows the same principles as Cas9 targeting for genome editing, but additional parameters need to be used. One key aspect to consider when deciding which repair technique to use (HDR or NHEJ) is whether the modification will need to take place in dividing or nondividing cells. HDR is restricted to the S and G2 phases of the cell cycle, and thus unsuitable for knock-ins in nondividing cells (Nami et al., 2018). We describe how to design a knock-in on CHOPCHOP using either HDR (see Basic Protocol 3) or NHEJ (see Alternate Protocol 2). For HDR, the donor sequence has long homology arms (Fig. 1C,D) that allow for more accurate DSB repair.
An important point to note is that CRISPR enzymes will continue to cleave DNA if the gRNA target site/PAM site remains intact. This could be problematic when a specific mutation or sequence is introduced. We recommend that the template be designed to block further Cas9 targeting after the initial DSB is repaired. This is commonly done by mutating either the PAM sequence or the gRNA seed sequence.
Knock-ins using Cas9 nickases
The three most important parameters for this experiment are the choice of homology arms, the choice of nickase (D10A or N863A; Mali et al., 2013), and the optimal spacing of gRNAs (Ran et al., 2013). D10A Cas9 mutants are considered to perform slightly better in generating knock-ins compared to N863A mutants. In either case, the gRNAs should target both DNA strands and should be designed in “PAM-out” configuration, facing away from each other. These parameters are set by default for the CHOPCHOP nickase design and substantially restrict the choice of possible gRNA pairs. The distance between the two gRNAs is optimal between 25 and 100 bp, but can be adjusted manually (Mali et al., 2013; Ran et al., 2013). For good overviews about these experimental considerations, see Internet Resources.
Validation
It is prudent to consider how you will validate your experiment. Primers flanking the targeted genomic region are useful for several validation approaches, and by default CHOPCHOP reports a set of primer pairs for each gRNA using Primer3 (Untergasser et al., 2012). Depending on your preferences, however, you might want to optimize the primer design parameters before searching for a gRNA. This can easily be accomplished by navigating to “Options” → “Primers”. For knock-out and other genetic ablation strategies, a common approach for evaluating experiment efficiency uses restriction enzymes to target the sequence around the expected mutation. To aid with this, CHOPCHOP highlights restriction enzyme sites around the gRNAs. The user can adjust the settings for these under “Options” → “General” → “Restriction Enzymes”. Together, these options empower the user to select gRNAs that are both efficient and amenable to validation.
Basic Protocol 1: DESIGN OF gRNAs FOR GENE KNOCK-OUT
Due to its versatility, efficiency, and precision, the CRISPR system has superseded previous tools to modify genomic information (Gaj, Sirk, Shui, & Liu, 2016). The most common application is creating gene knock-outs through the introduction of double-strand breaks followed by repair through the NHEJ pathway. This protocol provides a basic outline of the considerations and steps necessary for the use of CHOPCHOP when targeting Cas9 to genes. For additional considerations, see Strategic Planning.
Materials
- Computer, tablet, or phone
- Modern browser with javascript enabled (e.g., Chrome, Firefox, Safari)
Perform search
1.Go to https://chopchop.cbu.uib.no.
2.Enter the gene of interest in the “Target” field.
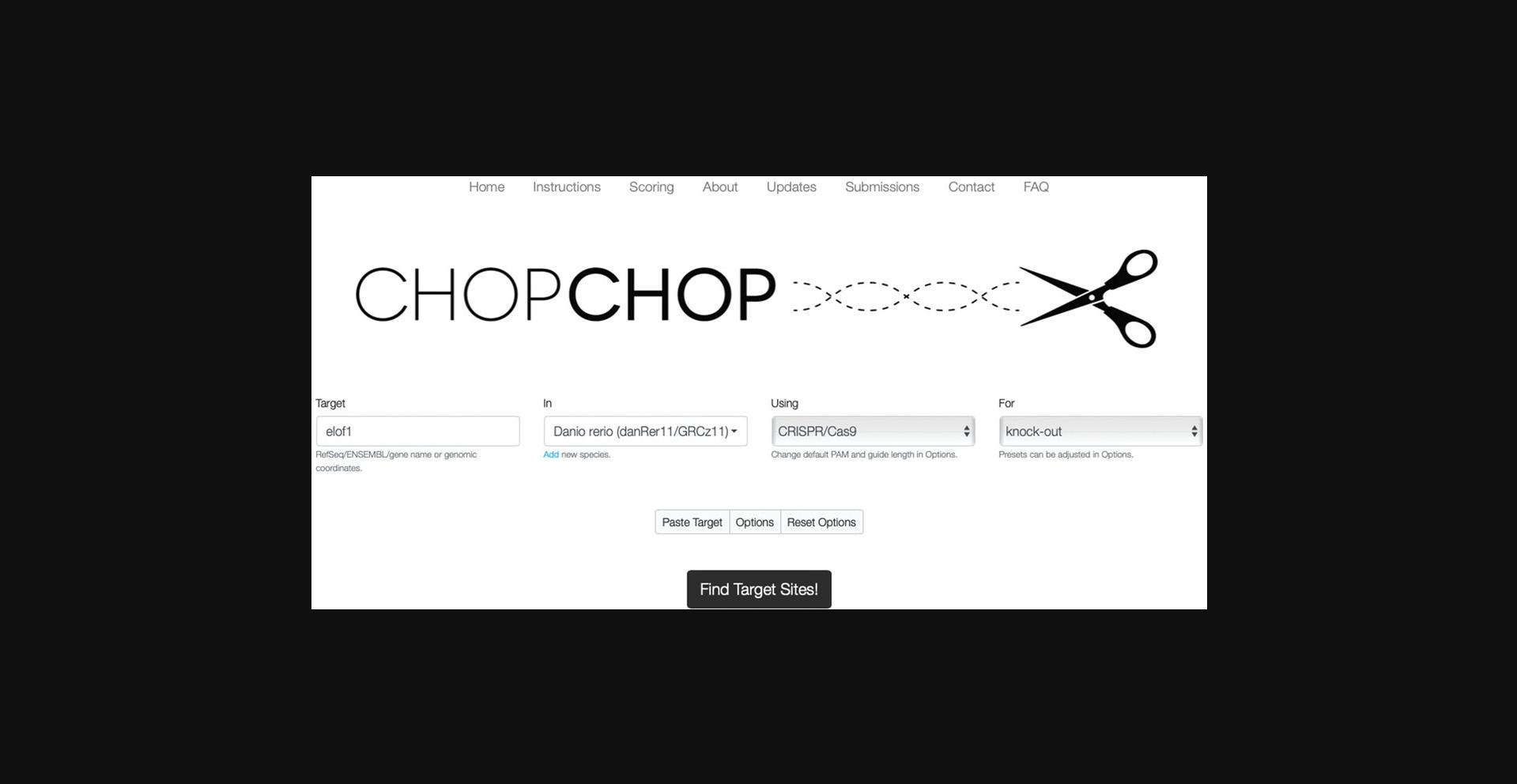
3.Choose the genome of interest in the “In” field.
4.Choose “CRISPR/Cas9” in the “Using” field.
5.Choose “knock-out” in the “For” field.
6.Optional : Modify the default search based on needs using the “Options” button. For example:
- Select a specific gene feature to target (e.g., 5′-UTR or coding region) under “Options” → “General” → “Target specific region of gene”.
- Specify a preferred set of restriction enzymes to validate mutations under “Options” → “General” → “Restriction enzymes”.
- Specify preferences for primer design under “Options” → “Primers”.
For information about other important options, see Critical Parameters.
Targeting effector proteins for chosen genetic loci follows the same ranking logic, but different “Options” for target locus definition (see Alternate Protocol 1).
7.Press “Find Target Sites!”.
Interpret results
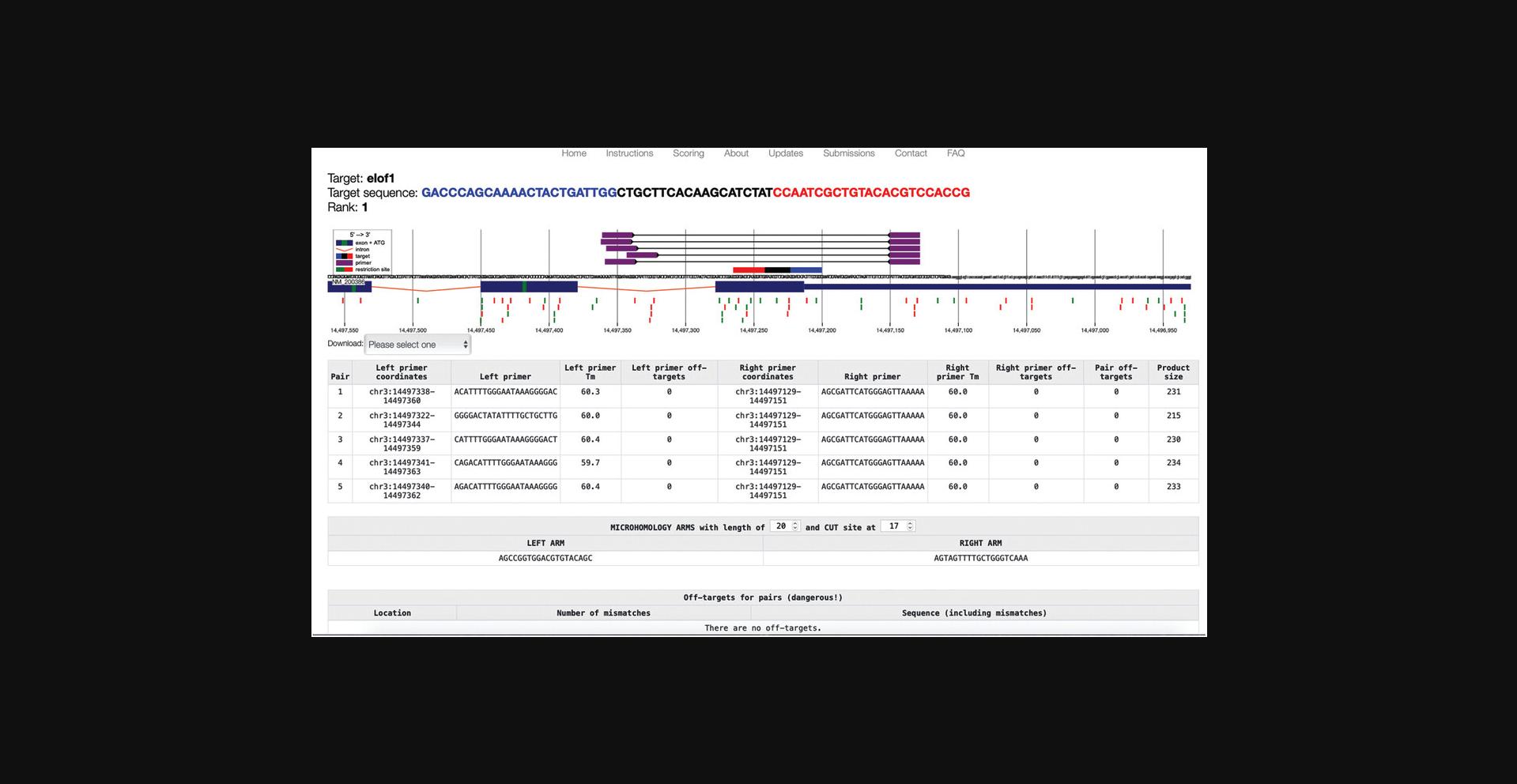
8.In the genome locus view of the results (Fig. 3, top), identify a gRNA that fits your purpose.
-
Consider your experimental needs. For example, choose a gRNA that targets splice sites, regions downstream of in-frame ATGs, or exons of important domains.
-
Consider the number of off-targets based on color coding (green indicates almost no off-targets, orange indicates some off-targets, and red indicates many off-targets).
This may change with future updates, but is specifically described under the “Scoring” link.
-
Consider the directionality of binding (indicated by black arrowheads).
-
Consider placement of in-frame ATG start codons (green lines) to avoid re-initiation.
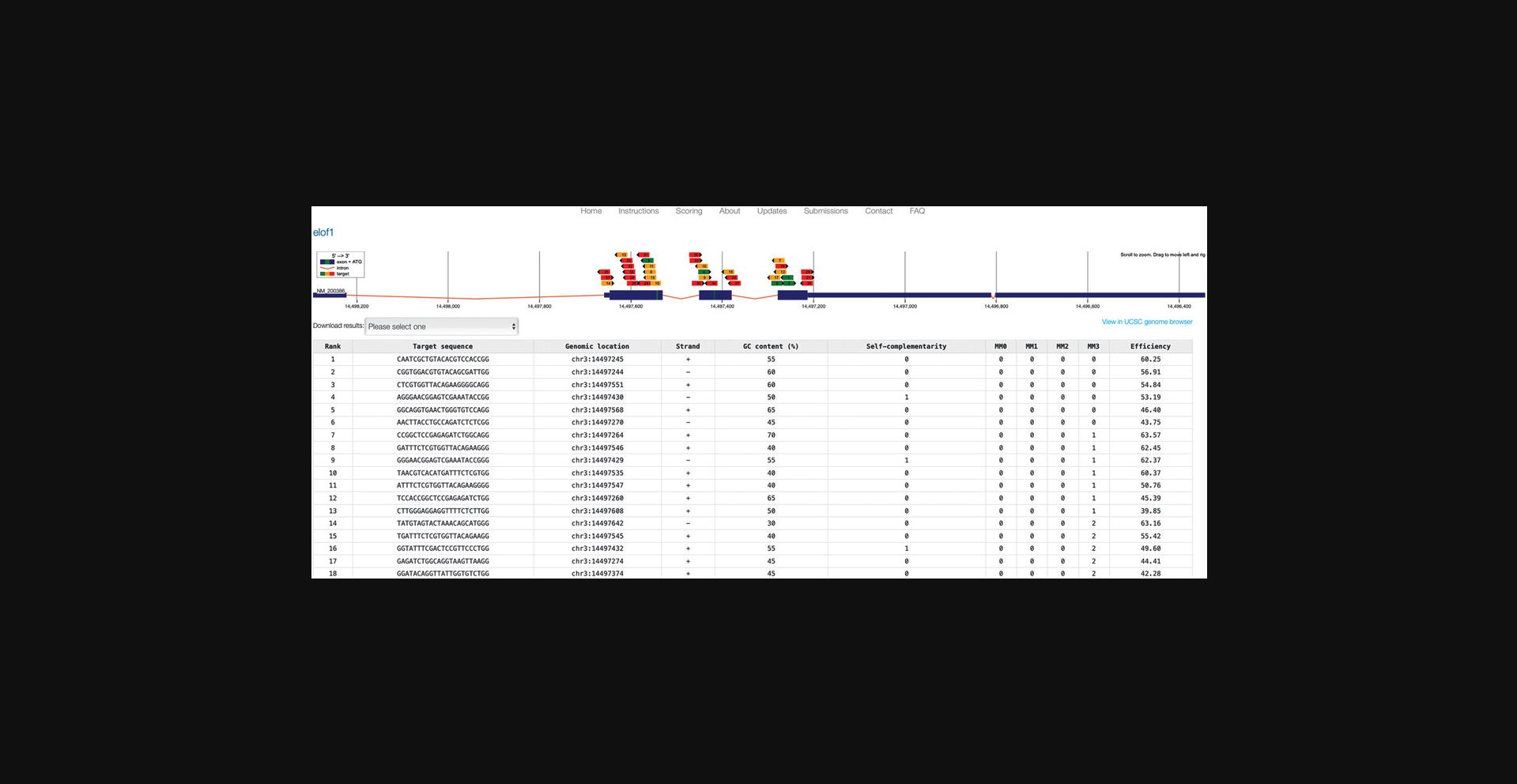
9.In the table view (Fig. 3, bottom), sort by clicking the header of any column.
10.Review all listed features of your preferred gRNAs. Make sure that the GC content fits your needs and that self-complementarity is not too high.
11.Optional : Download the complete list of proposed targets by choosing a preferred format in the “Download results” box (Fig. 3, top-left). Make sure your browser allows pop-up windows to open.
12.Optional : Transfer results to the UCSC Genome Browser by clicking on the link (Fig. 3, top-right).
13.When a suitable candidate is found, click on the graphical representation or the table entry to open a detailed view.
14.Inspect the detailed view (Fig. 4) for important considerations.
- Consider primer pairs that can be used for validation experiments (shown in violet).
In the table view, each primer pair is listed with coordinates, sequences, melting temperatures, and product size. This information can be downloaded in Genbank or .tsv format.
- Consider restriction sites (green and red) boxes.
Moving the cursor over a restriction site will display the name of the enzyme. Before choosing a gRNA that modifies a restriction site, verify that the enzyme can be retrieved from your local source, and that it does not cut too often within the PCR product, making analysis impossible. To be used efficiently for experiment validation, the restriction site should be as close as possible to the expected mutation site.
- Consider off-targets.
Any possible off-targets are listed at the bottom of the table view, with genomic location and possible off-target mismatches highlighted in red (there are no off-targets in this example).
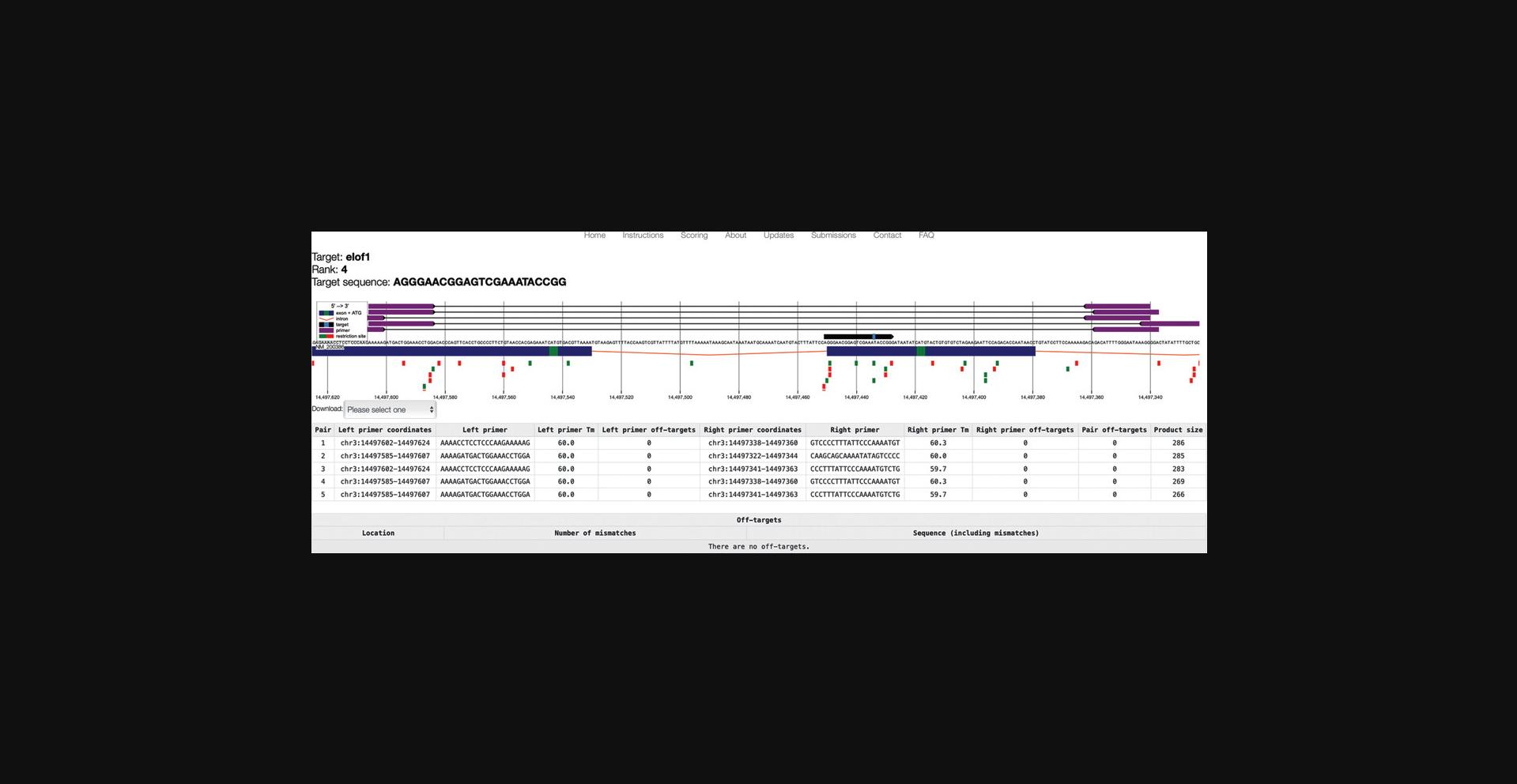
15.Once a preferred target has been found, copy the target sequence from the top left corner of the results page.
Alternate Protocol 1: DESIGN OF gRNAs FOR dCas9 FUSION/EFFECTOR TARGETING
The CRISPR/Cas9 system can be used not only for creating breaks in the genome, but also to recruit effectors to specific genomic locations by fusing effector domains to Cas9 (Adli, 2018). The superior target specificity and the modular setup of the CRISPR system make it an ideal tool for fusion protein targeting. In these approaches, the Cas9 protein is mutated to be enzymatically inactive and not produce DNA breaks. This so-called dead Cas9 (dCas9) is fused with an effector domain targeted to a specific genomic location. These effectors can be transcription activators (Konermann et al., 2015), repressors (Yeo et al., 2018), fluorescent tags (Ye, Rong, & Lin, 2017), or other proteins (Berckman & Chen, 2019; Lei et al., 2017; Xu et al., 2016; Ribeiro, Ribeiro, Barreto, & Ward, 2018; Tsui et al., 2018; Vora, Tuttle, Cheng, & Church, 2016). Combining these Cas9 effectors with specific gRNAs will elicit the desired effect at the targeted genomic locus and not elsewhere.
1.Follow steps 1-4 as in Basic Protocol 1.
2.Select “activation” or “repression” in the “For” field.
3.Define a specific genomic range for recruiting the effector under “Options” → “General”.
4.Optional : Modify other advanced options under “Options” (see Strategic Planning).
5.Press “Find Target Sites!” and proceed with interpretation as in Basic Protocol 1.
Support Protocol: DESIGN OF gRNAs FOR TARGETING TRANSGENIC OR PLASMID SEQUENCES
In the specific cases of targeting transgenic sequences in living organisms or designing gRNAs against plasmids, the user provides the sequence to search for targets and the choice of genome only applies to the off-target search.
1.Prepare the target sequence (plasmid or transgenic sequence) as a single FASTA file.
2.Go to https://chopchop.cbu.uib.no.
3.Press the “Paste Target” button (Fig. 2).
4.Copy the FASTA entry (including header) into the “Target” field.
5.Choose a genome for off-target search in the “In” field.
6.Proceed as in Basic Protocol 1 beginning with step 4.
Basic Protocol 2: DESIGN OF gRNAs FOR RNA TARGETING
Design for RNA targeting with Cas13 (formerly C2C2) has proven useful for applications such as viral detection (Freije et al., 2019; Gootenberg et al., 2017) and temporary alteration of pathogenic mutations (Cox et al., 2017). CHOPCHOP supports identification of optimal gRNAs for targeting RNA through its RNA knock-down mode. In this mode, off-targets are identified not on the genome, but rather the transcriptome. CHOPCHOP still optimizes for GC content, self-complementarity, and off-targets, but also optimizes for local RNA structure. In these cases, the PAM sequence is replaced by its RNA equivalent, the protospacer flanking sequence (PFS).
Perform search
1.Visit the website https://chopchop.cbu.uib.no.
2.Enter the gene or transcript name or ID in the “Target” field.
3.Choose the genome of interest in the “In” field.
4.Choose “CRISPR/Cas13” in the “Using” field.
5.Optional : Modify Cas effector settings under “Options” → “Cas13”.
6.Optional : Modify general options under “Options” → “General” (see Critical Parameters).
7.Press the “Find Target Sites!” button.
Interpret results
8.In the genome locus view of the results (Fig. 3, as in Basic Protocol 1), identify a gRNA that fits your purpose.
- Consider the number of off-targets based on color coding (green indicates almost no off-targets, orange indicates some off-targets, and red indicates many off-targets).
This may change with future updates, but is specifically described under the “Scoring” link.
- Consider the “Constitutive” column of the results table to determine whether a guide targets all isoforms of the gene.
The constitutive column can have value 0 or 1, where 1 means the guide is targeting all isoforms of the gene. The columns IsoformsMM0 to IsoformsMM3 show which isoforms of the gene are being targeted by gRNA, with increasing number of mismatches. In our example, all three isoforms are targeted by the gRNA with 0 mismatch, indicated also by constitutive value 1.
- Consider the MM0, MM1, MM2, and MM3 columns.
These count how many isoforms of the gene are targeted by a gRNA with an increasing number of mismatches (from 0 to 3). These columns do not count off-targets that belong to the family of isoforms of the targeted gene; these can be found in the IsoformMM0-MM3 columns.
- Consider the local structure score.
Local structure influences on-target efficiency, as Cas13 binds preferentially to unstructured regions. The CHOPCHOP local structure score is computed using RNAfold from the ViennaRNA package. Accessibility in the RNA is calculated in windows of 70 nt and indicates the probability that a given nucleotide position in the transcript is unpaired and therefore accessible to a gRNA. For each target, we take the mean probability across each position. The smaller the value, the less likely there is secondary structure in the transcript at that gRNA target site.
9.In the table view, sort the gRNAs by clicking on any column header.
10.Review all listed features of your preferred gRNAs. Make sure that the GC content fits your needs and that self-complementarity is not too high.
11.Optional : Download the complete list of proposed targets by choosing a preferred format in the “Download results” box. Make sure your browser allows pop-up windows to open.
12.Optional : Transfer results to the UCSC Genome Browser by clicking on the link.
13.When a suitable candidate is found, click on the graphical representation or results table entry to open a detailed view.
14.Inspect the detailed view for a full list of off-targets.
15.Once a preferred gRNA has been identified, copy the sequence next to “Guide (no PFS)” and use it when ordering gRNAs.
Basic Protocol 3: DESIGN OF gRNAs FOR SEQUENCE KNOCK-IN
The CRISPR/Cas9 system is commonly used to insert new sequences into a locus of interest. CRISPR knock-in allows site-specific gene engineering via programmable nuclease-induced DNA DSBs and subsequent repair by cellular repair pathways (both HDR and NHEJ). In order for the machinery to introduce a knock-in, a donor sequence must be provided. For additional considerations, see Strategic Planning.
Perform search
1.Go to https://chopchop.cbu.uib.no.
2.Enter the gene of interest in the “Target” field.
3.Choose the genome of interest in the “In” field.
4.Choose “CRISPR/Cas9” in the “Using” field.
5.Choose “knock-in” in the “For” field.
6.Optional : Modify the default search based on needs using the “Options” button (see Strategic Planning and Critical Parameters).
7.Press “Find Target Sites!”.
Interpret results
8.Identify a gRNA that fits your purpose and open the detailed view (see Basic Protocol 1, steps 8-13).

9.Inspect the detailed view for important considerations as described (see Basic Protocol 1, step 14).
10.Use the toggles (Fig. 5) to adjust the cut site respective to the PAM and the length of homology arms (20-800 bp).
11.Once a preferred target has been found, copy the target sequence from the top left corner of the results page.
Alternate Protocol 2: DESIGN OF gRNAs FOR KNOCK-IN USING NON-HOMOLOGOUS END JOINING
NHEJ relies not on DNA sequence homology, but rather ligation of broken DNA ends (Cubbon, Ivancic-Bace, & Bolt, 2018). This can be leveraged by introducing a plasmid containing a donor DNA construct to be integrated in the genomic DNA. In eukaryotes, NHEJ occurs predominantly in G1 phase and is inhibited during mitosis to prevent undesirable chromosome fusions at telomeres (Nami et al., 2018; Rees & Liu, 2018; Suzuki et al., 2016). NHEJ is an error-prone pathway that often creates insertions or deletions of nucleotides (indels). Indels can cause frameshifts in protein coding regions, leading to truncation and inactivity after translation. This is the repair pathway used for knock-outs when no donor sequence is introduced.
For an NHEJ knock-in, the gRNA is designed as normal to target the site of interest. The donor DNA, however, is designed with flanking sites identical to the target so that Cas9 cleaves the genomic target sequence at one location and the donor plasmid at two locations (Fig. 1E-G). This releases a donor sequence between the flanking sites that contains the desired insert. The sequence should also lack homology arms so that repair of the Cas9-induced genomic DSB cannot occur through the HDR pathway. Instead, cleavage generates blunt ends at the target site and at both ends of the donor sequence. The linearized donor DNA sequence can then be used for repair by the NHEJ pathway, allowing for its integration into the genomic target site. Once incorporated into the genome, the donor DNA disrupts the Cas9 target sequence and prevents further Cas9-induced DSBs. If the target DSB is rejoined by error-free NHEJ without donor DNA insertion, the Cas9 target sequence will remain intact and a second round of Cas9 cutting will ensue.
1.Follow steps 1-9 as in Basic Protocol 3.
2.Unless HMEJ or MMEJ is desired, design your template with your donor DNA without homology arms but with the same Cas9 target sequence. Remember to also include PAM sequences. We recommend following one the following for design of donor sequences:
- For homology-independent targeted insertion (HITI), flank the donor sequence by one (for circular donor) or two (for linear donors) gRNA target sites.
A third identical sequence in the reverse orientation is in the target locus in which the donor DNA is to be pasted. This allows for knock-in in the correct orientation, as intact gRNA target sequence persists if inserted in the reverse orientation (Suzuki & Izpisua Belmonte,2018; Suzuki et al.,2016).
- For microhomology-mediated end joining (MMEJ), flank the donor sequence by the two microhomology arms. To avoid promoter interference, ensure that integration is in the opposite orientation of the endogenous gene.
MMEJ relies on 5-25 bp of sequence complementarity (microhomology) between DNA strands. It can occur in most of the cell cycle, which leads to higher knock-in efficiency than HDR knock-in (Nakamae et al.,2017). The short arm lengths make it very easy to add in a single PCR without amplification from genomic DNA. MMEJ can perform seamless knock-in without carrying any of the target sequence from the gRNA. It requires three gRNAs, and the donor vector must include two different gRNA target sites.
Basic Protocol 4: DESIGN OF gRNAs FOR KNOCK-IN USING Cas9 NICKASES
Whether using HDR or NHEJ, knock-in approaches relying on DSB repair can have adverse effects on off-target loci. To reduce the risk of off-target genome edits and increase the chance of specific knock-in sequence insertions, one approach is to introduce SSBs flanking the site of insertion. Single nicks are usually efficiently corrected through cellular repair mechanisms without the introduction of mutations; however, two nicks in close vicinity can activate DSB repair pathways that can be leveraged for HDR-mediated knock-in (see Basic Protocol 3; Mali et al., 2013). This strategy can be implemented using Cas9 nickases (Trevino & Zhang, 2014). Cas9 nickases have one of the nuclease domains inactivated. The Cas9 D10A mutant cuts the DNA strand complementary to the gRNA (Fig. 1C), whereas the Cas9 N863A mutant cuts the opposite DNA strand. By using either of the nickases and designing a pair of gRNAs at optimal distance (Ran et al., 2013), one can exploit HDR-driven knock-in insertion. This strategy has the advantage of only allowing knock-in if both gRNAs introduce nicks at the same time on the same DNA. Any single gRNA cut event (whether on- or off-target) is efficiently repaired, reducing the risk of off-target mutations. For additional considerations, see Strategic Planning.
1.Follow steps 1-3 as in Basic Protocol 3.
2.Choose “CRISPR/Cas9 nickase” in the “Using” field.
3.Choose “knock-in” in the “For” field.
4.Optional : Modify the default search based on your needs using the “Options” button. To modify the optimal distance between two gRNAs (and possible off-targets shown), go do “Options” → “Cas9 nickase” → “Distance between guides”.
5.Press “Find Target Sites!”.
6.In the genome locus view (Fig. 6), identify a gRNA pair that fits your purposes.
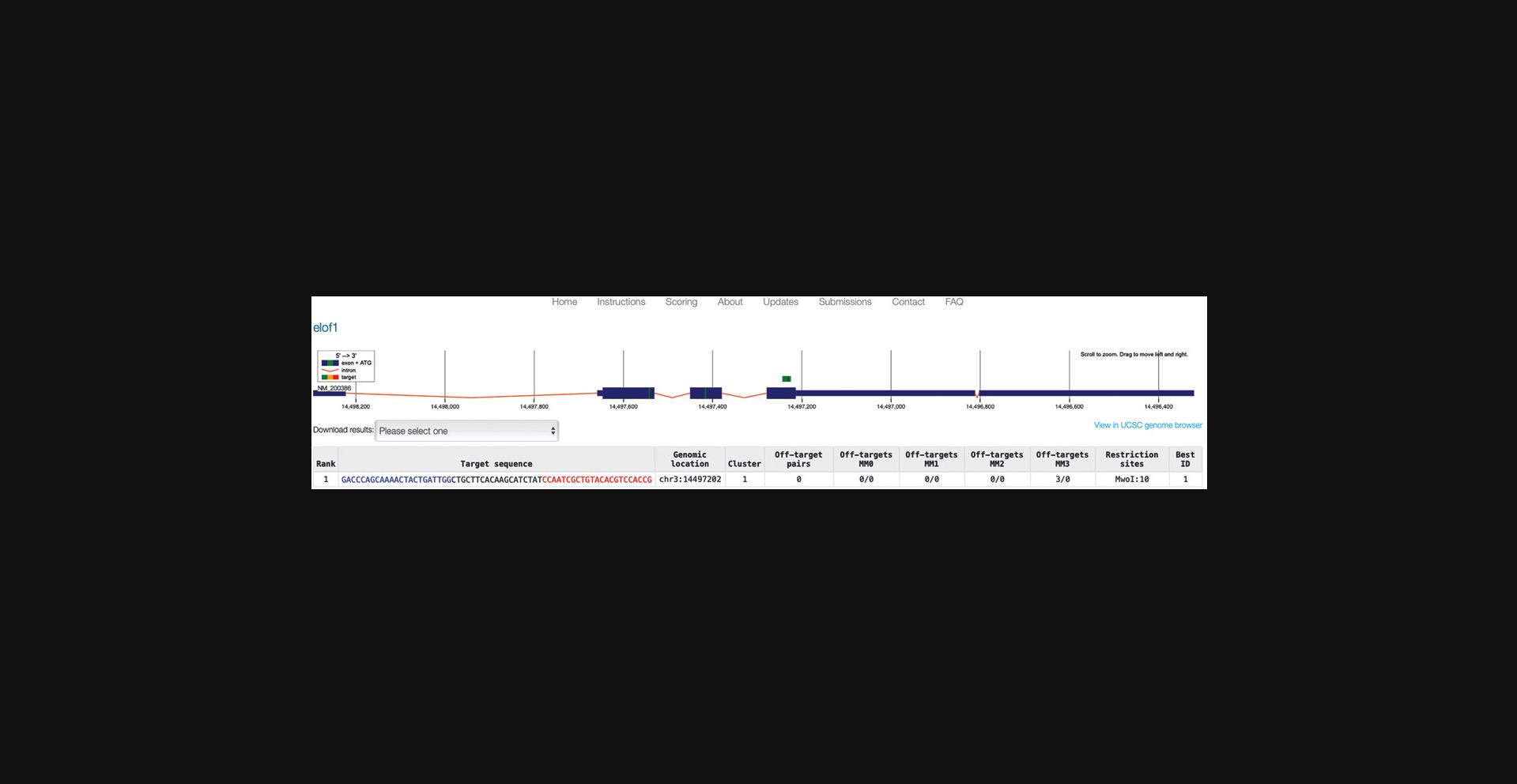
7.After clicking on your preferred target, adjust the HDR homology arms according to your needs (Fig. 7).
COMMENTARY
Background Information
CHOPCHOP was one of the first tools for computational design of guide RNAs for CRISPR genome editing (Montague et al., 2014) and is now one of the most cited tools for design of gRNAs. Over time, support has been added for an increasing number of genomes, and the tool has been updated to stay current with developments in the field. CHOPCHOP supports a range of Cas proteins, targeting modes, and efficiency scoring methods (Labun, Montague, Gagnon, Thyme, & Valen, 2016, 2019).
Critical Parameters
CHOPCHOP offers a large number of options. Most will not be of use for many users and are therefore hidden under the “Options” button on the home page. Nonetheless, users are advised to look through those options and familiarize themselves with the default settings so that results can be fine tuned if needed.
Each query on the website is required: genome/annotation (“In”), protein (“Using”), and specific application (“For”). CHOPCHOP applies a set of default parameters for each application (e.g., knock-out, knock-in, nanopore enrichment). If the settings are changed, they are saved to reflect the latest query run by the user. They can be returned to default by clicking the “Reset Options” button. This is also useful when you encounter an error on the website and need a fresh start for testing purposes.
CHOPCHOP currently supports more than 300 genomes. If your genome of interest is not provided or you prefer to use your own genomic sequence, you can submit your genome and annotations to the authors. For details, see the “Submissions” or “Add new species”. Alternatively, to avoid waiting for the genome update, a local version of CHOPCHOP is available at https://bitbucket.org/valenlab/chopchop (requires basic bioinformatics skills). Local installation also supports bulk designs for many genes at once, unlike the main website where each query has to be run separately.
CHOPCHOP provides a list of the most popular PAM for the respective CRISPR protein selected. It also supports any custom PAM, written with IUPAC code. The PAM can be configured under “Options”. For example, to use Staphylococcus aureus Cas9 targeting 21-bp-NNGRRT, select “CRISPR/Cas9” under “Using”, to go “Options” → “Cas9”, and change “PAM-3′” to NNGRRT and “sgRNA length without PAM” to 21. Similarly, for the alternative PAM located on the 5′ end of the gRNA, select “CRISPR/Cpf1 or CasX” and adjust the “5′-PAM” under “Options” → “Cpf1”.
When targeting genes with multiple isoforms (e.g., ATF3 on hg38 has six isoforms), CHOPCHOP will by default compute the intersection of all isoforms and scan it for gRNAs. The intersection mode ensures that all isoforms will be targeted, but restricts the regions searched to exons that are common to all isoforms. If this is too restrictive, one can switch to the union mode under “Options” → “General” → “Isoform consensus determined by”. The union mode will take the region of all exons of all isoforms of a gene. Although this is the most inclusive approach, it does not guarantee that the designed gRNA will target all of the isoforms at the same time, because exons represented in only one isoform are considered. In cases where annotations are ambiguous (e.g., isoforms are not on the same chromosome), CHOPCHOP will return an error. In that case, specific isoform of the desired gene target (specified as a transcript ID) should be used.
In addition to gene name or gene identifiers, FASTA input genomic coordinates are also supported using the “Paste Input” button. This is provided in the form of chromosome:start-end (e.g., chr1:4512312-4512512). In cases where CHOPCHOP does not find a gene/isoform name, you can instead use sequence or location inputs. For multi-FASTA files, CHOPCHOP makes use of only the first sequence entry. When using FASTA input from transcripts, it is important to realize that, if exons are concatenated, gRNAs that cross over exon-exon borders will be reported. Therefore, one must make sure that the selected guide maps to the genome. When using FASTA from a sequence that is also in the genome, it is expected that the gRNA will have one perfect off-target at the position of the target site. To address this, CHOPCHOP will by default subtract one zero mismatch off-target for each gRNA. This can be adjusted by going to “Options” → “General” → “Fasta input” and checking “Color scoring should ignore one off-target without mismatches”.
Troubleshooting
The PAM sequence (e.g., NGG) is always included in the general results table under “Target sequence” (hence, it is not named “gRNA sequence”). When ordering gRNA for your experiment, the PAM must be excluded so that only the gRNA sequence is ordered.
Finding working gene names depends on the annotations, and CHOPCHOP does not yet support auto-complete. If your gene is not found in our database of all genes, CHOPCHOP will output the following error: “Error 501: Your gene Id: yourgeneid has not been found in our database”. Additionally, examples of gene names will be listed, e.g., SLC41A3 (ENST00000512470), VPS28 (ENST00000642606). In that case, consult Ensembl or RefSeq gene sets or genome browsers to find an identifier for your gene of interest.
Generally, CHOPCHOP includes instructions on what to do in case of an error. When an error is encountered, do not be discouraged by the red color. Simply read the error message and follow the instructions provided.
Time Considerations
CHOPCHOP deliver results for relatively short gene queries quickly: typically at most a couple of minutes. Additionally, CHOPCHOP keeps queries and their results for a couple days and returns results immediately when possible. If many users use the website at the same time, however, long waiting times may result: as long as an hour, depending on the size of the genes, the analyses performed, and demand during peak hours. In this case, you can bookmark the results page and return at a later date. Results will be waiting under the same link.
Acknowledgments
This work was supported by the Research Council of Norway (#250049) and the Norwegian Cancer Society (project #190290).
Author Contributions
Kornel Labun: Conceptualization, Writing-original draft, Writing-review & editing, Maximilian Krause: Writing-original draft, Writing-review & editing, Yamilla Torres Cleuren: Writing-original draft, Writing-review & editing, Eivind Valen: Funding acquisition, Writing-review & editing
Conflict of Interest
The authors declare no conflict of interest.
Open Research
Data Availability Statement
Data sharing not applicable – no new data generated.
Literature Cited
- Adli, M. (2018). The CRISPR tool kit for genome editing and beyond. Nature Communications , 9(1), 1911. doi: 10.1038/s41467-018-04252-2.
- Berckman, E. A., & Chen, W. (2019). Exploiting dCas9 fusion proteins for dynamic assembly of synthetic metabolons. Chemical Communications , 55(57), 8219–8222. doi: 10.1039/c9cc04002a.
- Cox, D. B. T., Gootenberg, J. S., Abudayyeh, O. O., Franklin, B., Kellner, M. J., Joung, J., & Zhang, F. (2017). RNA editing with CRISPR-Cas13. Science , 358(6366), 1019–1027. doi: 10.1126/science.aaq0180.
- Cubbon, A., Ivancic-Bace, I., & Bolt, E. L. (2018). CRISPR-Cas immunity, DNA repair and genome stability. Bioscience Reports , 38(5), BSR20180457. doi: 10.1042/bsr20180457.
- Freije, C. A., Myhrvold, C., Boehm, C. K., Lin, A. E., Welch, N. L., Carter, A., … Sabeti, P. C. (2019). Programmable inhibition and detection of RNA viruses using Cas13. Molecular Cell , 76(5), 826–837.e11. doi: 10.1016/j.molcel.2019.09.013.
- Gaj, T., Sirk, S. J., Shui, S.-L., & Liu, J. (2016). Genome-editing technologies: Principles and applications. Cold Spring Harbor Perspectives in Biology , 8(12), a023754. doi: 10.1101/cshperspect.a023754.
- Gootenberg, J. S., Abudayyeh, O. O., Lee, J. W., Essletzbichler, P., Dy, A. J., Joung, J., … Zhang, F. (2017). Nucleic acid detection with CRISPR-Cas13a/C2c2. Science , 356(6336), 438–442. doi: 10.1126/science.aam9321.
- Jang, H.-K., Song, B., Hwang, G.-H., & Bae, S. (2020). Current trends in gene recovery mediated by the CRISPR-Cas system. Experimental & Molecular Medicine, 52(7), 1016–1027. doi: 10.1038/s12276-020-0466-1.
- Konermann, S., Brigham, M. D., Trevino, A. E., Joung, J., Abudayyeh, O. O., Barcena, C., … Zhang, F. (2015). Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature , 517(7536), 583–588. doi: 10.1038/nature14136.
- Labun, K., Montague, T. G., Gagnon, J. A., Thyme, S. B., & Valen, E. (2016). CHOPCHOP v2: A web tool for the next generation of CRISPR genome engineering. Nucleic Acids Research , 44(W1), W272–W276. doi: 10.1093/nar/gkw398.
- Labun, K., Montague, T. G., Krause, M., Torres Cleuren, Y. N., Tjeldnes, H., & Valen, E. (2019). CHOPCHOP v3: Expanding the CRISPR web toolbox beyond genome editing. Nucleic Acids Research , 47(W1), W171–W174. doi: 10.1093/nar/gkz365.
- Lei, Y., Zhang, X., Su, J., Jeong, M., Gundry, M. C., Huang, Y.-H., … Goodell, M. A. (2017). Targeted DNA methylation in vivo using an engineered dCas9-MQ1 fusion protein. Nature Communications , 8(1), 16026. doi: 10.1038/ncomms16026.
- Mali, P., Aach, J., Benjamin Stranges, P., Esvelt, K. M., Moosburner, M., Kosuri, S., … Church, G. M. (2013). CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nature Biotechnology , 31(9), 833. doi: 10.1038/nbt.2675.
- Montague, T. G., Cruz, J. M., Gagnon, J. A., Church, G. M., & Valen, E. (2014). CHOPCHOP: A CRISPR/Cas9 and TALEN web tool for genome editing. Nucleic Acids Research , 42(W1), W401–W407. doi: 10.1093/nar/gku410.
- Nakamae, K., Nishimura, Y., Takenaga, M., Nakade, S., Sakamoto, N., Ide, H., … Yamamoto, T. (2017). Establishment of expanded and streamlined pipeline of PITCh knock-in – a web-based design tool for MMEJ-mediated gene knock-in, PITCh designer, and the variations of PITCh, PITCh-TG and PITCh-KIKO. Bioengineered , 8(3), 302–308. doi: 10.1080/21655979.2017.1313645.
- Nami, F., Basiri, M., Satarian, L., Curtiss, C., Baharvand, H., & Verfaillie, C. (2018). Strategies for in vivo genome editing in nondividing cells. Trends in Biotechnology , 36(8), 770–786. doi: 10.1016/j.tibtech.2018.03.004.
- Ran, F. A., Ann Ran, F., Hsu, P. D., Lin, C.-Y., Gootenberg, J. S., Konermann, S., … Zhang, F. (2013). Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell , 155(2), 479–480. doi: 10.1016/j.cell.2013.09.040.
- Rees, H. A., & Liu, D. R. (2018). Base editing: Precision chemistry on the genome and transcriptome of living cells. Nature Reviews Genetics , 19(12), 770–788. doi: 10.1038/s41576-018-0059-1.
- Ribeiro, L. F., Ribeiro, L. F. C., Barreto, M. Q., & Ward, R. J. (2018). Protein engineering strategies to expand CRISPR-Cas9 applications. International Journal of Genomics , 2018, 1–12. doi: 10.1155/2018/1652567.
- Suzuki, K., & Izpisua Belmonte, J. C. (2018). In vivo genome editing via the HITI method as a tool for gene therapy. Journal of Human Genetics , 63(2), 157–164. doi: 10.1038/s10038-017-0352-4.
- Suzuki, K., Tsunekawa, Y., Hernandez-Benitez, R., Wu, J., Zhu, J., Kim, E. J., … Belmonte, J. C. I. (2016). In vivo genome editing via CRISPR/Cas9 mediated homology-independent targeted integration. Nature , 540(7631), 144–149. doi: 10.1038/nature20565.
- Trevino, A. E., & Zhang, F. (2014). Genome editing using Cas9 nickases. Methods in Enzymology , 161–174. doi: 10.1016/b978-0-12-801185-0.00008-8.
- Tsui, C., Inouye, C., Levy, M., Lu, A., Florens, L., Washburn, M. P., & Tjian, R. (2018). dCas9-targeted locus-specific protein isolation method identifies histone gene regulators. Proceedings of the National Academy of Sciences of the United States of America , 115(12), E2734–E2741. doi: 10.1073/pnas.1718844115.
- Untergasser, A., Cutcutache, I., Koressaar, T., Ye, J., Faircloth, B. C., Remm, M., & Rozen, S. G. (2012). Primer3—new capabilities and interfaces. Nucleic Acids Research , 40(15), e115. doi: 10.1093/nar/gks596.
- Vora, S., Tuttle, M., Cheng, J., & Church, G. (2016). Next stop for the CRISPR revolution: RNA-guided epigenetic regulators. FEBS Journal , 283(17), 3181–3193. doi: 10.1111/febs.13768.
- Xu, X., Tao, Y., Gao, X., Zhang, L., Li, X., Zou, W., … Hu., R. (2016). A CRISPR-based approach for targeted DNA demethylation. Cell Discovery , 2, 16009. doi: 10.1038/celldisc.2016.9.
- Ye, H., Rong, Z., & Lin, Y. (2017). Live cell imaging of genomic loci using dCas9-SunTag system and a bright fluorescent protein. Protein & Cell, 8(11), 853–855. doi: 10.1007/s13238-017-0460-0.
- Yeo, N. C., Chavez, A., Lance-Byrne, A., Chan, Y., Menn, D., Milanova, D., … Church, G. M. (2018). An enhanced CRISPR repressor for targeted mammalian gene regulation. Nature Methods , 15(8), 611–616. doi: 10.1038/s41592-018-0048-5.
Internet Resources
M. Gearing. Cas9 nickase design and homology directed repair. Retrieved December 9, 2020.
Integrated DNA Technologies. When and how to use nickases for efficient genome editing. Retrieved December 9, 2020.
Citing Literature
Number of times cited according to CrossRef: 13
- Juan M Angueyra, Vincent P Kunze, Laura K Patak, Hailey Kim, Katie Kindt, Wei Li, Transcription factors underlying photoreceptor diversity, eLife, 10.7554/eLife.81579, 12 , (2023).
- Reza Mohammadhassan, Sara Tutunchi, Negar Nasehi, Fatemeh Goudarziasl, Lena Mahya, The Prominent Characteristics of the Effective sgRNA for a Precise CRISPR Genome Editing, CRISPR Technology - Recent Advances, 10.5772/intechopen.106711, (2023).
- Heather M. Terron, Derek S. Maranan, Luke A. Burgard, Frank M. LaFerla, Shelley Lane, Malcolm A. Leissring, A Dual-Function “TRE-Lox” System for Genetic Deletion or Reversible, Titratable, and Near-Complete Downregulation of Cathepsin D, International Journal of Molecular Sciences, 10.3390/ijms24076745, 24 , 7, (6745), (2023).
- Rikiya Imamura, Mizuki Saito, Mikio Shimada, Junya Kobayashi, Masamichi Ishiai, Yoshihisa Matsumoto, APTX acts in DNA double-strand break repair in a manner distinct from XRCC4, Journal of Radiation Research, 10.1093/jrr/rrad007, 64 , 3, (485-495), (2023).
- Maria Czarnek, Jakub Kochan, Mateusz Wawro, Rafał Myrczek, Joanna Bereta, Construction of a Set of Novel Transposon Vectors for Efficient Silencing of Protein and lncRNA Genes via CRISPR Interference, Molecular Biotechnology, 10.1007/s12033-023-00675-5, (2023).
- Saumya Mary Mathew, Strategies for generation of mice via CRISPR/HDR-mediated knock-in, Molecular Biology Reports, 10.1007/s11033-023-08278-8, 50 , 4, (3189-3204), (2023).
- Lynn C. Thomason, Nina Costantino, Xintian Li, Donald L. Court, Recombineering: Genetic Engineering in Escherichia coli Using Homologous Recombination, Current Protocols, 10.1002/cpz1.656, 3 , 2, (2023).
- Krishnagowdu Saravanan, Kumar Praveenkumar, Nandakumar Vidya, Kumaraguru Gowtham, Mohanasundaram Saravanan, Enhancement of Agricultural Crops: A CRISPR/Cas9-Based Approach, Vegetable Crops - Health Benefits and Cultivation, 10.5772/intechopen.100641, (2022).
- Kanishk Goel, Jonathan E. Ploski, RISC-y Business: Limitations of Short Hairpin RNA-Mediated Gene Silencing in the Brain and a Discussion of CRISPR/Cas-Based Alternatives, Frontiers in Molecular Neuroscience, 10.3389/fnmol.2022.914430, 15 , (2022).
- Shuliang Chen, Melissa A. Roberts, Chun-Yuan Chen, Sebastian Markmiller, Hong-Guang Wei, Gene W. Yeo, James G. Granneman, James A. Olzmann, Susan Ferro-Novick, VPS13A and VPS13C Influence Lipid Droplet Abundance, Contact, 10.1177/25152564221125613, 5 , (251525642211256), (2022).
- Ben C. King, Anna M. Blom, Intracellular complement: Evidence, definitions, controversies, and solutions, Immunological Reviews, 10.1111/imr.13135, 313 , 1, (104-119), (2022).
- Lauren E. Limbach, Rocky L. Penick, Rudy S. Casseday, Maddelyn A. Hyland, Erika A. Pontillo, Afomia N. Ayele, Kristen M. Pitts, Sarah D. Ackerman, Breanne L. Harty, Amy L. Herbert, Kelly R. Monk, Sarah C. Petersen, Peripheral nerve development in zebrafish requires muscle patterning by tcf15/paraxis, Developmental Biology, 10.1016/j.ydbio.2022.07.001, 490 , (37-49), (2022).
- Arun S Rajkumar, John P Morrissey, Protocols for marker-free gene knock-out and knock-down in Kluyveromyces marxianus using CRISPR/Cas9 , FEMS Yeast Research, 10.1093/femsyr/foab067, 22 , 1, (2021).