An X-HTDC method for estimating particulate phosphorus from microalgae
Ying-Yu Hu, Zoe V. Finkel
particulate phosphorus
intracellular phosphorus
phosphomolybdenum-ascorbic reduction
orthophosphate
oxalate reagent
adsorbed phosphorus
X-HTDC
High temperature dry combustion
Abstract
Total particulate phosphorus (TPP) is often determined using the High Temperature Dry Combustion (HTDC) method followed by hydrolysis of the ash and then molybdenum colorimetry. Here we show that a higher than traditionally-used combustion temperature, 800 °C vs. 450 - 550 °C, improves phosphorus recovery from several organic phosphorus compounds, marine phytoplankton cultures and particulate samples from the field. The ashing auxiliary MgSO4 further improves P recovery by improving decomposition, reducing volatilization during combustion, and increasing the efficiency of hydrolysis. A 0.2 M HCl hydrolysis, under 90 °C for 30 minutes yields a higher P recovery compared with hydrolysis at room temperature or 60 °C. In aggregate these improvements to the method double the P recovery from phospholipids to 97%. TPP recovery from laboratory phytoplankton cultures and field samples increased an average of 11%, primarily due to the improvements in P recovery from phospholipids, polyphosphates, and nucleic acids. We refer to this new method as the eXtra high temperature dry combustion ash/hydrol method (X-HTDC) and recommend its application for measuring particulate phosphorus from organic compounds in aquatic systems.
The working range of this assay is 1.22 to 500 uM orthophosphate. Minimum sampling biomass is 0.19 ug P/filter.
In order to assess the intracellular phosphorus in microalgae, we recommend an oxalate reagent (Tovar-Sanchez 2003) to wash the microalgae collected on the filter to remove surface adsorbed phosphorus.
Before start
We have found that crucibles may lose their temperature resistance after acid-washing or long soaks in alkaline detergent. Crucibles tended to shatter in the oven during the initial increase in temperature from room temperature to 500 °C, even when the ramp rate was carefully controlled at 150 °C/h. We recommend not soaking crucibles in acid but instead we suggest the crucibles be filled with 0.2 M HCl and then incubated at 90 °C for 30 minutes as the acid-washing step. It is necessary to inspect the temperature resistance of newly acquired crucibles by combusting them at 500 °C for 6 h (ramp rate: 150 °C/h) after acid-washing. We found that crucibles that pass this inspection do not usually shatter when heated to 800 °C.
Steps
Sampling
Sampling microalgae for total particulate phosphorus (i.e. intracellular phosphorus and adsorbed phosphorus)
Filter microalgae in liquid media onto polycarbonate filters, using gentle vacuum pressure (130 mmHg). Equipment
| Value | Label |
|---|---|
| Filter forceps | NAME |
| blunt end, stainless steel | TYPE |
| Millipore | BRAND |
| XX6200006P | SKU |
| http://www.emdmillipore.com/ | LINK |
Rinse filter tunnel with filtered artificial seawater (phosphate free) to avoid sample loss.
Place sample filters in 2 mL Cryogenic Vials.
Equipment
| Value | Label |
|---|---|
| Cryogenic Vials with Closures | NAME |
| Polypropylene, 2 mL | TYPE |
| Corning® | BRAND |
| 66021-974 | SKU |
Filter blank media (without cells) through polycarbonate filter as blank.
Flash freeze filters and store at -20°C.
Sampling microalgae for intracellular particulate phosphorus
Filter microalgae in liquid media onto polycarbonate filters , using gentle vacuum pressure (5 inches Hg). Equipment
| Value | Label |
|---|---|
| Filter forceps | NAME |
| blunt end, stainless steel | TYPE |
| Millipore | BRAND |
| XX6200006P | SKU |
| http://www.emdmillipore.com/ | LINK |
Add 5mL oxalate reagent onto the filter, and let oxalate reagent sit in the filter funnel for 0h 5m 0s
Drain and then rinse filter funnel with filtered artificial seawater (phosphate free).
Place sample filters in 2 mL Cryogenic Vials.
Equipment
| Value | Label |
|---|---|
| Cryogenic Vials with Closures | NAME |
| Polypropylene, 2 mL | TYPE |
| Corning® | BRAND |
| 66021-974 | SKU |
Filter blank media (without cells) through polycarbonate filter as blank.
Flash freeze filters and store at -20°C.
X-HTDC-ing
Mark number at the bottom of each crucible with pencil, log the following information:(1) The number of crucible(2) The code of sample in the crucible Equipment
| Value | Label |
|---|---|
| Porcelain crucibles | NAME |
| 40 mL | TYPE |
| VWR | BRAND |
| 89037-996 | SKU |
Equipment
| Value | Label |
|---|---|
| Crucible cover | NAME |
| VWR | BRAND |
| 71000-146 | SKU |
Transfer sample to crucible with clean filter forceps and lay filter at the bottom.
0.17M MgSO4reagent:Dissolve 1.023g MgSO4 in 50 mL MilliQ water
Add 200µL 0.17M MgSO4 directly onto each sample and blank filter.
Cover the crucibles and place in the oven at 90°C until samples are completely dry.
Equipment
| Value | Label |
|---|---|
| Forced air oven | NAME |
| VWR | BRAND |
| 89511-410 | SKU |
Combust dried samples at 800°C for 9h 0m 0s
Equipment
| Value | Label |
|---|---|
| Muffle furnace | NAME |
| F30428C | TYPE |
| Thermo | BRAND |
| 10-505-13 | SKU |
| https://www.fishersci.com/us/en/home.html | LINK |
Allow samples to gradually cool down in the muffle furnace.
Pencil mark on crucibles should be still visible, however, it can be easily removed by water. Therefore, when removing samples out of the furnace, label the lid and crucible with sharpie immediately.
Digesting
0.2M HCl reagent:In a reagent bottle, dissolve one part of 12N HCl in 59 parts of MilliQ water
Preheat oven to 90°C
Add 5 mL 0.2 M HCl to each crucible.
Gently swirl the crucible.
Cover the crucibles and place crucibles in the muffin tin pan for easier-handling.
Incubate in the oven for 0h 30m 0s
Cool samples down to Room temperature
Gently swirl the crucible and then transfer 500 uL solution to 2 mL microtube. Duplicate each sample and blank.
Equipment
| Value | Label |
|---|---|
| Maxymum Recovery® Snaplock Microcentrifuge Tube | NAME |
| 2.0 mL, Polypropylene, Clear, Nonsterile, | TYPE |
| Axygen® | BRAND |
| MCT-200-L-C | SKU |
Preparing standard working solutions
Standard working solutions and reagents can be prepared during sample digestion.
KH2PO4 primary standard stock solution (≈ 1 mM)
Transfer about 1 g KH2PO4 into a beaker, cover the beaker with foil
Place the beaker into an oven, dry KH2PO4 at 110°C for at least 2h 0m 0s
Move KH2PO4 into a vacuum desiccator, allow KH2PO4 to cool to room temperature
Dissolve around 0.136 g dried KH2PO4 in 1 L milliQ water.* Use 1 L volumetric flask
- Take notes of the actual weight of KH2PO4 for final concentration of standard stock solution
Transfer standard stock solution into a 1 L bottle and store in the fridge.
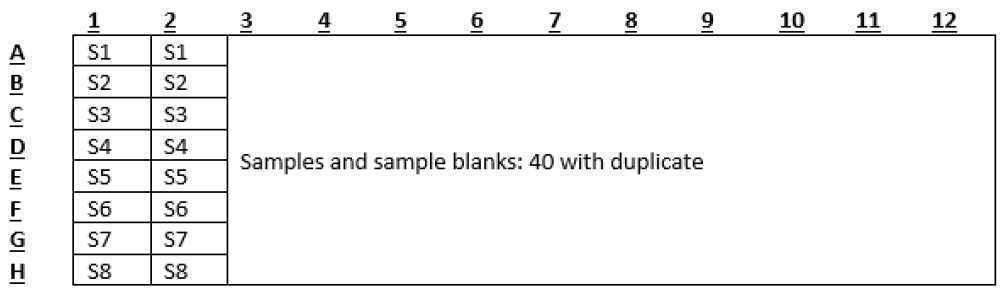
Standard working solution
| A | B | C |
|---|---|---|
| KH2PO4 | Primary (ul) | MilliQ (ul) |
| S1 | 0 | 1000 |
| S2 | 5 | 995 |
| S3 | 10 | 990 |
| S4 | 20 | 980 |
| S5 | 50 | 950 |
| S6 | 100 | 900 |
| S7 | 150 | 850 |
| S8 | 200 | 800 |
Transfer 500 uL of each standard working solution to 2 mL microtube.
Preparing working reagents
6N (3 M) sulfuric acid reagent:Carefully add 1 part 18M concentrated sulfuric acid into 5 part MilliQ water
2.5% ammonium molybdate reagent:
Weigh 0.25g ammonium molybdate in a Falcon tube and top to 10g with MilliQ water.
Cap and shake until totally dissolved.
10% ascorbic acid reagent:Weigh 1g ascorbic acid in a Falcon tube and top to 10g with MilliQ water;Cap and shake until all dissolved.
Calculate the volume of molybdate-ascorbic reagent:
Total volume of reagent_mL = (0.5 mL) X (#standard working solution + #samples + #blanks)
Mix the reagents into Falcon tube:
| A | B |
|---|---|
| Reagent | Part(s) as in volume |
| MilliQ | 2 |
| 6N sulphuric acid | 1 |
| 2.5% ammonium molybdate | 1 |
| 10% ascorbic acid | 1 |
Colorimetric measurement
Preheat incubator/shaker to 37°C
Equipment
| Value | Label |
|---|---|
| SHAKING INCUBATOR | NAME |
| 71L | TYPE |
| Corning® LSE™ | BRAND |
| 6753 | SKU |
Add 500µL reagent to each standard, sample and blank, starting from blanks, including blank for standards and blank for samples.
Equipment
| Value | Label |
|---|---|
| Finntip Stepper Tips | NAME |
| 5 mL | TYPE |
| Thermo Scientific | BRAND |
| 9404200 | SKU |
| https://www.fishersci.com/us/en/home.html | LINK |
Vortex each tube.
Incubate at 37°C for 3h 0m 0s while shaking at 200 rpm
Read plate in microplate reader
| A | B |
|---|---|
| Shake duration | 00:00:05 |
| Shaking type | Continuous |
| Shaking force | High |
| Shaking speed [rpm] | 600 |
| Wavelength [nm] | 820 |
| Use transmittance | No |
| Pathlength correction | No |
| Measurement Time [ms] | 100 |
Equipment
| Value | Label |
|---|---|
| Varioskan LUX Multimode Microplate Reader | NAME |
| Thermo Fisher | BRAND |
| VL0L00D0 | SKU |
Calculating
Subtract the average absorbance at 820 nm of the blank standard replicates from the absorbance at 820 nm of all other standard working solutions.
Subtract the average absorbance at 820 nm of the blank sample (i.e. blank filter) replicates from the absorbance at 820 nm of all other individual samples.
Prepare a standard curve by plotting the average blank-corrected 820 nm absorbance for each standard working solution versus its concentration in uM.
Use the standard curve to determine the orthophosphate concentration of each unknown sample by using its blank-corrected 820 nm absorbance.
(P per sample)_ug = (orthophosphate)_uM X (V_HCl)_mL X (0.001) X (30.97)