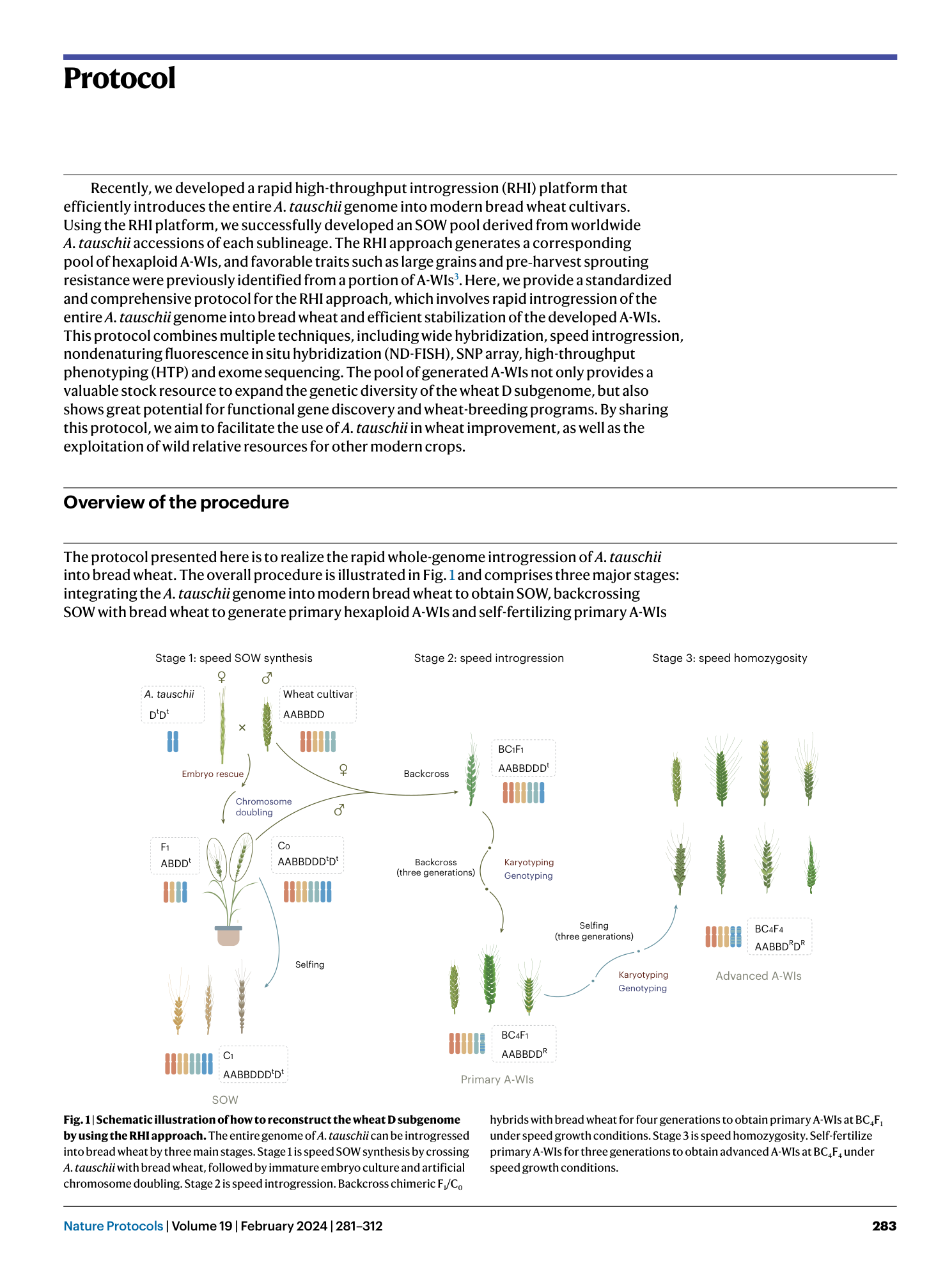
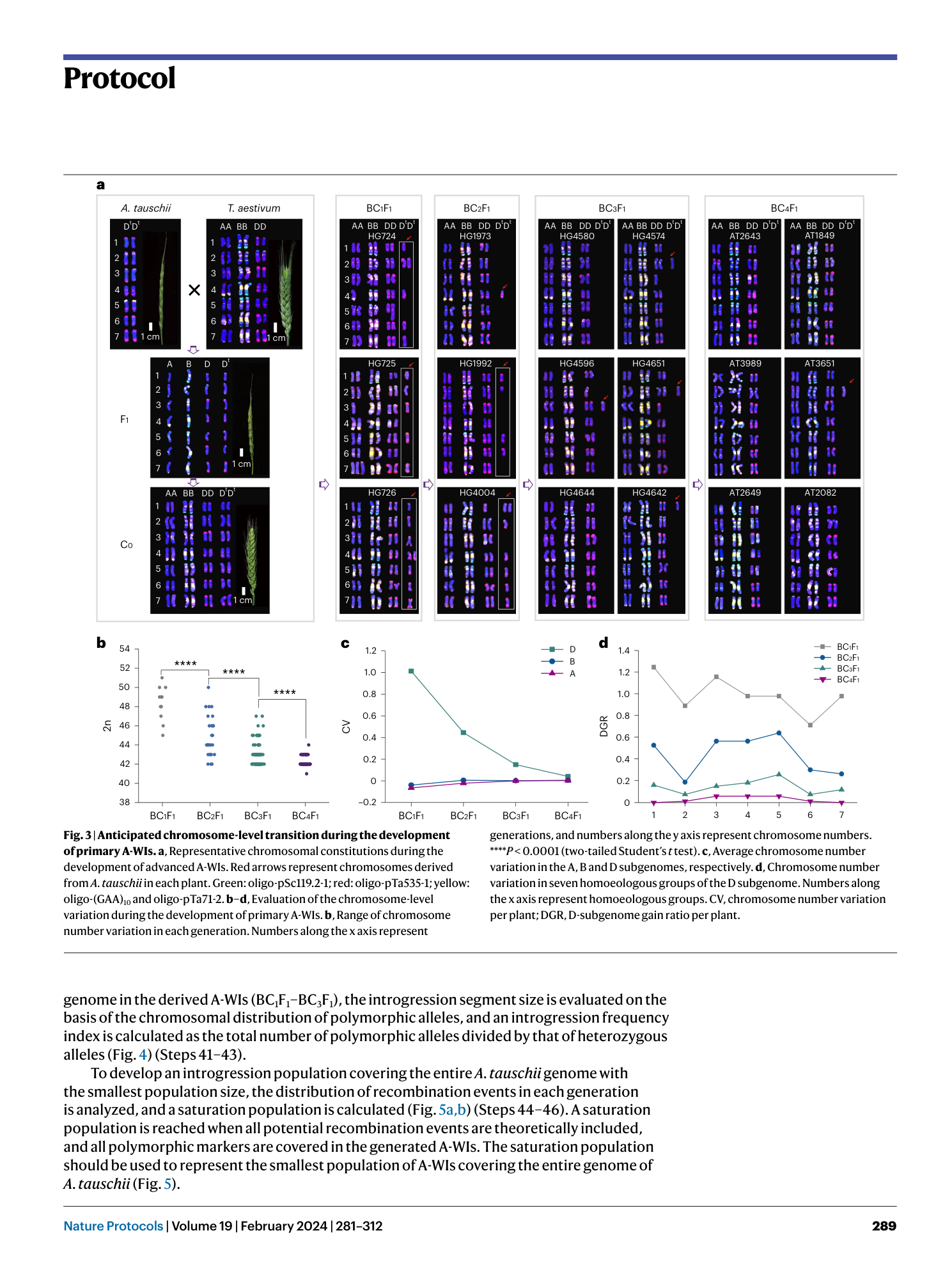
A platform for whole-genome speed introgression fromAegilops tauschiito wheat for breeding future crops
Zheng Li, Can Li, Hao Li, Lele Zhu, Ruixiao Fan, Yifan Liu, Aaqib Shaheen, Fang Nie, Xuqin Liu, Yuanyuan Li, Wenjuan Liu, Yingying Yang, Tutu Guo, Yu Zhu, Mengchen Bu, Chenglin Li, Huihui Liang, Shenglong Bai, Feifei Ma, Guanghui Guo, Zhen Zhang, Jinling Huang, Yun Zhou, Chun-Peng Song









































Extended
Extended Data Fig. 1 An SOW pool developed by crossing 85 A. tauschii accessions from all five sublineages with bread wheat cultivar.
a , Sublineage information of 85 A. tauschii accessions. The accession number of each sublineage is indicated in parentheses. b , Spike and grain phenotype of 18 SOWs and parent lines. AK58 and T093 represent wheat cultivar and A. tauschii , respectively. c , ND-FISH karyotypes of 12 SOWs. Most of the SOWs carried a complete chromosome set of A. tauschii , while several SOWs occasionally lose chromosomes. Green: oligo-pSc119.2; red: oligo-pTa535; yellow: oligo-(GAA) 10 . Adapted with permission from ref. 3 , Springer Nature Limited.
Extended Data Fig. 2 Meiotic pairing of A. tauschii T093 × common wheat AK58 F 1 hybrids.
a – c , The averaged univalent (I) and bivalent (II) number in each meiotic cell of A. tauschii T093 ( n = 85 cells), common wheat AK58 ( n = 50 cells) and their F 1 hybrids ( n = 72 cells). d , A meiotic cell of A. tauschii T093 × common wheat AK58 F 1 hybrids showing seven bivalents and 14 univalents. Arrows represent the bivalents of 1D with 1Dt, and 2D with 2Dt, respectively. Green: oligo-(GAA) 10 ; red: oligo- pTa535-1. Bar = 20 μm. ANPC, average number per cell.
Extended Data Fig. 3 Graphical representation of the whole-genome coverage in BC 1 F 1 , BC 2 F 1 and BC 3 F 1 in seven homologous groups based on a 55K SNP array.
a – c , Whole-genome coverage in BC 1 F 1 ( a ), BC 2 F 1 ( b ) and BC 3 F 1 ( c ) by using original population size. d – f , Whole-genome coverage in BC 1 F 1 ( d ), BC 2 F 1 ( e ) and BC 3 F 1 ( f ) by using calculated saturation population size. Each row represents an A-WI, and each column represents a SNP locus. Pink: heterozygous for A. tauschii T093 alleles; blue: homozygous for bread wheat AK58 alleles; white: missing data. 0: genotype consistent with bread wheat cultivar AK58; 1: missing data; 2: genotype consistent with A. tauschii T093.
Extended Data Fig. 4 Distribution of introgressed fragment size in different generations.
a and b , Histogram of a foreign fragment size of BC 4 F 1 ( a ) and BC 4 F 2 ( b ) in seven homologous groups with 10 Mb as the length interval. c and d , count of introgressed fragments with length <10 Mb. The width of each column represents a span of 1 Mb. chr, chromosome.
Supplementary information
Supplementary Information
Supplementary Figs. 1 and 2 and Table 2
Reporting Summary
Supplementary Video 1
A video file showing how to emasculate the spikes of A. tauschii
Supplementary Table 1
The inheritance frequencies of A. tauschii T093 alleles evaluated by 55K SNP array in derived BC 1 F 1 , BC 2 F 1 and BC 3 F 1 generations
Supplementary Table 3
Statistics of introgressed fragment sizes in derived BC 4 F 1 , BC 4 F 2 and BC 4 F 4 generations
Supplementary Data 1
Supplementary Data

