Non-destructively barcoding hundreds of freshwater macroinvertebrates with a MinION
Elise C. Knobloch, Ray C Schmidt
Abstract
This project aimed to optimize protocols needed to produce CO1 barcodes for 1000s of African freshwater macroinvertebrates, from many different orders, in the most cost-efficient way possible. Since many of these specimens represent undescribed or poorly-known taxa we also wanted to utilize a non-destructive method of DNA extraction. To do so, we modified the methods detailed by Srivathsan et al. (2021). Here we present the protocol from specimen preparation and DNA extraction to sequence generation. In addition to the methods outlined by Srivathsan et al. (2021) we also pulled together the protocols from Oxford Nanopore Technologies and other vendors. We have added some tips and comments to these procedures that we found helpful in the process. We used these protocols to produce barcodes for hundreds of freshwater macroinvertebrates that were collected in Gabon. This project was funded by the National Science Foundation's Research Experience for Post-Baccalaureate Students (REPS) program (DEB #1920116).
Before start
Steps
Specimen preparation and DNA extraction
Specimen preparation and DNA extraction
Specimen preparation
Fill two Petri dishes with ddH2O
After heating, remove the
DNA Dilution
Put 10µL of ddH2O in each well of a 96-well plate
Put 1µL of DNA extraction in each coinciding well
Remove specimen/s from ethanol and place in one dish of ddH2O for0h 10m 0s
Remove specimen/s from water, and rinse again in the second dish for 0h 5m 0s
Place specimen/s on a paper towel to dry for about 0h 5m 0s
DNA extraction
Put 10µL
Place one rinsed and dried specimen in each well head-first
Cover with TempPlate sealing foil or reusable TempPlate pressure-fit sealing mat
Place the covered 96-well plate in the thermal cycler
Heat at 65°C for 0h 18m 0s ,98°C for 0h 2m 0s
PCR Amplification
PCR Amplification
PCR Setup
Combine ddH2O,
| A | B |
|---|---|
| Per 20µL Reaction | |
| ddH20 | 7 |
| 2x Buffer-APEX red | 10 |
| Primer F | 0.5 |
| Primer R | 0.5-not in master mix |
| DNA Template | 2-not in master mix |
Distribute 17.5µL of master mix into each tube of a strip tube
Distribute DNA template and reverse primer into each tube
Thermal Cycler Protocol:94°C for 0h 1m 0s, then 94°C for 0h 0m 30s, 42°C for 0h 1m 0s, 72°C for 0h 1m 30s, go back to step 2 39x, 72°C for 0h 7m 0s, infinite hold at 10°C
Equipment
| Value | Label |
|---|---|
| T100 Thermal Cycler | NAME |
| Thermal Cycler | TYPE |
| BioRad | BRAND |
| 1861096 | SKU |
| http://www.bio-rad.com/ | LINK |
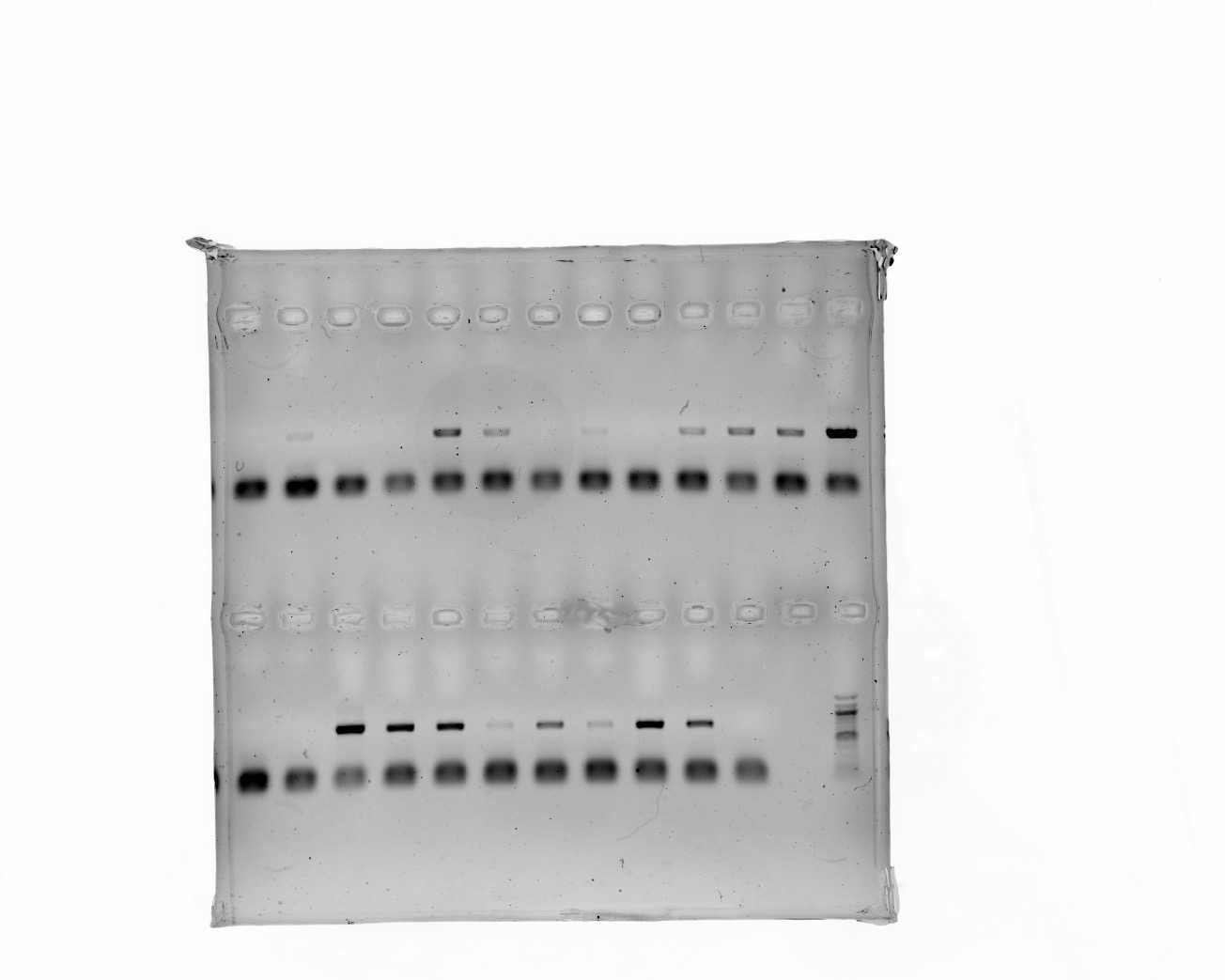
Gel Electrophoresis
Run a gel on at least 6 specimens from each master mix batch. Including a subset of the reaction on the gel allows you to confirm that there were no issues with the PCR master mix and also estimates how many reactions were successful.
Pooling and Cleaning
Pooling and cleaning
Take 4µL from each PCR-This will yield approximately 1.152mL of PCR product
Add another 250µL of 70% ethanol and let sit for 0h 0m 30s
Remove all ethanol
Let dry for 0h 5m 0s to0h 10m 0s with the cap open so the ethanol can evaporate
Add 40µL of ddH2O and elute the pellet (remove from the rack)
Let sit for 0h 2m 0s
Quantify cleaned PCR with
Equipment
| Value | Label |
|---|---|
| Quantas Fluorometer | NAME |
| Fluorometer | TYPE |
| Promega | BRAND |
| E6150 | SKU |
| http://www.promega.com | LINK |
Qubit Fluorometer (QuantasTMFluorometer protocol listed below)
-
Mix
1µLof DNA sample with200µLofin a 0.5ml PCR tube. -
Vortex
-
Place tube into the tube holder and close the lid
-
Vortex sample again and repeat to be sure you got an accurate reading
Add 125µL of
Let sit for 0h 10m 0s at room temperature
Let sit on magnetic rack for 0h 2m 0s
Remove supernatant, leaving about 5µL on the bottom of the tube
Add 250µL of freshly made 70% ethanol and pipette gently up and down
Let sit on rack for 0h 0m 30s
Remove all ethanol
Library preparation
Library preparation
Dilute DNA-->concentration of DNA should be 100-200fmol amplicon DNA
Add 100µLof 70% ethanol (do not touch pellet)
Remove ethanol
Add 100µL of 70% ethanol, take off magnet rack and spin down, put back on magnetic rack
Remove ethanol, allow to dry for about 0h 0m 30s
Add 30.5µL of ddH2), remove from magnetic rack and resuspend pellet
Incubate off rack for 0h 2m 0s
Place back on magnetic rack until clear
Remove liquid and put in clean tube
Combine in a strip-tube: 22.5µL of diluted DNA, 3.5µL of 1.5µL of 2.5µL of H2O
Heat at 20°C for 0h 5m 0s, 65°C for 0h 5m 0s
Transfer mix into new 1.5 ml tube
Vortex
Add 30µL of
Incubate on Hula mixer for 0h 5m 0s
Spin down, place on magnet until clear
Keeping the tube on the magnet, pipette off supernatant
Library preparation: Ligation
Ligation
Spin down
Allow to dry for about 0h 0m 30s
Add 15µL of0h 10m 0s
Place on magnetic rack and allow pellet to form, remove liquid and transfer to a clean tube
Quantify
Dilute so the concentration of the DNA library is 3-20fmol
Combine in a new tube in this order: 30µL of DNA from previous step, 12.5µL of LNB, 5µL of 2.5µL of AMX
Flick and spin mixture, incubate at room temp for 0h 10m 0s
Vortex
Add 20µL of
Incubate at room temp on Hula mixer for 0h 5m 0s
Spin sample, put on magnetic rack, let pellet form, pipette of supernatant
Add 125µL of
Add 125µL of
Setting Nanopore Parameters
Setting Nanopore Sequencing Parameters
We largely followed the default settings for the sequencing run. W set the sequencing kit to SQK-LSK-109, run time of 17 hours, and changed the FastQ so that it didn't compress the files.
Loading flow cell
Loading flow cell
Priming buffer: mix 117µL of 3µL of
Mix 15µL of 10µL 5µL of DNA library in a separate tube
.Insert 120µL of the priming buffer into the flow cell
Insert 30µL of prepped DNA library into flow cell
Demultiplexing
Demultiplexing
Creating Demultiplexing file
Create an excel sheet with 5 columns* First column: sample/specimen ID
- Second column: forward tag
- Third column: reverse tag
- Fourth column: forward primer sequence
Save as a .txt file
Merge resulting Fastq files
Run ONTBarcoder https://github.com/asrivathsan/ONTbarcoder using default parameters.
View resulting CO1 sequences for further analyses.