MERS-CoV Mpro large scale purification protocol
Korvus Wang, michael fairhead, Eleanor Williams
Disclaimer
Research was supported in part by NIAID of the U.S National Institutes of Health under award number U19AI171399. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Abstract
This protocol details the expression and purification of MERS Mpro construct bearing a N-terminal His-SUMO tag at large scale (>6L).
Attachments
Steps
Abbreviations
CV - column volume, total volume of resin in a column
IMAC - immobilised metal affinity chromatography
FT - flow through
Plasmid Transformation
MERS Mpro N-terminal His-SUMO tagged construct was inoculated from its BL21(DE3)-RR glycerol stock.
Protein Expression
See (Nathan's protocol DOI) for MERS-MPro large scale expression protocol
Protein Purifcation
Lyse cell pellet
Thaw and resuspend the pellet in ~7mL of lysis buffer per g of pellet. Stir gently with magnetic stir bar at Room temperature for 0h 30m 0s to allow lysozyme and bezonase to start breaking down
cell components.
Lyse by sonication 0h 0m 4s 0h 0m 12s for a total 'on' time of 0h 7m 0s at 50% amplitude to fully rupture the cells. Ensure pellet is °C during sonication to prevent overheating.
Centrifuge the lysed cells for 38000x g,4°C to remove insoluble cell debris, and collect supernatant in a bottle 4°C
Perform IMAC to extract target protein from the lysed cell mixture
Dispense 5mL Nickle affinity resin Ni Sepharose 6 FF - Cytiva into a gravity flow column. Equilibrate resin by first rinsing with ~ 10CV distilled water, then ~ 10CV binding buffer to remove the storage solution.
Resuspend the equilibrated resin with some binding buffer and add to the supernatant bottle. Incubate the resin with the supernatant for 0h 30m 0s while rotating or otherwise mixing gently at 4°C
Load the resin/supernatant mix back onto the gravity flow column, retaining the FT separately for SDS-PAGE analysis.
Wash the column with 10CV of wash buffer twice. Allow wash buffer to pass through completely between washes. This is to remove non-specific, weak binding of contaminant proteins from the resin for a cleaner elution.
Collect washes separately for SDS-PAGE analysis.
Elute the protein with 1.5CV of elution buffer.
Repeat step 5.5 once more, collecting a total of 2 separate elution fractions. This is to ensure maximum retrieval of protein from the resin.
The total protein concentration of the elutions are measured by Nanodrop. Although still a mixture, A280 value can give an estimate of the protein content, which will determine how much protease need to be added to remove the affinity tag.
Wash used IMAC resin with 10CV of base buffer, and leave in the column submerged in a small amount of base buffer such that the resin is kept moist.
This washed IMAC resin will later be reused for reverse IMAC (rIMAC)
Run SDS-PAGE of all samples from total lysis supernatant to final elution. Stain gel with protein staining solution Coomasssie Blue and determine which fractions contain the target protein by finding the band corresponding to the target molecular weight.
Elution de-salting, tag cleavage and reverse IMAC
The three elutions are pooled and desalted using HiPrep 26/10 deasalting columns, run on AKTA pure at the maximum flow rate of 10mL/min.
For tag removal, His-SENP1 is added in 1:100 ratio to the total protein content of the desalted sample, as determined by nanodrop. The mixture is left at 4°C
In morning, pour the cleavage mixture over the washed resin and collect FT.
Wash rIMAC resin with 2CVwash buffer to remove any target protein still bound to the resin.
Take samples of the FT and wash, characterise content by SDS-PAGE
(Optional) elute rIMAC resin with 2CV elution buffer to confirm if the protein shows non-specific binding to the resin used.

Purify sample further by size exclusion chromatography.
Concentrate all fractions of the rIMAC containing the target protein in spin concentrators of the appropriate MWCO, to a final volume of under 5mL .
Remove any solid aggregates from the sample by centrifugation at 17200x g,4°C , then immediatly draw up the supernatant with a 5mL syringe and a blunt-tip fill needle, taking care not to disturb the pellet.
Using the AKTA Pure system:
Inject the sample onto a 5mL sample loop.
Run the sample down HiLoad 16/60 Superdex 200 pg gel filtration column at 1mL/min in gel filtration buffer, collecting 1mL aliquots.
From the chromatogram, fraction E1-F8 analyse by SDS-PAGE.
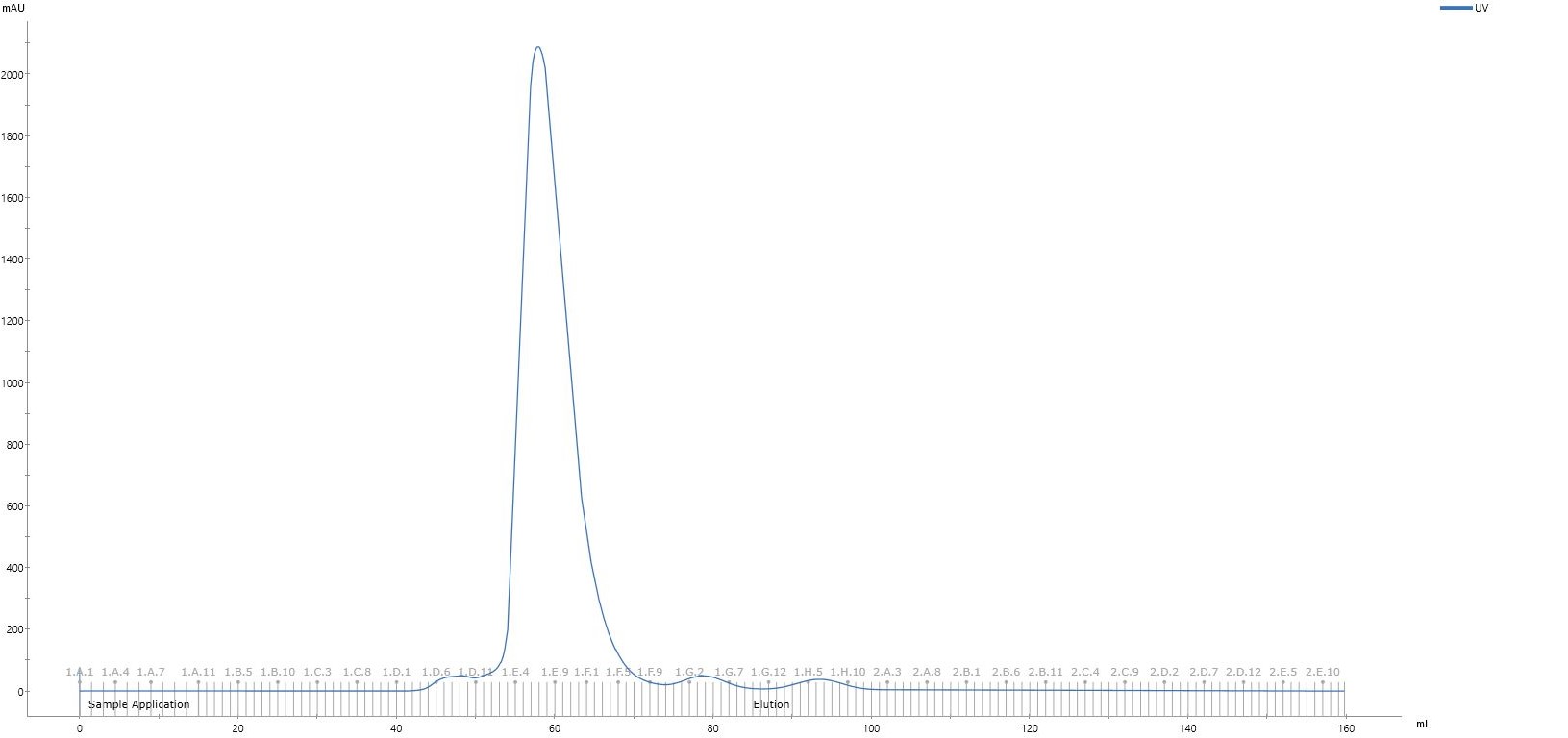
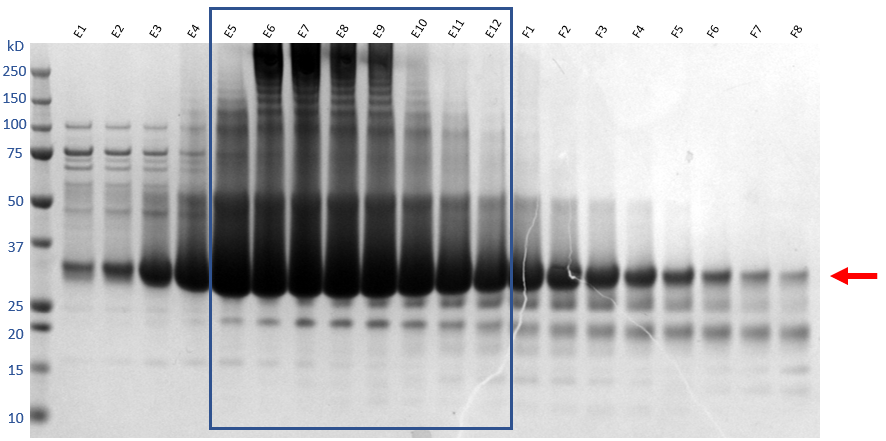
Take the fractions that contain the target protein, which in this case are fraction E1-F6. Concentrate the final sample in Vivaspin 6 10kda MWCO centrifugal concentrator until the concentration reaches >33mg/mL or 1millimolar (mM) .
Take 1µL of the final sample for SDS-PAGE, and another for mass spectroscopy (MS).
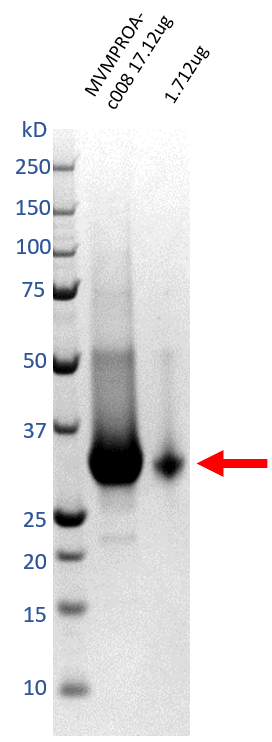
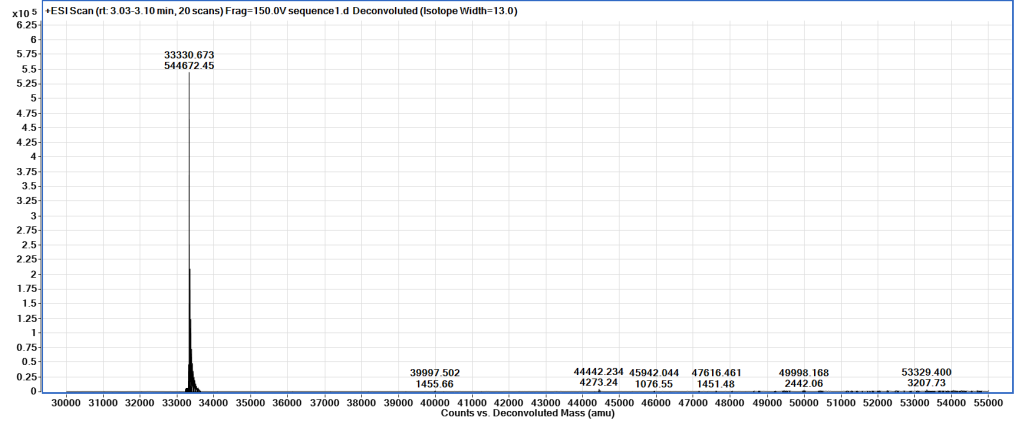
Aliquot into appropriate volumes for future usage to minimise freeze/thaw cycles. Flash-freeze in liquid nitrogen, and store at -80°C until required.

