HILIC-MS analysis of central carbon metabolites in gram negative bacteria
Bashar Amer, Ramu Kakumanu, Edward EK Baidoo
Abstract
Central carbon metabolites are essential for energy metabolism, secondary metabolite and macromolecule biosynthesis in prokaryotes. Phosphorylated intermediates such as sugar phosphates are, however, difficult to resolve with conventional reversed phase (RP) chromatography. In addition, coenzyme A (CoA) compounds and nucleotide cofactors, which are also phosphorylated are difficult to resolve by RP chromatography. To this end, we have developed a hydrophilic interaction liquid chromatography quadrupole time-of-flight mass spectrometry (HILIC-QTOF-MS) method that to quantify these metabolic intermediates and cofactors in Escherichia coli DH1 and Pseudomonas putida K2440.
Attachments
Steps
Sample preparation
Calibration curves: Pure analytical standards were dissolved in 50:50 methanol:water (v/v) to make a stock solution of 100micromolar (µM) concentration. The stock solution was diluted 4-fold to make a standard solution of 25micromolar (µM)(with 50:50 methanol:water, v/v). A seven-point calibration curve was produced via a series of 2-fold serial dilutions (with 50:50 methanol:water, v/v), which were conducted from the 25micromolar (µM) standard solution to a standard solution ending in a concentration of 0.390625micromolar (µM); 2-fold serial dilutions for CoA standards, however, were conducted from a 20micromolar (µM) standard solution to standard solution ending in a concentration of 0.3125micromolar (µM). All solutions were prepared on ice and stored at -20°C until LC-MS data acquisition.
Extraction of intracellular metabolites: Wild type Escherichia coli DH1 ( E. coli ) and Pseudomonas putida K2440 ( P. putida) were grown on 0.5% glucose in LB medium at 37°C. Growth was monitored during mid exponential phase to early stationary phase. E. coli and P. putida reached average optical densities at 600 nm wavelength (OD 600 nm) of 1.67 and 2.16, respectively. 1.5mL of the cell culture was harvested (in triplicate) and transferred to a 2mL centrifuge tube. The cell culture was centrifuged at 14000x g,0h 0m 0s for 2 minutes at room temperature. After removing the supernatant, metabolic activity from the cell pellet was quenched by adding 250 µL of ice-cold methanol and mixing thoroughly by vortexing for 2 minutes at 4°C. 250µL of water was then added to the methanol lysate, mixed by vortexing, and centrifuged at 14000x g,0h 0m 0s for 5 minutes at 4°C. 450µL``,0h 0m 0s of the lysate was filtered via a 3k Da molecular weight cut-off centrifuge filter (Amicon-Ultra, part number UFC500396, MilliPoreSigma, St. Louis, MO, USA) at 13000x g,0h 0m 0s at 4°C, for 60 minutes prior to LC-MS analysis.
Ultra high performance liquid chromatography (UHPLC) conditions
Chromatographic separation was performed via an Agilent Technologies 1290 Infinity II UHPLC system (Table 1). The sample tray and column compartment were set to 5°C and 30°C, respectively. A sample injection volume of 1 µL was used throughout. The UHPLC mobile phases were composed of solvents (A) 10millimolar (mM) ammonium acetate, 0.2% volume ammonium hydroxide, and 5micromolar (µM) medronic acid in water and (B) 10millimolar (mM) ammonium acetate, 0.2% volume ammonium hydroxide, and 5micromolar (µM) medronic acid in 90% volume acetonitrile, and water as the remaining solvent. The mobile phases were degassed by sonication for 5 minutes. The 1290 Infinity II UHPLC system pump was purged with the aforementioned mobile phases for up to 5 minutes (at 50% volumesolvent A and 50% volume solvent B) prior to LC column installation. The analytes were separated on an Agilent Technologies InfinityLab Poroshell 120 HILIC-Z PEEK lined (2.1 mm internal diameter, 100 mm length, and 2.7 µm stationary phase particle size) column, which was connected to an Agilent Technologies InfinityLab Poroshell 120 HILIC-Z guard column (2.1 mm internal diameter and 2.7 µm stationary phase particle size). The LC column was then equilibrated with the starting mobile phase composition at a flow rate of 0.1 mL/min until the system backpressure was stable. The UHPLC gradient is described in table 2.
| A | B | C |
|---|---|---|
| Component | Description | Part number |
| 1260 Infinity II Isocratic Pump | Isocratic pump for internal reference lock mass delivery | G7110B |
| 1290 Infinity II MCT | Column compartment | G7116B |
| 1290 Infinity II Multisampler | Autosampler for automatic sample injection | G7167B |
| 1290 Infinity II High Speed Pump | UHPLC pump | G7120A |
| 6545 Q-TOF LC/MS | Quarupole-time-of-flight mass spectormeter | G6545B |
Table 1. The Agilent Technologies UHPLC-QTOF-MS system.
| A | B | C | D |
|---|---|---|---|
| Time (min) | Percentage of mobile phase B (%B) | Flow rate (mL/min) | Maximum system pressure (Bar) |
| 0 | 90 | 0.250 | 600 |
| 2 | 90 | 0.250 | 600 |
| 11.5 | 61.5 | 0.250 | 600 |
| 11.7 | 60 | 0.330 | 600 |
| 12.7 | 60 | 0.400 | 600 |
| 12.9 | 90 | 0.550 | 600 |
| 14.3 | 90 | 0.550 | 600 |
Table 2. UHPLC gradient. The maximum allowable system backpressure for the column was 600 bar.
QTOF-MS method parameters
The Agilent Technologies Infinity II UHPLC system was coupled to an Agilent Technologies 6545 QTOF-MS system. The LC column effluent was delivered to the Agilent Jet Stream (AJS) ion source . An Agilent Technologies Isocratic Pump was used to deliver the reference mass correction solution. AJS facilitated the production of gas-phase [M - H]- or [M - 2H]2- ions in the negative ion mode. High mass accuracy was achieved via reference mass correction. Reference mass correction was performed with the trifluoroacetic acid (anion) [M - H]- at 112.98559 m/z (from the LC mobile phase) and the trifluoroacetic acid anion adduct of HP-0921 (i.e. hexakis(1H, 1H,3H-tetrafluoropropoxy)phosphazine (C18H18F24N3O6P3,921.23 Da, CAS NO. 58943-98-9) at 1033.98811 m/z . The reference ions were diluted to 2 µM in 80:20 acetonitrile:water (v/v) and delivered to an electrospray ionization sprayer at a flow rate of 5 µL/min via the 1260 Infinity II Isocratic Pump (Table 1). Prior to analysis, the Agilent 6545 QTOF-MS system was tuned via the ESI-L-Low calibration solution for an acquisition range of up to 1700 m/z . MS data acquisition parameters are described in table 3.
| A | B |
|---|---|
| QTOF-MS parameters | Values |
| Acquisition range (m/z) | 70-1100 m/z |
| Acquisition rate (spectra/s) | 0.86 |
| Nebulizer pressure (*Psi) | 20 |
| Drying gas temperature (ºC) | 300 |
| Drying gas flow rate (L/min) | 10 |
| Sheath gas temperature (ºC) | 350 |
| Sheath gas flow (L/min) | 12 |
| Capillary voltage (V) | 3500 |
| Fragmentor (V) | 100 |
| Skimmer (V) | 50 |
| OCT 1 RF Vpp (V) | 300 |
| Nozzle voltage [Expt] (V) | 2000 |
Table 3. QTOF-MS parameters. *Psi is lb/in2.
LC-MS data acquisition and analysis software
LC-MS data acquisition was performed via the Agilent MassHunter Workstation software (version 8). Data processing and analysis were performed via Agilent MassHunter Qualitative Analysis (version 6) and Profinder (version 8) software.
Method validation
![Figure 2. Method validation table. The mass-to-charge ratios shown are theoretical values. The CoA compounds were detected via [M - 2H]2- ions. All other compounds were detected via [M - H]- ions. Average standard and metabolite retention times are used in the last two columns of fig. 2. Phosphoglycerate data represents 2- and 3-phosphoglycerate, which could not be resolved by the method. ATP, NAD, and NADH are abbreviations for adenosine triphosphate, nicotinamide adenine dinucleotide, and nicotinamide adenine dinucleotide, reduced, respectively. Figure 2. Method validation table. The mass-to-charge ratios shown are theoretical values. The CoA compounds were detected via [M - 2H]2- ions. All other compounds were detected via [M - H]- ions. Average standard and metabolite retention times are used in the last two columns of fig. 2. Phosphoglycerate data represents 2- and 3-phosphoglycerate, which could not be resolved by the method. ATP, NAD, and NADH are abbreviations for adenosine triphosphate, nicotinamide adenine dinucleotide, and nicotinamide adenine dinucleotide, reduced, respectively.](https://static.yanyin.tech/literature_test/protocol_io_true/protocols.io.4r3l2opzxv1y/Table_v2.jpg)
Biological data
Conclusions
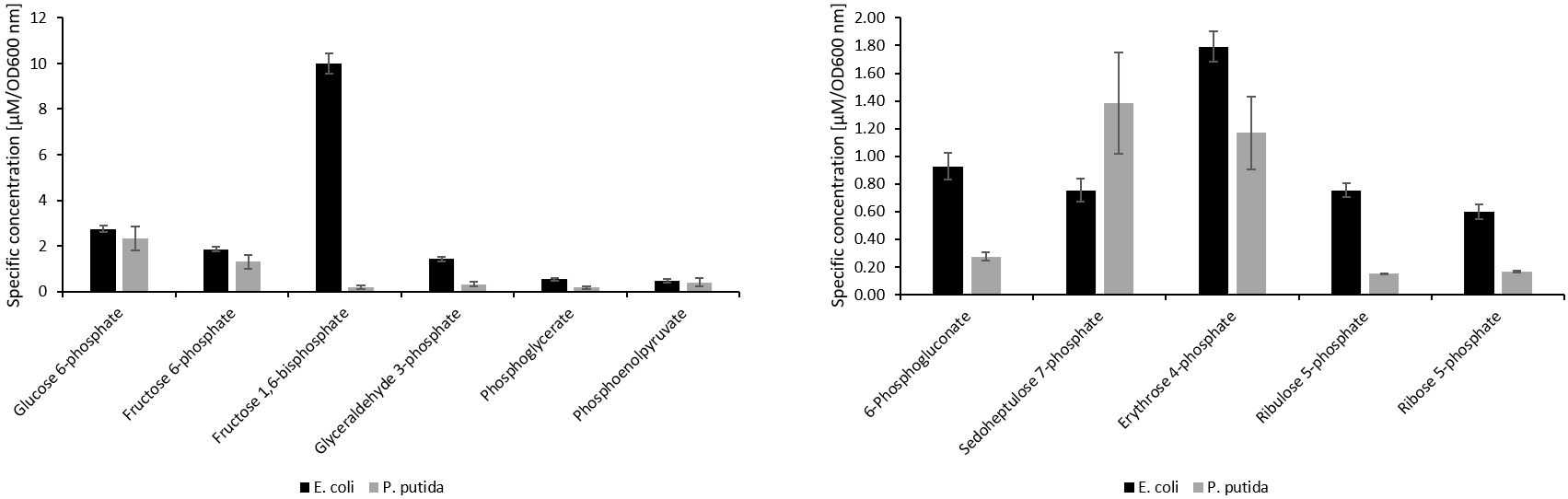
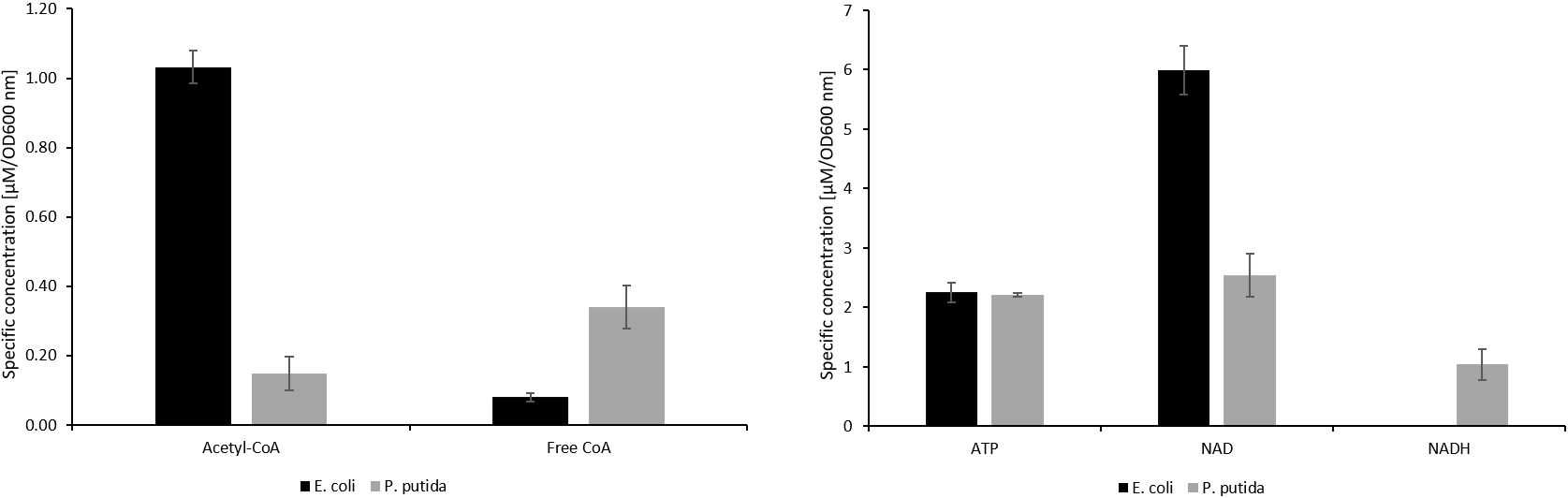
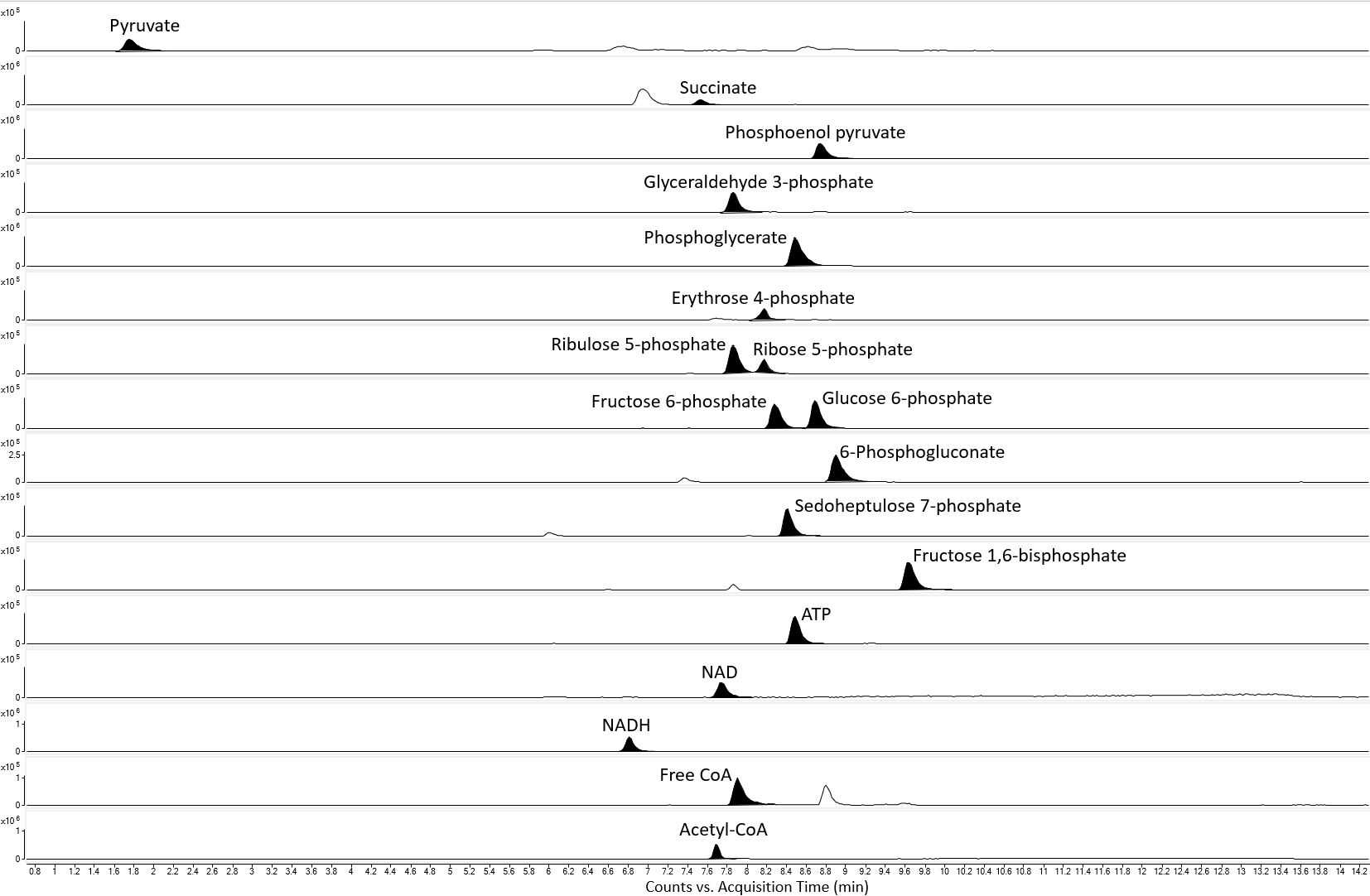
The HILIC-QTOF-MS method was able to separate the analytes tested, except for the isomers 2- and 3-phosphoglycerate (fig. 1). The method showed good linearity for all the analytes tested, with R2 values of ≥0.99 . The limit of detection ranged from 20nanomolar (nM) to 1.67micromolar (µM) for the analytes detected. Retention time repeatability measurements were ≤ 0.49 %RSD for the analytes tested, except for pyruvate, which had a %RSD value of 1.09. There was very little difference between standard and metabolite retention times, suggesting a minimal effect of the sample matrix on retention time stability (fig. 2). The method successfully quantified intracellular sugar phosphate (fig. 3), acetyl-CoA and free CoA (fig. 4), and organic acid metabolites (fig. 5) in E. coli and P. putida metabolite extracts. Additionally, the method was able to quantify the intracellular nucleotide cofactors ATP, NAD, and NADH (fig. 4) in these microbial metabolite extracts. The metabolite concentration data suggests that the method may be applicable for other phosphorylated metabolites and organic acids.