Estimation of Microbial Viability Using Flow Cytometry
Hazel Davey, Hazel Davey, Stéphane Guyot, Stéphane Guyot
Abstract
For microorganisms in particular, viability is a term that is difficult to define and a state consequently difficult to measure. The traditional (and gold standard) usage equates viability and culturability (i.e., the ability to multiply) but the process of determining culturability is often too slow. Flow cytometry provides the opportunity to make rapid and quantitative measurements of dye uptake in large numbers of cells and we can therefore exploit the flow cytometric approach to evaluate so-called viability stains and to develop protocols for more routine assessments of microbial viability. This article provides a commentary and several protocols have been included to ensure that users have a firm basis for attempting these reasonably difficult assays on traditional flow cytometer instruments. What is clear is that each assay must be carefully validated with the particular microorganism of interest before being applied in any research, clinical, or service form. © 2020 The Authors.
INTRODUCTION
One of the most basic questions that a microbiologist might ask of a microorganism is whether it is alive or not, and in microbiology, it is often necessary to determine the number of living (viable) cells in a sample or culture of interest. However, perhaps surprisingly, this is a question that is not always easily answered (Davey, 2011). The gold standard for determining the number of viable microbial cells in a sample is usually achieved by plating a 0.1- to 1-ml sample of cells (diluted as required) onto an agar plate (Hattori, 1988; Postgate, 1969) and scoring as viable (a posteriori) those cells that were able to form visible colonies. The culture viability is then defined as the ratio of these cells to the total cell count in the original sample, which is determined microscopically. However, there are several problems associated with this technique, not the least of which is the length of time required to obtain the results. For example, a plate count actually only indicates how many of the cells can replicate under the conditions provided for growth. In the case of environmental samples, the laboratory media, the temperature, and other factors may differ substantially from those in the original sample; thus, the proportion of cells that can divide and form colonies may be much lower than the number of cells that would score as viable using a more rapid method. Nevertheless, the plate count method has remained the gold standard, in part due to the fact that traditional microscopic analyses of stained cells are time consuming and can lead to operator fatigue; thus, conclusions are normally drawn from the analysis of at best a few hundred cells. Furthermore, microscopic examination is largely a qualitative technique, wherein a judgment of alive or dead is all that is possible, and the interpretation of the extent of a cell's staining may vary among operators. Also, for some slowly growing organisms (e.g., mycobacteria), it may take several weeks to determine how many cells were viable (as defined by the above technique) in the original sample. Even when the sample contains quickly growing organisms and the plates are incubated under optimal growth conditions, a minimum of overnight growth is usually required before the resulting colonies can be counted. In clinical situations and for economic reasons, such a delay is often unacceptable. Thus, many rapid methods have been developed to allow a speedier assessment of the viable microbial load in a sample.
These alternative, rapid viability measurements include a variety of stain-based methods. The so-called vital stains that have been used in attempts to estimate microbial viability fall into three broad categories: (1) some dyes are excluded by intact membranes of viable cells but enter freely into cells where the permeability barrier has been lost; therefore, the presence of the dye within the cell may be expected to be correlated with cell death; (2) other dyes are actively accumulated by viable cells; thus, the stained cells are deemed to be viable; (3) membrane-permeant nonfluorescent precursors can be converted by the activity of intracellular enzymes of living cells to membrane-impermeant fluorescent molecules; again, fluorescent cells are deemed viable. Each of these dye-based approaches is discussed in more detail below.
Flow cytometry offers an alternative method of determining the amount of fluorescent dye taken up by each cell in a population. Since quantitative measurements can be made very rapidly on a large number of individual cells, an accurate picture of the distribution of dye uptake by many thousands of cells is possible within a few minutes. This article begins with a discussion of the various advantages and disadvantages of classical (proliferative) versus cytochemical (dye-based) viability assays. It discusses the three classes of cytochemical methods in greater detail and provides instructions for three protocols. Finally, it discusses the use of cell sorting in conjunction with tests for microbial viability. (For a previous version of this work see Current Protocols article: Davey, Kell, Weichart, & Kaprelyants, 2004.)
THE PROBLEM OF DETERMINING VIABILITY
In classical terms, a microbial cell is generally considered viable if it possesses all the components and mechanisms necessary for sustained proliferation (Greenwood & Peutherer, 1992). According to Postgate (1976): “At present one must accept that the death of a microbe can only be discovered retrospectively: a population is exposed to a recovery medium, incubated, and those individuals which do not divide to form progeny are taken to be dead…. There exist at present no shortcuts which would permit assessment of the moment of death: vital staining, optical effects, leakage of indicator substances and so on are not of general validity…. The term ‘viability’ applies to populations, not individuals (except in an all-or-none sense: an individual is either viable or nonviable).”
The definition of viability (Davey, 2011) of an individual microorganism has evolved over the intervening years to a more nuanced spectrum of physiological states (Table 1). Culturability is evidently best determined by the classical method of assessing cellular proliferation directly and scoring only those cells that have visibly multiplied. However, the underpinning assumption of rapid microbiology is that one can estimate something that might correlate with this by assessing the presence and functionality of individual vital factors and processes. The cytochemical approach gives an insight into the physiology of individual cells by providing data on parameters such as membrane energization and enzyme activity. Both approaches have advantages and disadvantages, as discussed below.
| State | Characteristic | Plate count result | Exclusion stain | Active-accumulation stain | Metabolic activity stain |
|---|---|---|---|---|---|
| Healthy | Capable of division after a lag phase at a rate close to µmax | +++ | +++ | +++ | +++ |
| Viable and culturable but damaged | Capable of division after a lag phase at a rate substantially below μmax | ++ | +++ | +++ | +++ |
| Limited culturability | Capable of a limited number of divisions producing invisible or micro-colonies | +/− | +++ | +++ | +++ |
| Dormant | Capable of division only after resuscitation | +/− | +++ | ++ | ++ |
| Leaky | Membrane is damaged but repairable | +/− | +/− | +/− | +/− |
| Detected activity but culture attempt unsuccessful | Incapable of division but has metabolic activity | − | ++ | ++ | ++ |
| Moribund | Incapable of division, no metabolic activity but appears microscopically intact | − | +/− | − | − |
| Dead | Cell integrity has been lost | − | − | − | − |
The Growth-Based Approach
The classical approach requires an a priori knowledge of the suitable growth media and conditions for the organism or organisms present in the sample, as well as the use of a suitable method of growth detection for the organisms. In practice, because of limited time, materials, and prior knowledge, the most convenient method is all too often chosen. High-nutrient, complex media such as Luria broth and trypticase soy broth (TSB) for bacteria are often used for these procedures to ensure growth. Growth detection is usually measured either as colony-forming units (cfu) on a solid agar plate or as turbidity in liquid media. There are several problems associated with this kind of approach.
Standard growth conditions
Many microorganisms have growth requirements that are very different from the standard conditions applied. For instance, standard conditions usually involve the aerobic incubation of a sample at a higher temperature than that at which it was collected (e.g., ≥30°C may be used even in the case of environmental samples). Many bacteria in the environment have not yet been cultured axenically by any method devised to date although the proportions of cultured and uncultured is a topic of debate (Martiny, 2019; Steen et al., 2019). In many cases where success has eventually been obtained, organisms defied efforts to culture them until some critical component was added to the medium. A well-known example is Legionella (Meyer, 1983).
Changes in physiological state
Microorganisms with known growth requirements may reside in a physiological state in which the (otherwise appropriate) standard culture conditions do not support growth, or do so only for a small fraction of the population, or only after long lag phases (Table 1). However, in using these (or other) definitions and descriptions it should be noted that viability of an individual cell can only be tested once and under a specific set of conditions. As an unsuccessful test for viability may cause further changes to the cell, we cannot know what may have happened had a different approach been tried. Of course, numerous studies have sought to compare populations but culture heterogeneity makes such assays difficult to interpret.
Growth determination method
In some cases, growth and division of viable cells can remain undetected due to the constraints of the growth determination method employed. Organisms displaying slow growth rates or long lag phases may not be capable of producing enough biomass to form visible colonies or detectable turbidity during the period of incubation allowed. In some cases, cessation of growth may occur after a limited number of divisions (Kell, Kaprelyants, Weichart, Harwood, & Barer, 1998) or the organism may be unable to form colonies on solid media. These factors, alone or in combination, may lead to false-negative results.
Thus, the main drawback of classical, growth-based viability assays is the possibility of false-negative results; false positives can be excluded by correct sterile technique.
The Cytochemical Approach
There are occasions in which it is the metabolic activity of the cells that is of concern, that is, whether they are capable of multiplication or not. Clearly a cell whose DNA has been damaged at the origin of replication could not multiply but the rest of its activities would probably be unaffected. If, for example, these activities included the production of a toxin, then a method that detects metabolic activity would be of more interest than one that requires proliferation to score for cellular presence and activity.
Cytochemical assays can have several advantages over proliferation-based assays. They are generally less time consuming, in some cases delivering instantaneous results. They facilitate (at least potentially) a method of measuring something that might be correlated with other measures of viability (such as culturability) in organisms for which suitable growth conditions have not been established. For organisms that display extremely slow growth rates, long lag phases, or low growth yields, proliferation-based methods are often impossible or impractical, and thus the cytochemical approach offers an attractive alternative. In some cases (e.g., flow cytometry), the cytochemical approach allows simultaneous analysis of other traits providing multidimensional snapshots of mixed populations.
However, these rapid assays also often have their own drawbacks that can make them difficult to interpret. There is a fundamental difference in that the cytochemical approach is normally based on single parameters such as membrane energization or intactness, enzyme activity, or the uptake of a substrate. In contrast, the ability to increase in size, replicate DNA, separate chromosomes into progeny and separate the resulting cells relies on many factors. Thus, assays based on a single factor can give rise to false-positive results. Negative control samples should therefore involve samples killed by a range of relevant treatments (e.g., heat, ethanol, chlorine). Additionally, and conceptually more difficult, negative controls produced by a more “natural” cell death such as starvation should be considered. However, in general, data used to demonstrate effectiveness of an assay result from growing cells as positive controls and aggressively killed cells for negative controls. In natural environments, starvation and/or stress may be long term, and the activity of cells may be reduced to extremely low levels (especially in the case of dormant cells), such that positive results might be below the limits of detection of the assay. Similarly, injured cells may have damaged membranes and score as nonviable whereas repair of the damage during cultivation on a rich medium would allow subsequent growth. The apparent paradox is avoided by the use of operational definitions (Barer, Kaprelyants, Weichart, Harwood, & Kell, 1998; Kell et al., 1998) in which viability is not in fact an innate property of a cell but is scored as a result of experimental measurements. Using these definitions, however, it should not be surprising if different experiments lead to different results.
Taken together, the cytochemical viability assays might be less time consuming and more convenient in many cases, but, their use should be predicated on an understanding of what is being measured and the consequences of that for conclusions that can be drawn.
FLUORESCENT STAINS FOR MICROBIAL VIABILITY DETERMINATION BY FLOW CYTOMETRY
Despite the problems associated with fluorescent staining protocols for viability measurements, useful information can be obtained providing one carefully selects and tests an appropriate protocol for the problem under investigation. To illustrate this, this section discusses the three general classes of molecules whose uptake and/or fluorescence may be expected to reflect the viability of the cells (see Table 2) and presents sample procedures for the determination of microbial viability.
| Stain | Mode of action | Excitation sourcea | References |
|---|---|---|---|
| BacLight kit (Thermo Fisher Scientific) | Exclusion of PI and staining with SYTO 9 | Argon (488 nm) | Nasrabadia, An, Kwon, and Hwang (2018); Vanhauteghem et al. (2019) |
| Bis-(1,3-dibutylbarbituric acid) trimethine oxonol: DiBAC4(3) | Uptake by dead cells | Argon (488 nm) | Guyot et al. (2015) |
| ChemChrome B/Y/V6 | Proprietary information | Argon (488 nm) | Helmi, David, Di Martino, Jaffrezic, and Ingrand (2018) |
| 3,3′-Dihexyloxacarbocyanine iodide: DiO6(3) | Membrane potential | Argon (488 nm) | Omajali, Mikheenko, Overton, Merroun, and Macaskie (2019) |
| Fluorescein diacetate (FDA) | Enzymic activity | Argon (488 nm) | Steadman Tyler et al. (2019) |
| Propidium iodide (PI) | Exclusion | Argon (488 nm) or He-Ne (544 nm) | Gudde, Hulce, Largen, and Franke (2019) |
| SYBR Green + PI | Exclusion | Argon (488 nm) | Feng, Wang, Zhang, Shi, and Zhang (2014) |
| SynaptoGreen C4 | Exclusion | Argon (488 nm) | Zahavy et al. (2018) |
- a He-Cd, helium/cadmium; He-Ne, helium/neon. Lamp-based flow cytometers may provide suitable alternative wavelengths.
It must be stressed that the examples given below are provided as a guide only. The large variety of microorganisms that one may wish to study and the different modes of death that may befall them preclude the development of a universal flow cytometric viability test. The most useful advice that can be given to an experimenter is to try a range of stains with sensibly designed control samples (i.e., exposed to the same types of stress that will be present in the experimental samples) and to choose the method that gives the most reliable results. For the reasons outlined above, the magnitude and duration of stress, in addition to the type, should be considered.
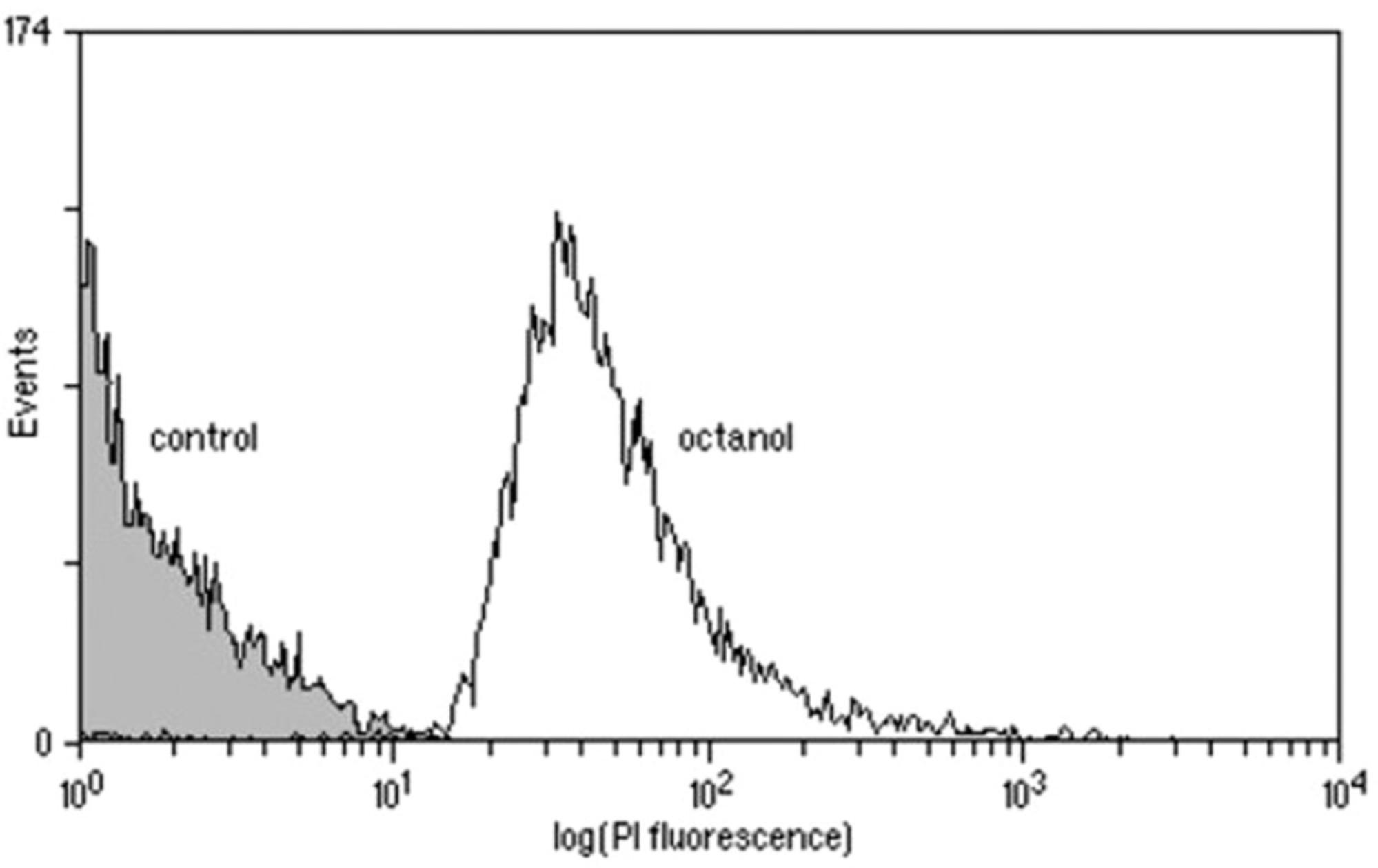
Under ideal conditions, complete separation of viable and dead cells will be achieved (see example in Fig. 1) and the analysis gates can easily be set to encompass live and/or dead cells. Under less favorable conditions, overlap may occur between the control samples on a single-parameter histogram. It may be possible to separate the two populations by using a dual-parameter dot plot of, for example, fluorescence versus forward light scatter or to use more complex gating approaches involving ratios of parameters. If this is not found to be the case, it may be necessary to choose a different staining protocol or to accept a “zone of uncertainty” in the results. In some cases, two or more fluorescent probes may be used. Here, compensation of fluorescence may be required and experimenters should check their emission spectra to minimize overlap.
As a general rule, the use of 0.22-μm (or smaller) filtered water is recommended for all aqueous solutions. Fluorescent reagents should generally be protected from light to avoid photobleaching and buffers should be protected from light to inhibit microbial growth. While advantageous in some assays, the use of microplates for high-throughput sample preparation should generally be avoided due to the delay this introduces in processing, during which time sample viability may change.
Specific details for preparing and storing dyes are given in the examples below. Some fluorescent probes used to measure microbial viability are stored in non-aqueous solutions such as DMSO or ethanol. The stock solution should therefore be highly concentrated to minimize the volume of solvent added to samples before flow cytometric analysis in order to avoid a second stress based on solvent addition.
Dye Exclusion
The exclusion of dye by an intact membrane is probably the most straightforward viability test to understand and perform. Fluorescent stains normally excluded by living cells are used to assess viability on the grounds that dead cells have leaky membranes that are permeable to the stains. Propidium iodide (PI) is generally excluded by intact plasma membranes and its uptake is often used to indicate cell death in eukaryotic and prokaryotic cells. PI is often the dye of choice for viability determinations in animal cells and has a long history of use for this purpose in both flow cytometry (Sasaki, Dumas, & Engleman, 1987) and fluorescence microscopy (Jones & Senft, 1985). It is not surprising therefore, that it was adopted as a viability stain for both yeast (Deere et al., 1998) and bacteria (López-Amorós, Comas, & Vives-Rego, 1995). There is, however, an inherent danger in blindly transferring protocols developed for one cell type to another, particularly when one cell type is eukaryotic and the other is prokaryotic. In the case of ethidium bromide, for instance, efficient efflux pumps capable of removing the dye from Escherichia coli have been demonstrated (Jernaes & Steen, 1994). More recently, the applicability of PI for analysis of environmental bacteria has been questioned (Shi et al., 2007) and the existence of stained but viable yeast in stressed populations has been demonstrated (Davey & Hexley, 2011). Thus, the applicability of excluded dyes for microbial viability determinations needs to be carefully considered for each type of organism and stress condition.
Example 1: Measuring Viability by Dye Exclusion Using TO-PRO-3
TO-PRO-3 is a nucleic acid stain that can be used for viability testing by dye exclusion. This series of dyes has an advantage over many other exclusion dyes in that its fluorescence is enhanced some 1,000-fold on binding to nucleic acids (Rye et al., 1992; Rye, Dabora, Queseda, Mathies, & Glazer, 1993; Rye, Yue, et al., 1993). Undiluted solutions of TO-PRO-3 (as obtained from the manufacturer) should be stored frozen and protected from light. The diluted solution is stable for several weeks when stored in the same manner. This procedure calls for a flow cytometer with a 633-nm helium/neon (He−Ne) laser or 635-nm laser diode as excitation source and a detector set to receive emission above 650 nm.
Staining
Place 999 μl of a cell sample or control, at 105 to 107/ml, into a tube suitable for use with the flow cytometer. Dilute 1 mM TO-PRO-3 (Thermo Fisher Scientific) to 0.1 mM in a buffer suitable for use with the sample of interest. Add 1 μl per sample (final 0.1 μM) and mix gently (e.g., with a pipet). Stain each sample and control immediately before use.
Calibration
Stain a dead control (e.g., ethanol-fixed or heat-treated cells) and analyze on the flow cytometer, adjusting the photomultiplier tube (PMT) voltage where applicable to ensure that the sample fluorescence (at >650 nm) is to the right-hand side of the display. Position an analysis gate to encompass the dead cells. Stain the live control (freshly harvested cells, ≥95% alive) and analyze on the flow cytometer. Position an analysis gate to encompass the live cells. Adjust the gates so that optimum separation of live and dead cells is achieved. If good separation is not achieved, vary the final stain concentration to between 0.05 and 1 μM, alter the PMT voltages where the instrument allows it, and/or alter the staining time according to the nature of the cell sample.
Analysis
Stain a sample and analyze using the settings determined during calibration. Record the percentages of viable and dead cells.
Dye Uptake
The mitochondria of eukaryotic cells have the ability to accumulate lipophilic cations such as rhodamine 123 concentratively and in an uncoupler-sensitive fashion (Chen et al., 1982). Nevertheless, some caution is required in interpreting the fluorescence of intact eukaryotic cells. Ludovico, Sansonetty, and Côrte-Real (2001) observed that dead cells displayed a diffuse fluorescence in the cytoplasm whereas live cells displayed mitochondrial staining together with some slight non-specific fluorescence at the cell envelope. Viable bacteria also accumulate rhodamine 123, while nonviable ones do not (Diaper, Tither, & Edwards, 1992). Under certain conditions, the extent to which individual bacteria take up rhodamine 123 quantitatively reflects the extent of their viability, i.e., whether they are immediately culturable, nonculturable, or dormant (Kaprelyants & Kell, 1992).
An alternative approach is the use of lipophilic anions, which in contrast to cations, bind preferentially to nonviable cells. The lipophilic anion bis-(1,3-dibutylbarbituric acid) trimethine oxonol, or DiBAC4(3), has been shown to enter eukaryotic membranes only if the membranes are de-energized (Wilson & Chused, 1985).
Example 2: Measuring Viability Using a Combination of Dye Exclusion and Dye Uptake
PI is a nucleic acid stain that is normally excluded by the intact membrane of viable cells. Uptake of DiBAC4(3) is dependent on de-energization rather than permeabilization of the cell membrane. Thus, these two dyes can be combined in a single protocol that provides a method for following de-energization as part of a “two-stage death” of microbial cells. Both dyes can be obtained in powder form. Stock solutions prepared as described below appear to be stable for many months when frozen and protected from the light.
It is possible to carry out this procedure on a flow cytometer with a single excitation source at 488 nm. Alternatively, the PI can be excited with a second laser at ∼544 nm (e.g., a green He−Ne laser). In this case, the fluorescence of DiBAC4(3) must be separated from the green scatter signal via temporally/spatially separated illumination of the stream of cells. Emissions of the DiBAC4(3) and PI are detected at ∼525 and >600 nm, respectively.
Staining
PI stock is prepared as an aqueous solution at a concentration of 3.33 mg/ml. DiBAC4(3) stock is prepared at a concentration of 1 mg/ml in acetone. A combined staining solution is prepared by adding 100 μl PI stock and 240 μl DiBAC4(3) to 24.66 ml filtered water. This working solution is combined with an equal volume of cell suspension at a concentration of ∼106 cells/ml. Samples should be incubated 30 min in the dark at room temperature prior to analysis on the flow cytometer.
Calibration
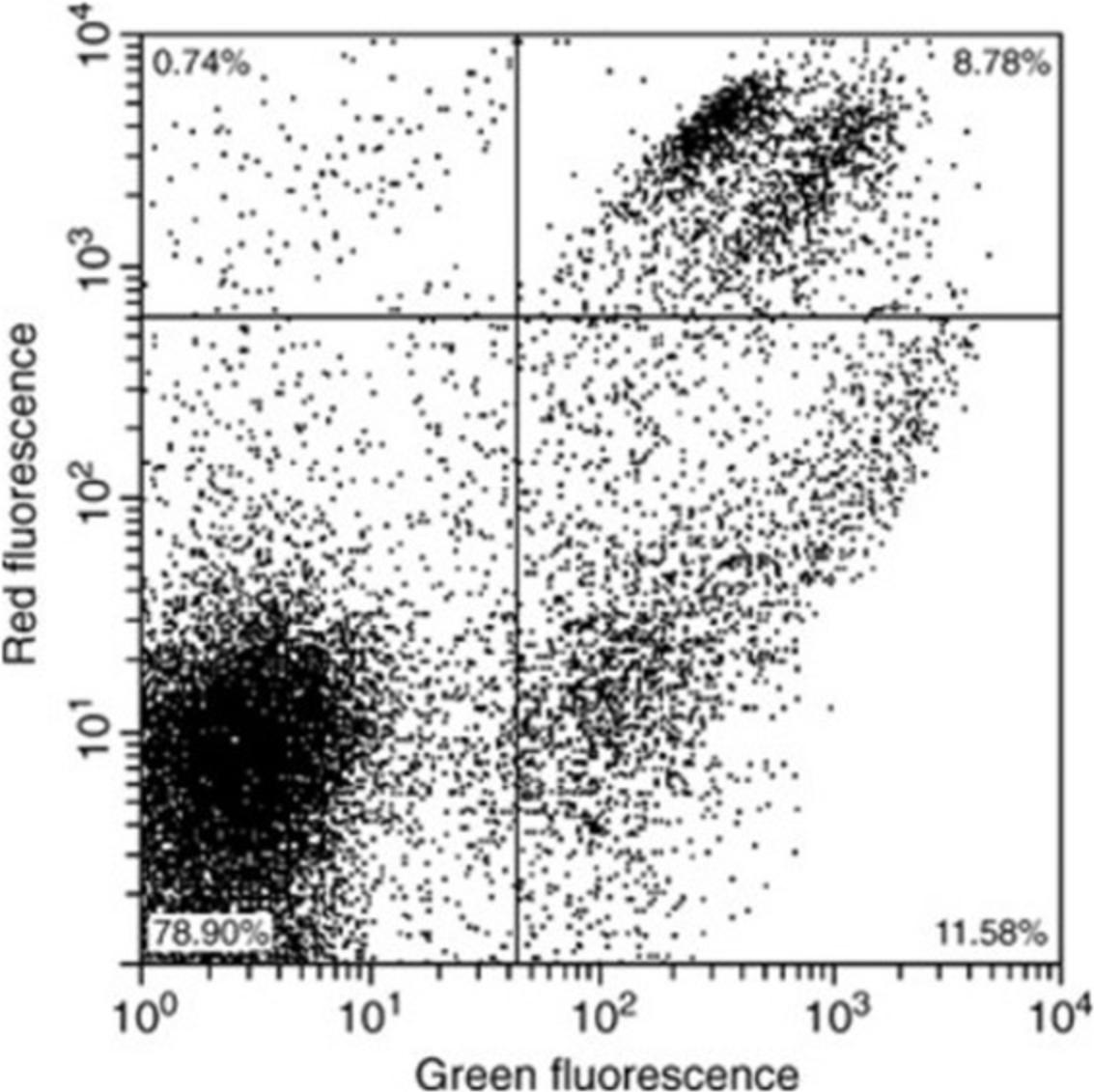
Set up a dual-parameter scatter plot of log red fluorescence (PI) versus log green fluorescence (DiBAC4(3)). If necessary, gate data plotted on this graph via forward and/or side scatter to exclude debris. Stain and analyze live and dead controls (as described in Example 1 above). Adjust PMT settings to give results similar to those shown in Figure 2. If this cannot be achieved, it may be necessary to vary the dye concentrations and/or the staining time according to the nature of the cell sample.
Analysis
Stain a sample and analyze using the settings determined during calibration. Record the percentage or absolute numbers of cells in each quadrant.
Metabolic Activity
In certain circumstances the activity of the cells may be of more interest than their membrane integrity. For this purpose, a third class of viability stains is used in mammalian cell biology, often as a positive marker in a dual-staining protocol with ethidium bromide or PI (Aeschbacher, Reinhardt, & Zbinden, 1986). These stains are themselves nonfluorescent and membrane permeant but are metabolically altered inside the cell to become fluorescent and, under ideal conditions, impermeant. One example is fluorescein diacetate (FDA), which is cleaved by intracellular esterases to produce fluorescein. Dead cells do not stain because they lack enzyme activity and/or the fluorescein diffuses freely through their damaged membranes. Although flow cytometric analyses of mammalian cells with this dye are well established, less success has been evident with microorganisms. Breeuwer, Drocourt, Rombouts, and Abee (1994) and Breeuwer et al. (1995) showed that FDA and carboxyfluorescein diacetate (CFDA) penetrated yeast rapidly but that esterase activity was limiting and an energy-dependent efflux of carboxyfluorescein from viable cells also impacted negatively on the efficacy of the staining method. Success has been reported with 5′-carboxyfluoroscein succidyl ester (CFDA-SE) with Neisseria gonorrhoeae (Thakur, Obradovic, Dillon, Ng, & Wilson, 2019).
Example 3: Measuring Viability by Assessing Metabolic Activity with CFDA
CFDA is cleaved by intracellular enzymes to produce fluorescent carboxyfluorescein. In using this approach, the experimenter must be aware that the operational definition of viability is different: It relates to (immediate) enzyme activity rather than membrane health. This procedure requires a flow cytometer with a 488-nm argon laser or other suitable excitation source and a detector set to collect emission at ∼525 nm.
Staining
For cell samples, use 106 cells/ml suspended gram-positive bacteria in 50 mM KH2PO4, pH 7.4 or gram-negative bacteria in TE buffer. For dead controls, use ethanol-fixed or heat-treated cells; for live controls, use freshly harvested cells, ≥95% alive. Place 990 μl of cell samples and controls into tubes suitable for use with the flow cytometer. Add 10 μl of 1 mM CFDA in dimethyl sulfoxide (Thermo Fisher Scientific; final 10 μM) to each and mix gently (e.g., with a pipet). Incubate for 30 min at the normal growth temperature of the microorganism.
Calibration
Analyze the dead control on the flow cytometer, adjusting the PMT voltage as necessary to ensure that the sample fluorescence (at ∼525 nm) is to the left-hand side of the display. Position an analysis gate to encompass the dead cells. Analyze the live control and position an analysis gate to encompass the live cells. Note that with CFDA, live cells are brightly fluorescent and dead cells are nonfluorescent or weakly fluorescent. Adjust the gates so that optimum separation of live and dead cells is achieved. If good separation is not achieved, adjust the stain concentration, alter the PMT voltages, and/or alter staining time according to the nature of the cell sample.
Analysis
Analyze cell samples using the settings determined during calibration. Record the percentages of viable and dead cells.
Commercial Kits
A variety of kits have been produced specifically for the measurement of viability of specific types of organisms. For example, the LIVE/DEAD Bac Light Bacterial Viability Kit (developed by Molecular Probes and now available via Thermo Fisher Scientific) gives a two-color viability assessment of both gram-negative and gram-positive bacteria, where live cells are labeled green with SYTO 9 and dead cells are labeled red with PI. However, as freely admitted by the manufacturer, “under certain conditions, bacteria having compromised membranes may recover and reproduce, even though such bacteria may be scored as ‘dead’ in this assay” (https://static.yanyin.tech/literature/current_protocol/10.1002/cpcy.72/original_pdf/cpcy.72.pdf). Despite this, the kit is widely used in microbiology for a wide range of species. The growing use of such kits reflects, at least in part, their ease of use. In the case of the Bac Light kit, the reagents are simultaneously added to the bacterial suspension, which is then incubated for a few minutes. The sample is then analyzed without washing, so “live” and “dead” bacteria can be distinguished and quantitated rapidly. There is a great danger that because of the name the uninitiated may use these tests blindly (despite the manufacturer's warnings) without checking the reliability of the dyes with the organisms and conditions used in their experiments. By their nature, a detailed protocol is provided by the manufacturers of such kits so it is considered unnecessary to provide one here.
Overall Principles and Recommendations
When selecting a stain for a particular application there are several factors that need consideration. Some, such as the extinction coefficient, quantum yield, and photostability, are of general applicability to flow cytometric fluorescence measurements and are discussed in detail elsewhere (Davey & Kell, 1996; Shapiro, 2003). The absence of overlap between the emission spectrum of the fluorescent probe and the autofluorescence of the cells and availability of a light source of appropriate wavelength are also important; a list of viability stains that are compatible with common flow cytometric light sources is shown in Table 2.
One factor that is of particular relevance in the measurement of viability is the toxicity of the stain and any solvent in which it is prepared (e.g., DMSO, ethanol, acetone). Protocols used to assess viability should clearly not perturb viability and using concentrated solutions of stain can avoid addition of excess solvent, although appropriate controls are of course required.
When the appropriate stain and excitation source have been selected, it is important to perform a series of experiments to determine the optimum concentration of the stain and the optimum length of time between addition of stain and subsequent analysis. The optimum concentration will inevitably be a compromise between a high one (for maximum signal) and a low one (for specificity). It may be necessary to measure and adjust the cell concentration to ensure that stain uptake is not limiting. In this case the use of a flow cytometer that allows determination of absolute cell numbers is an ideal approach.
The simultaneous use of at least two fluorescent probes can add reliability or enhance the information obtained from an assay (Buysschaert, Byloos, Leys, Van Houdt, & Boon, 2016). This approach requires the investigator first to make sure (1) that the various fluorescent signals emitted are each predominantly detected by a single photomultiplier tube and (2) make a minimal impact on the signal received by other detectors. Secondly, the probes should be used simultaneously so as to optimize the signal measured by each detector. This step may require performing compensation when two emission spectra are superimposed.
USE OF CELL SORTING IN VIABILITY STUDIES
While flow cytometric analysis allows the investigator to perform a rapid and quantitative version of experiments that could otherwise be performed by fluorescent microscopy, flow cytometric cell sorting allows the process to be taken one very important step further. With flow cytometric analysis one can simply say that the distribution of dye uptake is correlated with a plate count of the same sample. However, providing that the staining protocol does not affect the viability of the cells (which may be determined by plate counts of stained and unstained samples), one can exploit the sorting capability of appropriate instruments to separate cells from the purported viable and nonviable fractions of the histogram. This allows determination of the culturability of exactly those cells whose cytological properties have already been determined directly (Kaprelyants, Mukamolova, Davey, & Kell, 1996).
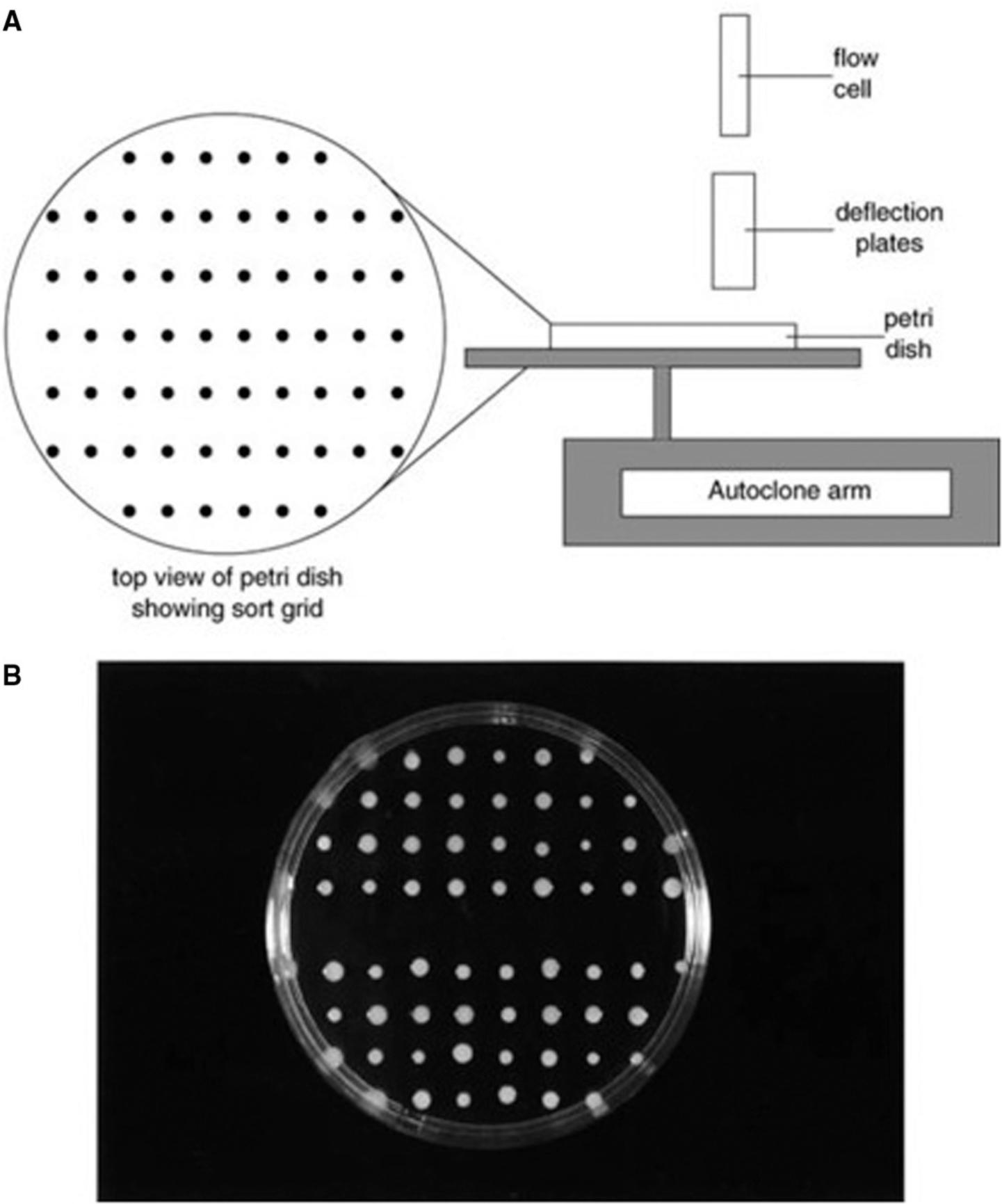
Some cell sorters have a motorized stage designed for the collection of cells into the wells of microtiter plates; it is also possible to modify the stage and collection protocol to allow microbial cells to be collected directly onto agar plates (Fig. 3). Thus, an event from a tightly defined region on a histogram can be correlated directly to the growth (or absence of growth) of a colony on an agar plate. Although the sorting approach offers many advantages, it is important to also be aware of its potential pitfalls. The sorting step, in most instruments, leads to a high level of cell dilution that causes practical problems for subsequent growth-based analysis. As microbial cells often form aggregates during growth or sample preparation. If one cell in an aggregate has a leaky membrane, it will take up an exclusion dye such as PI and the group will collectively score as dead in the viability assay. However, if this group also contains one or more live cells, a colony will result when the clump is sorted onto an agar plate. This problem can be overcome to some extent by using the forward scatter signal as an indicator of size. However, because the size of microbial cells, even within a single species, can vary greatly with growth conditions, a more robust approach may be to use two or more viability stains to give a broader picture of the physiological status of the cells. An additional problem may arise with damaged cells. The process of flow cytometric analysis followed by sorting onto an agar plate may in its own right be considered an additional stress that may convert an injured cell into a nonviable one. Such stresses can be quantified to some extent by plating injured cells before and after sample preparation. The effect, if any, of the sheath fluid on injured cells should also be determined.
CONCLUSIONS
The rapid cytological estimation of true microbial viability is extremely difficult (if not impossible in principle), not least because of the problems in defining viability in microbial cells. Although the flow cytometric approach has much to offer for the determination of microbial viability, it must be emphasized that no single stain nor even cocktail is likely to be a universal indicator of viability.
A cell that is killed by exposure to environmental extremes such as heat, pH, and so forth is likely to be very different from a cell that is killed by exposure to an antibiotic or other chemical, and different again from a cell that dies (loses culturability) owing to a lack of nutrients in its environment. Thus, the flow cytometric properties of a cell and the distribution of dye uptake within a population will depend on how the cells die and more generally on their entire physiological state and its history.
In conclusion, although there are as yet no perfect stains, careful protocol development currently allows valuable information to be obtained regarding specific problems. In the case of organisms that have not been exposed to excessive stress (e.g., laboratory cultures under normal conditions and in some cases clinical samples), substantial progress is being made towards the rapid and routine flow cytometric assessment of microbial viability or vitality.
LITERATURE CITED
- Aeschbacher, M., Reinhardt, C. A., & Zbinden, G. (1986). A rapid cell membrane permeability test using fluorescent dyes and flow cytometry. Cell Biology and Toxicology , 2, 247–255. doi: 10.1007/BF00122693.
- Barer, M. R., Kaprelyants, A. S., Weichart, D. H., Harwood, C. R., & Kell, D. B. (1998). Microbial stress and culturability: Conceptual and operational domains. Microbiology , 144, 2009–2010. doi: 10.1099/00221287-144-8-2009.
- Breeuwer, P., Drocourt, J. L., Rombouts, F. M., & Abee, T. (1994). Energy-dependent, carrier-mediated extrusion of carboxyfluorescein from Saccharomyces cerevisiae allows rapid assessment of cell viability by flow cytometry. Applied and Environmental Microbiology , 60, 1467–1472. doi: 10.1128/AEM.60.5.1467-1472.1994.
- Breeuwer, P., Drocourt, J. L., Bunschoten, N., Zwietering, M. H., Rombouts, F. M., & Abee, T. (1995). Characterization of uptake and hydrolysis of fluorescein diacetate and carboxyfluorescein diacetate by intracellular esterases in Saccharomyces cerevisiae , which result in accumulation of fluorescent product. Applied and Environmental Microbiology , 61, 1614–1619. doi: 10.1128/AEM.61.4.1614-1619.1995.
- Buysschaert, B., Byloos, B., Leys, N., Van Houdt, R., & Boon, N. (2016). Reevaluating multicolor flow cytometry to assess microbial viability. Applied Microbiology and Biotechnology , 100(21), 9037–9051. doi: 10.1007/s00253-016-7837-5.
- Chen, L. B., Summerhayes, I. C., Johnson, L. V., Walsh, M. L., Bernal, S. D., & Lampidis, T. J. (1982). Probing mitochondria in living cells with rhodamine 123. Cold Spring Harbor Symposia on Quantitative Biology , 46, 141–155. doi: 10.1101/SQB.1982.046.01.018.
- Davey, H. M. (2011). Life, death, and in-between: Meanings and methods in microbiology. Applied and Environmental Microbiology , 77, 5571–5576. doi: 10.1128/AEM.00744-11.
- Davey, H. M., & Hexley, P. (2011). Red but not dead? Membranes of stressed Saccharomyces cerevisiae are permeable to propidium iodide. Environmental Microbiology , 13, 163–171. doi: 10.1111/j.1462-2920.2010.02317.x.
- Davey, H. M., & Kell, D. B. (1996). Flow cytometry and cell sorting of heterogeneous microbial populations—The importance of single-cell analyses. Microbiological Reviews , 60, 641–696. doi: 10.1128/MMBR.60.4.641-696.1996.
- Davey, H. M., Kell, D. B., Weichart, D. H., & Kaprelyants, A. S. (2004). Estimation of microbial viability using flow cytometry. Current Protocols in Cytometry , 29(1), 11.3.1–11.3.21. doi: 10.1002/0471142956.cy1103s29.
- Deere, D., Shen, J., Vesey, G., Bell, P., Bissinger, P., & Veal, D. (1998). Flow cytometry and cell sorting for yeast viability assessment and cell selection. Yeast , 14, 147–160. doi: 10.1002/(SICI)1097-0061(19980130)14:2<147::AID-YEA207>3.0.CO;2-L.
- Diaper, J. P., Tither, K., & Edwards, C. (1992). Rapid assessment of bacterial viability by flow cytometry. Applied Microbiology and Biotechnology , 38, 268–272. doi: 10.1007/BF00174481.
- Feng, J., Wang, T., Zhang, S., Shi, W., & Zhang, Y. (2014). An optimized SYBR Green I/PI assay for rapid viability assessment and antibiotic susceptibility testing for Borrelia burgdorferi. Plos One , 9, e111809. doi: 10.1371/journal.pone.0111809.
- D. R. S. Greenwood, & J. Peutherer (eds.). (1992). Medical microbiology. London: Churchill Livingston.
- Gudde, L. R., Hulce, M., Largen, A. H., & Franke, J. D. (2019). Sterol synthesis is essential for viability in the planctomycete bacterium Gemmata obscuriglobus. FEMS Microbiology Letters , 366(3), p.fnz019. doi: 10.1093/femsle/fnz019.
- Guyot, S., Gervais, P., Young, M., Winckler, P., Dumont, J., & Davey, H. M. (2015). Surviving the heat: Heterogeneity of response in Saccharomyces cerevisiae provides insight into thermal damage to the membrane. Environmental Microbiology , 17(8), 2982–2992. doi: 10.1111/1462-2920.12866.
- Hattori, T. (1988). The viable count: Quantitative and environmental aspects. Berlin: Springer-Verlag.
- Helmi, K., David, F., Di Martino, P., Jaffrezic, M., & Ingrand, V. (2018). Assessment of flow cytometry for microbial water quality monitoring in cooling tower water and oxidizing biocide treatment efficiency. Journal of Microbiological Methods , 152, 201–209. doi: 10.1016/j.mimet.2018.06.009.
- Jernaes, M. W., & Steen, H. B. (1994). Staining of Escherichia coli for flow cytometry: Influx and efflux of ethidium bromide. Cytometry , 17, 302–309. doi: 10.1002/cyto.990170405.
- Jones, K. H., & Senft, J. A. (1985). An improved method to determine cell viability by simultaneous staining with fluorescein diacetate-propidium iodide. Journal of Histochemistry and Cytochemistry , 33, 77–79. doi: 10.1177/33.1.2578146.
- Kaprelyants, A. S., & Kell, D. B. (1992). Rapid assessment of bacterial viability and vitality using rhodamine 123 and flow cytometry. Journal of Applied Bacteriology , 72, 410–422. doi: 10.1111/j.1365-2672.1992.tb01854.x.
- Kaprelyants, A. S., Mukamolova, G. V., Davey, H. M., & Kell, D. B. (1996). Quantitative analysis of the physiological heterogeneity within starved cultures of Micrococcus luteus using flow cytometry and cell sorting. Applied and Environmental Microbiology , 62, 1311–1316. doi: 10.1128/AEM.62.4.1311-1316.1996.
- Kell, D. B., Kaprelyants, A. S., Weichart, D. H., Harwood, C. L., & Barer, M. R. (1998). Viability and activity in readily culturable bacteria: A review and discussion of the practical issues. Antonie Van Leeuwenhoek , 73, 169–187. doi: 10.1023/A:1000664013047.
- López-Amorós, R., Comas, J., & Vives-Rego, J. (1995). Flow cytometric assessment of Escherichia coli and Salmonella typhimurium starvation-survival in seawater using rhodamine 123, propidium iodide, and oxonol. Applied and Environmental Microbiology , 61, 2521–2526. doi: 10.1128/AEM.61.7.2521-2526.1995.
- Ludovico, P., Sansonetty, F., & Côrte-Real, M. (2001). Assessment of mitochondrial membrane potential in yeast cell populations by flow cytometry. Microbiology , 147, 3335–3343. doi: 10.1099/00221287-147-12-3335.
- Martiny, A. C. (2019). High proportions of bacteria are culturable across major biomes. ISME Journal , 4, 2125–2128. doi: 10.1038/s41396-019-0410-3.
- Meyer, R. D. (1983). Legionella infections—A review of 5 years of research. Reviews of Infectious Diseases , 5, 258–278. doi: 10.1093/clinids/5.2.258.
- Nasrabadia, A. M., An, S., Kwon, S., & Hwang, J. (2018). Investigation of live and dead status of airborne bacteria using UVAPS with LIVE/DEAD® BacLight Kit. Journal of Aerosol Science , 115, 181–189. doi: 10.1016/j.jaerosci.2017.10.012.
- Omajali, J. B., Mikheenko, I. P., Overton, T. W., Merroun, M. L., & Macaskie, L. E. (2019). Probing the viability of palladium-challenged bacterial cells using flow cytometry. Journal of Chemical Technology and Biotechnology , 94, 295–301. doi: 10.1002/jctb.5775.
- Postgate, J. R. (1969). Viable counts and viability. Methods in Microbiology , 1, 611–628. doi: 10.1016/S0580-9517(08)70149-1.
- Postgate, J. R. (1976). Death in microbes and macrobes. In T. R. G. Gray & J. R. Postgate (Eds.), The survival of vegetative microbes (pp. 1–19). Cambridge: Cambridge University Press.
- Rye, H. S., Yue, S., Wemmer, D. E., Queseda, M. A., Haugland, R. P., Mathies, R. A., & Glazer, A. N. (1992). Stable fluorescent complexes of double-stranded DNA with bis-intercalating asymmetric cyanine dyes: Properties and applications. Nucleic Acids Research , 20, 2803–2812. doi: 10.1093/nar/20.11.2803.
- Rye, H. S., Dabora, J. M., Queseda, M. A., Mathies, R. A., & Glazer, A. N. (1993). Fluorometric assay using dimeric dyes for double- and single-stranded DNA and RNA with picogram sensitivity. Analytical Biochemistry , 208, 144–150. doi: 10.1006/abio.1993.1020.
- Rye, H. S., Yue, S., Quesada, M. A., Haugland, R. P., Mathies, R. A., & Glazer, A. N. (1993). Picogram detection of stable dye-DNA intercalation complexes with two-color laser-excited confocal fluorescence gel scanner. Methods in Enzymology , 217, 414–431. doi: 10.1016/0076-6879(93)17080-O.
- Sasaki, D. T., Dumas, S. E., & Engleman, E. G. (1987). Discrimination of viable and non-viable cells using propidium iodide in two color immunofluorescence. Cytometry , 8, 413–420. doi: 10.1002/cyto.990080411.
- Shapiro, H. M. (2003). Practical flow cytometry ( 4th ed.). Hoboken, NJ: John Wiley & Sons.
- Shi, L., Gunther, S., Hubschmann, T., Wick, L. Y., Harms, H., & Muller, S. (2007). Limits of propidium iodide as a cell viability indicator for environmental bacteria. Cytometry , 71A, 592–598. doi: 10.1002/cyto.a.20402.
- Steadman Tyler, C. R., Sanders, C. K., Erickson, R. S., Dale, T., Twary, S. N., & Marrone, B. L. (2019). Functional and phenotypic flow cytometry characterization of Picochlorum soloecismus. Algal Research , 43, 101614. doi: 10.1016/j.algal.2019.101614.
- Steen, A. D., Crits-Christoph, A., Carini, P., DeAngelis, K. M., Fierer, N., Lloyd, K. G., & Cameron Thrash, J. (2019). High proportions of bacteria and archaea across most biomes remain uncultured. ISME Journal , 13(12), 3126–3130. doi: 10.1038/s41396-019-0484-y.
- Thakur, S. D., Obradovic, M., Dillon, J. R., Ng, S. H., & Wilson, H. L. (2019). Development of flow cytometry based adherence assay for Neisseria gonorrhoeae using 5′-carboxyfluorosceinsuccidyl ester. BMC Microbiology , 19, 67. doi: 10.1186/s12866-019-1438-2.
- Vanhauteghem, D., Audenaert, K., Demeyere, K., Hoogendoorn, F., Janssens, G. P., & Meyer, E. (2019). Flow cytometry, a powerful novel tool to rapidly assess bacterial viability in metal working fluids: Proof-of-principle. PLoS One , 14(2), e0211583. doi: 10.1371/journal.pone.0211583.
- Wilson, H. A., & Chused, T. M. (1985). Lymphocyte membrane potential and Ca2+ sensitive potassium channels described by oxonol dye fluorescence measurements. Journal of Cellular Physiology , 125, 72–81. doi: 10.1002/jcp.1041250110.
- Zahavy, E., Rotem, S., Gur, D., Aloni-Grinstein, R., Aftalion, M., & Ber, R. (2018). Rapid antibiotic susceptibility determination for Yersinia pestis using flow cytometry spectral intensity ratio (SIR) fluorescence analysis. Journal of Fluorescence , 28(5), 1151–1161. doi: 10.1007/s10895-018-2279-3.
KEY REFERENCES
- Buysschaert et al. (2016). See above.
Optimization of staining protocols for flow cytometry applications to assess microbial viability.
- Davey (2011). See above.
Provides definitions of microbial viability and direction in the choice and interpretation of viability testing methods.
- Shapiro, H. M. (2003). See above. Retrieved from https://www.mybeckman.uk/resources/reading-material/ebooks/practical-flow-cytometry?country=GB&language=en-GB (Accessed January 2020).
Classical reference that describes the science and wide range of applications of flow cytometry.
Citing Literature
Number of times cited according to CrossRef: 24
- E. P. Efremova, O. M. Zemlyanko, G. A. Zhouravleva, Application of Flow Cytometry for Viability Assessment of Mutants for Translation Termination Factors in the Yeast Saccharomyces cerevisiae, Микробиология, 10.31857/S0026365624020268, 93 , 2, (239-243), (2024).
- E. P. Efremova, O. M. Zemlyanko, G. A. Zhouravleva, Application of Flow Cytometry for Viability Assay of Mutants for Translation Termination Factors in the Yeast Saccharomyces cerevisiae, Microbiology, 10.1134/S0026261723604244, 93 , 2, (236-239), (2024).
- Christina Thompson, Connor Waldron, Sierra George, Zhiming Ouyang, Assessment of the hypothetical protein BB0616 in the murine infection of Borrelia burgdorferi , Infection and Immunity, 10.1128/iai.00090-24, 92 , 6, (2024).
- Daniel McGuire, Telma Costa, Ana Tenreiro, Joana Cruz, Rui Sousa, Miguel Leão de Sousa, Carmo Martins, Francisco Pinto, Margarida Gama-Carvalho, Rogério Tenreiro, Leonor Cruz, Use of immuno-flow cytometry and real-time PCR disclose the epidemiological behaviour of Erwinia amylovora populations during the winter in Portuguese pear orchards, Journal of Plant Pathology, 10.1007/s42161-023-01561-4, 106 , 3, (937-951), (2024).
- Elodie Hoch, Romain Briandet, Bernard Hezard, Adrienne Lintz, Valérie Stahl, Lysiane Omhover-Fougy, Assessment of Viability of Listeria monocytogenes by Flow Cytometry, Foodborne Bacterial Pathogens, 10.1007/978-1-0716-4100-2_7, (105-122), (2024).
- Emre İlpars, Štěpánka Titlová, Katarína Hanzalíková, Ivana Křížová, Tomáš Brányik, Alcohol-Free Beer Produced Using Maltose-Negative Wine Yeast Saccharomyces cerevisiae with Probiotic Potential, Fermentation, 10.3390/fermentation9090805, 9 , 9, (805), (2023).
- Michael J. Fairhurst, Jochem N. A. Vink, Julie R. Deslippe, Monica L. Gerth, A Method for Quantifying Phytophthora Oospore Viability Using Fluorescent Dyes and Automated Image Analysis , PhytoFrontiers™, 10.1094/PHYTOFR-11-22-0140-TA, 3 , 3, (497-505), (2023).
- Eun-Sil Yu, Byoung-Hoon Kang, Myeong-Su Ahn, Jik Han Jung, Ji-Ho Park, Ki-Hun Jeong, Highly Efficient On-Chip Photothermal Cell Lysis for Nucleic Acid Extraction Using Localized Plasmonic Heating of Strongly Absorbing Au Nanoislands, ACS Applied Materials & Interfaces, 10.1021/acsami.3c01856, 15 , 29, (34323-34331), (2023).
- Ilija Djekić, Branko Velebit, Branimir Pavlić, Predrag Putnik, Daniela Šojić Merkulov, Anica Bebek Markovinović, Danijela Bursać Kovačević, Food Quality 4.0: Sustainable Food Manufacturing for the Twenty-First Century, Food Engineering Reviews, 10.1007/s12393-023-09354-2, 15 , 4, (577-608), (2023).
- Kieu The Loan Trinh, Nae Yoon Lee, Recent Methods for the Viability Assessment of Bacterial Pathogens: Advances, Challenges, and Future Perspectives, Pathogens, 10.3390/pathogens11091057, 11 , 9, (1057), (2022).
- Himanshu Kakkar, Nalini Chaudhary, Devashish Mehta, Varsha Saini, Shallu Maheshwari, Jitender Singh, Preeti Walia, Avinash Bajaj, N-methyl Benzimidazole Tethered Cholic Acid Amphiphiles Can Eradicate S. aureus-Mediated Biofilms and Wound Infections, Molecules, 10.3390/molecules27113501, 27 , 11, (3501), (2022).
- Hamzah Al-madani, Hui Du, Junlie Yao, Hao Peng, Chenyang Yao, Bo Jiang, Aiguo Wu, Fang Yang, Living Sample Viability Measurement Methods from Traditional Assays to Nanomotion, Biosensors, 10.3390/bios12070453, 12 , 7, (453), (2022).
- Verónica A. El Mujtar, Fernando Chirdo, Antonio Lagares, Luis Wall, Pablo Tittonell, Soil bacterial biodiversity characterization by flow cytometry: The bottleneck of cell extraction from soil, Methods in Ecology and Evolution, 10.1111/2041-210X.13876, 13 , 7, (1388-1401), (2022).
- Anca Manuela Mitchell, Valentina Gogulancea, Wendy Smith, Anil Wipat, Irina Dana Ofiţeru, Recombinant Protein Production with Escherichia coli in Glucose and Glycerol Limited Chemostats, Applied Microbiology, 10.3390/applmicrobiol1020018, 1 , 2, (239-254), (2021).
- Takahiro Sawada, Masayuki Katayama, Shogo Takatani, Yoshiyuki Ohiro, Early detection of drug-resistant Streptococcus pneumoniae and Haemophilus influenzae by quantitative flow cytometry, Scientific Reports, 10.1038/s41598-021-82186-4, 11 , 1, (2021).
- Sheena M. Feist, Richard F. Lance, Genetic detection of freshwater harmful algal blooms: A review focused on the use of environmental DNA (eDNA) in Microcystis aeruginosa and Prymnesium parvum, Harmful Algae, 10.1016/j.hal.2021.102124, 110 , (102124), (2021).
- Jossana Pereira de Sousa Guedes, Evandro Leite de Souza, Evaluation of Physiological Characteristics of Bacterial Cells in Foods and Water with Flow Cytometry, Detection and Enumeration of Bacteria, Yeast, Viruses, and Protozoan in Foods and Freshwater, 10.1007/978-1-0716-1932-2_3, (17-26), (2021).
- Jossana Pereira de Sousa Guedes, Evandro Leite de Souza, Enumeration of Viable Cells of Bacteria in Food and Water with Flow Cytometry, Detection and Enumeration of Bacteria, Yeast, Viruses, and Protozoan in Foods and Freshwater, 10.1007/978-1-0716-1932-2_2, (9-16), (2021).
- Joy P. Dunkers, Hariharan Iyer, Brynna Jones, Charles H. Camp, Stephan J. Stranick, Nancy J. Lin, Toward absolute viability measurements for bacteria, Journal of Biophotonics, 10.1002/jbio.202100175, 14 , 12, (2021).
- Nur Nasyifa Mohd Maidin, Muhamad Ramdzan Buyong, Ruslinda A. Rahim, Mohd Ambri Mohamed, Dielectrophoresis applications in biomedical field and future perspectives in biomedical technology, ELECTROPHORESIS, 10.1002/elps.202100043, 42 , 20, (2033-2059), (2021).
- V. Sekova, E. Bobrova, E. Isakova, Yu. Deryabina, The Antioxidant Enzymes Activity From the Poly-extromophilic Yarrowia lipolytica Yeast Under Oxidative Stress During Long-lasting Cultivation, Bulletin of Science and Practice, 10.33619/2414-2948/61/02, 6 , 12, (23-35), (2020).
- M. V. Roshchina, O. V. Gunar, Alternative Microbiological Methods for Drug Quality Testing and their Implementation in Pharmacy Practice, Pharmaceutical Chemistry Journal, 10.1007/s11094-020-02283-y, 54 , 8, (834-837), (2020).
- Senem Kamiloglu, Gulce Sari, Tugba Ozdal, Esra Capanoglu, Guidelines for cell viability assays, Food Frontiers, 10.1002/fft2.44, 1 , 3, (332-349), (2020).
- Sierra George, Connor Waldron, Christina Thompson, Zhiming Ouyang, Analysis of bb0556 Expression and Its Role During Borrelia burgdorferi Mammalian Infection, Molecular Microbiology, 10.1111/mmi.15319.

