Purification of human cortex excitatory neuron nuclei from fetal and postnatal tissue using fluorescent activated nuclei sorting (FANS) in combination with a SATB2 antibody.
Barry Chioza, Joe Burrage, Jonathan Mill, Emma L Dempster, Jonathan P Davies, Stefania S S Policicchio, Gina Commin
FANS
post-mortem brain
nuclei
flow cytometry
anti-NeuN
nuclei sorting
anti-SATB2
fetal
neurodevelopment
FACS
neuron
development
Abstract
Increased understanding of the functional complexity of the genome has led to growing recognition about the role of epigenetic/transcriptional variation in health and disease. Current analyses of the human brain, however, are limited by the use of “bulk” tissue, comprising a heterogeneous mix of different neural cell types. Because epigenetic processes play a critical role in determining cell type-specific patterns of gene regulation it is important to consider cellular composition in regulatory genomic studies of human post-mortem tissue, and there is a need for methods to purify populations of specific cell-types. This protocol builds on a previous protocol that uses fluorescence-activated nuclei sorting (FANS) to isolate and profile nuclei from multiple different human brain cell-types from frozen post-mortem tissue. Because NeuN is not an optimal marker for neuronal nuclei from fetal cortex, we have optimized a method using a SATB2 antibody to purify nuclei from excitatory neurons in both fetal and postnatal cortex. Purified populations of nuclei are amenable to simultaneous profiling of i) DNA modifications (via bisulfite sequencing / array), ii) histone modifications (via CUT&Tag), iii) open chromatin analysis (via ATAC-seq), and iv) gene expression (via RNA-seq).
Steps
Nuclear prep for FACS separation (using SATB2 and Hoechst)
The protocol below yields approximately 660,000 SATB2+ve, 800,000 SATB2-ve nuclei per 200mg of frozen human fetal post-mortem cortex tissue. Recovery might vary from sample to sample due to high inter-sample variability (developmental stage and density brain collection, cortex sub-areas, fat content of tissue sectioned, and white to grey matter ratio). Yield will also vary for NeuN against developmental age, and we have shown NeuN is not suitable in pre-natal samples, but can be used in post-natal and adult cortex to collect a comparable population.
Refer to Materials- Table 1 for details about the equipment required and to Materials- Table 2 for specifications of reagents required.
Solution and buffer preps
- Lysis Buffer (LB)
- Sucrose Solution (SS)
- Staining Buffer (SB)
Solutions should be kept at 4°C or 4On ice. Refer to Materials- Table 3 for recipes of solutions and buffers.
Nuclei isolation
- Pre-cool the ultracentrifuge, including the rotor and swing buckets, to
4°C0h 30m 0sbefore starting this stage of the protocol.2. All buffers and the Dounce homogenisers should be pre-cooled on ice.4. Add DTT (1millimolar (mM)) to the SS and LB (i.e.17µL3M DTT per 50 mL of SS/LB)5. Transfer2mLLB to the homogeniser per200mghuman brain tissue6. Add the dissected tissue sample into the homogeniser7. Wait0h 5m 0sbefore douncing the tissue to allow the sample to defrostTo reduce heat caused by friction, the Dounce homogenisation step should be performed on ice with gentle strokes, and care should be taken to avoid foaming.NoteUsing the "TIGHT" pestle helps reduce the number of strokes required to reach full tissue disruption.
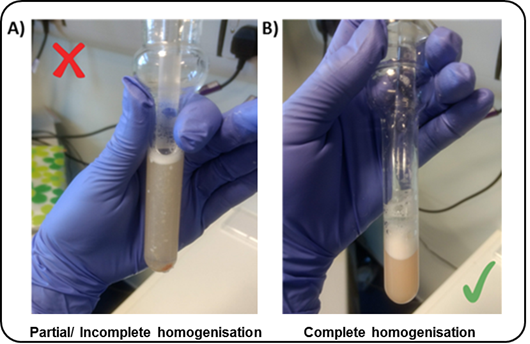
8.Transfer8mL SS (1.8M) to PA thin-walled ultracentrifuge tubes
9.Carefully overlay with tissue homogenate (1 mL per tube) - using a P1000 pipette, releasing slowly down the side of the tube
10.Overlay with another 1mL LB – do not worry about disrupting the homogenate phase
11.Balance opposite tubes by weight with 1x PBS using a fine microbalance
12.Perform ultracentrifugation for 0h 45m 0s(see Table 4 for centrifuge specification and conditions)
After Ultracentrifugation step
- Pour off any the supernatant, taking care not to dislodge the pellet (90-degree inclination of the tube). If the pellet is hard to see, it is okay to leave 100-200 µL solution in the ultracentrifuge tube
- Re-suspend pellet in SB (
750µL), gently pipette up and down - Let samples sit on ice for
0h 15m 0s( Blocking step ) - Transfer volume into 2 mL tubes
- Rinse out ultracentrifuge tubes in order to maximise nuclei collection by adding
750µLof SB per tube, pipetting up and down several times, and transferring into the 2 mL tubes - Centrifuge step: for
0h 5m 0s,4°C - Discard supernatant (pipetting off, or pouring off gently)
- Re-suspend each nuclei pellet in fresh SB (500 µL)
- If the sample was split then pool together pellets from the same sample (Final Volume =
1mL) - Add DNA dye (Hoechst, 1:1000) and mix thoroughly via inversion.
- Pipette out
100µLof nuclei solution for the Unstained Control (Hoechst dye only) and transfer to a new 2 mL tube - Bring the volume up to
500µLfor the Unstained tube with fresh SB - Replace the 100µL taken from the Stained" tube with 100 µL of fresh SB (Final Volume =
1mL``1mL)
Immunostaining
-
Add your antibody of choice (Ab) to staining tube:
SATB2 Alexa488 (1:1000)
or
NeuN Alexa488 (1:1000)
Refer to Table 5 Table 5 for specifications of the antibodies used
-
Incubate tubes for
1h 30m 0son the rotor (speed=14 max) at4°C, keeping the tubes in the dark -
Washing step:
1000x g,0h 0m 0sfor0h 5m 0s,4°C(both "Stained" and "Unstained" tubes) -
Discard supernatant (by pipetting off)
-
Re-suspend in fresh SB (
1mLfor the Unstained tube,1.5-2mLfor the Stained tube - depending on pellet size)
Fluorescence-Activated Nuclei Sorting (FANS)
For machine start-up, CST and Accudrop calibrations refer to BD FACSAria III User’s Guide for guidance and troubleshooting. The following instructions describe FANS using BD FACSAria III. Other FACS platforms can be used but might require modifications to the protocol.
General Gating Parameters For each sample, load stained and unstained tubes individually for data acquisition. A preliminary qualitative analysis of the acquired data is essential to select the appropriate gating strategy to maximize the nuclei capture while excluding unnecessary debris and to ensure optimal signal/noise ratio.
Gating Parameters (X-axis:Y-axis):
FSC-A:SSC-A (Size, cell granularity or internal complexity)
SSC-W:SSC-A (to gate out doublets)
FSC-A:DAPI-A (to gate the single nuclei population)
DAPI-A:FITC-A (to gate SATB2 or NeuN stained nuclei)
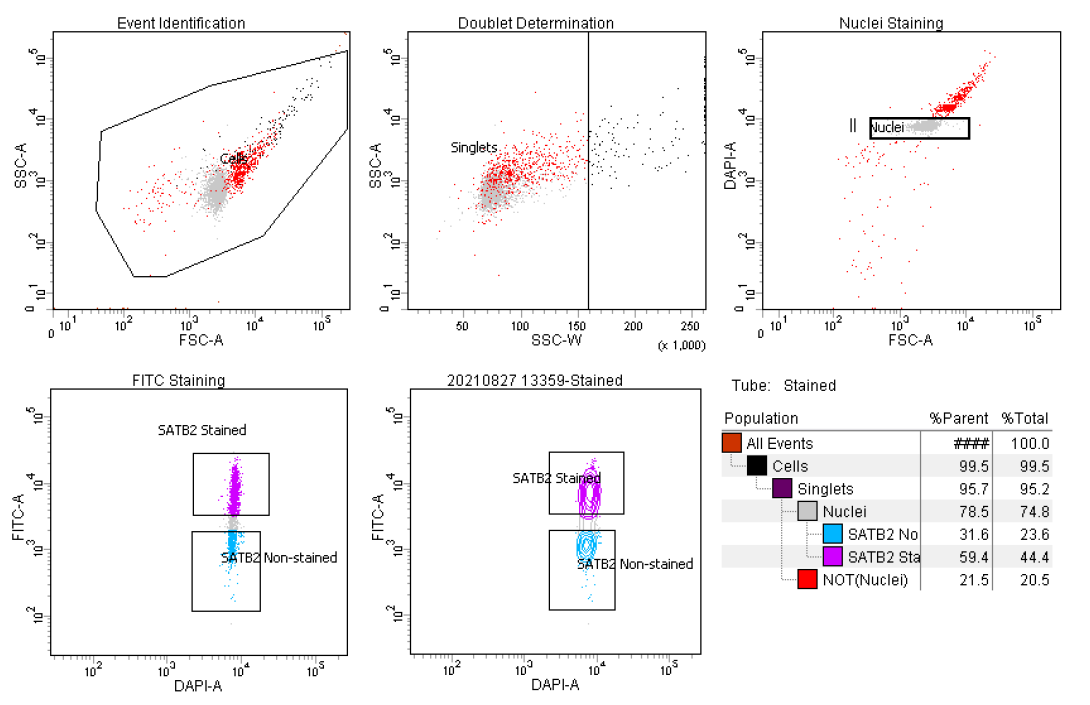
Data recording settings
In line with the experiment design, FSC, SSC, DAPI, FITC, are the parameters for which voltage values may need to be slightly adjusted due to experiment/ inter-sample variability.
It is advisable to set the threshold value at 200 during data recording. Moreover, in the acquisition dashboard tab, we recommend setting Events to Record ≤3000, Event to display ≤1000 and Flow Rate = 1.0 (1,000 events per second) in order to increase the accuracy of signal detection.
The flow rate can be increased during sample collection to reduce the sort speed (ideally max events per second =1,500 for a 100-micron nozzle). However higher flow rates impact the data resolution and accuracy of events detection, and subsequent sorting of cellular fractions (see BD FACSAria III User’s Guide for details).
During analysis, recorded data is displayed in plots, while gates are used to define populations of interest for selection. Figure 3 shows a representative example of the two most common outcomes we often observe.
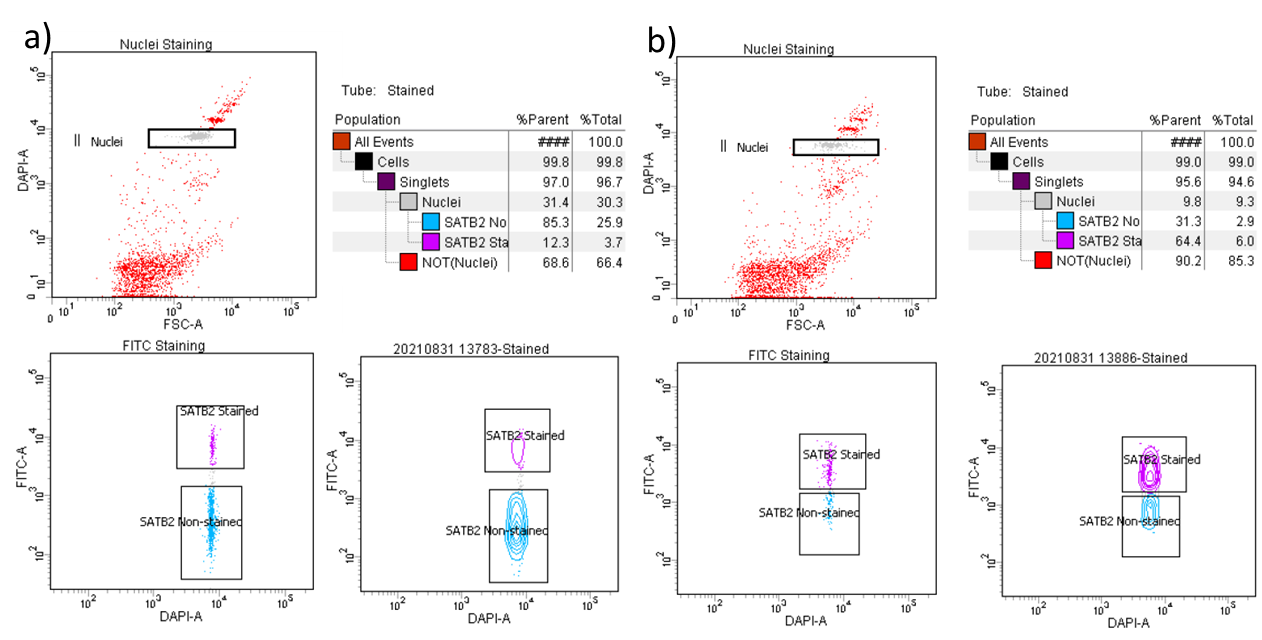
Sample Collection
-
- LoBind Tubes (Eppendorf, Cat No:30108051) are required to collect nuclei (to maximize sample recovery of nucleic acids by significantly reducing sample–to–surface binding).
-
- Collected fractions can be used directly for downstream applications (e.g. DNA/RNA extraction, chromatin shearing).
-
- If you are collecting for DNA or RNA, as soon as the number of events desired is reached, transfer the tubes
On ice, do not hold them atRoom temperature.NoteDuring collection, it is crucial to regularly pause the sorting to mix the two phases in order to preserve the integrity of resulting RNA preparations.
- If you are collecting for DNA or RNA, as soon as the number of events desired is reached, transfer the tubes
-
Keep samples
-80On iceand in the dark for the entire duration of the sorting. -
Lightly vortex sample tubes to make the mixture homogeneous (not clumped) before loading the tube into the FACS chamber.
-
Load the UNSTAINED control tube into the chamber first to postion Unstained gating boxes.
-
Proceed by collecting STAINED tube by simultaneously sorting for SATB2+ve (positive) or SATB2-ve(negative) and proceed with nuclei collection.
1000x g,4°C,0h 0m 0s for 0h 5m 0s
-
Carefully remove supernatant.
-
Add
100µLBAMBANKER to the tube. -
Gently resuspend.
-
Store in
-80°C
General Recommendations for the user
For every new experiment we recommend performing the following steps:
When loading your tube into the FACS machine, run the unstained / IgG control sample first as this aids in setting the baseline parameters
Check your event rate in the Acquisition Dashboard window. If it is greater than 1500 evt/s turn down the “ flow rate ” or unload and dilute the sample further. If less than 100 evt/s, turn up the “ flow rate ” (don’t exceed a flow rate of 5.0 if possible, as the instrument is less focused and more inaccurate at higher flow rates)
In the Acquisition Dashboard window choose the appropriate “ stopping gate ” and “ storage gate ” (when working with nuclei, set as “Nuclei” and “All events” respectively)
Choose the range of “ events to record ” and “ events to display ” that best suits your purpose (≥ 5000 for both is advised)
Under the “threshold” tab in the Cytometer window, change the threshold (should be set for FSC) so that any small events in the bottom corner of the FSC vs SSC graph (caused by general cell debris and dust) are no longer shown. The threshold should not be set too high so that it causes an arbitrary, artificial cut-off through the left side of your population but not so low that small events caused by debris/dust are visible (ideally between a threshold 200-500).
Under the “parameters” tab in the Cytometer window, adjust the “ FSC ” and “ SSC” values to get your population sitting in the centre of the FSC vs SSC graph (a re-adjustment of the “ threshold ” may be required at this point). It is essential to select “restart ” each time any of the parameters are changed to update the events being displayed to ensure only events are recorded under the new settings.
Adjust or draw a new gate in the FSC vs SSC plot to encompass the population of interest.
Look in the scatter graph of SSC-A vs SSC-W (if you opened a blank experiment you will need to draw one). Right-click on the graph and check it is only displaying the events encompassed by your previous FSC vs SSC gate. Adjust or draw a gate for SSC A vs SSC W to encompass all of the main population to the left of the graph and exclude outliers to the right (these are doublets and other cell debris clumps)
Under the “parameters” tab in the Cytometer window adjust parameters for the fluorochromes selected so the unstained / IgG control sample sits close to 0 for the fluorochrome on a graph of FSC vs fluorochrome.
Load the stained samples and check the stained population has a clear increase in signal for the fluorochrome in comparison to the unstained (signal generally should not exceed 10x4). Several minor re-adjustments of the fluorochrome’s “parameters" may be necessary for the stained sample at this stage. If so, the unstained / IgG control has to be reset and re-recorded.
-
Select the correct option for the collection device in the Sort Layout window ( we recommend “ 4-Way Purity ” for general collection)
-
Regularly check your “ Efficiency ” in the Sort Layout window value. Between 80-100% is ideal, 70% is acceptable if less than 70% either the sample is too concentrated or you are sorting a rare population. Although the “ flow rate ” in the Acquisition Dashboard window can be increased to make the sort quicker, faster flow rates will negatively impact the efficiency.
-
Check the “Electronic abort rate” (N° errors /sec) and “Electronic abort count” (Tot N° of errors) at the bottom of the Acquisition Dashboard window. These parameters measure potential miss-sorts (different from efficiency as efficiency measures undetermined drops which are directed to the “ Waste ” and therefore lost but do not contaminate). “Electronic abort rate” should be <1% of total events per second.
-
For long sorts, gate positions should be regularly monitored, especially for stained populations as fluorochromes lose intensity over time and the population can shift towards the unstained. Gates can be moved during long sorts to compensate.
