Protocol for coarse grained simulation of protein ligand system using GROMACS
Vidya Niranjan, Akshay Uttarkar, M Purushotham Rao
Abstract
Coarse-grained (CG) simulations are a powerful tool for studying the behavior of biomolecular systems. They are becoming increasingly important tools for drug discovery, as they can be used to study a wide variety of systems over long timescales.
CG simulations are faster than all-atom MD simulations, which allows researchers to study larger systems over longer timescales. They can also be used to study systems that are too large or too complex to be studied with all-atom MD simulations. Additionally, CG simulations can be used to study systems that are difficult or impossible to study experimentally. CG simulations are typically 100-1000 times faster than all-atom MD simulations.
This protocol provides steps along with a video tutorial perform CG simulation for protein ligand system. The advantages and test cases are but not limited to identification of active site, detection of cryptic pockets on protein, competitive binding between two or more ligands and many more
The link the video tutorial is available in https://youtu.be/xjfbA1G3PIM
Before start
A basic understanding on gromacs and simulations.
For visual assistance refer to https://youtu.be/xjfbA1G3PIM
PART-1
The tutorial on Protocol for the development of coarse-grained structures for macro molecular simulation using GROMACS is available at https://protocols.io/view/protocol-for-the-development-of-coarse-grained-str-cp64vrgw.html and visual assistance for the same at https://youtu.be/QMR4f4eRSbs
Steps
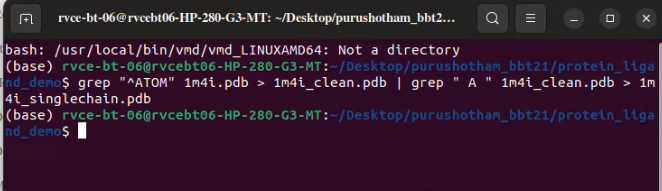
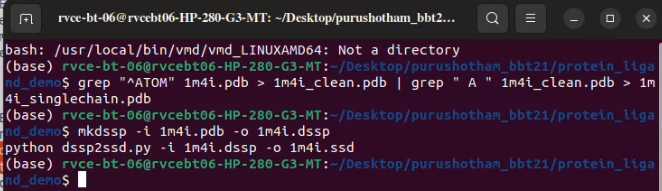
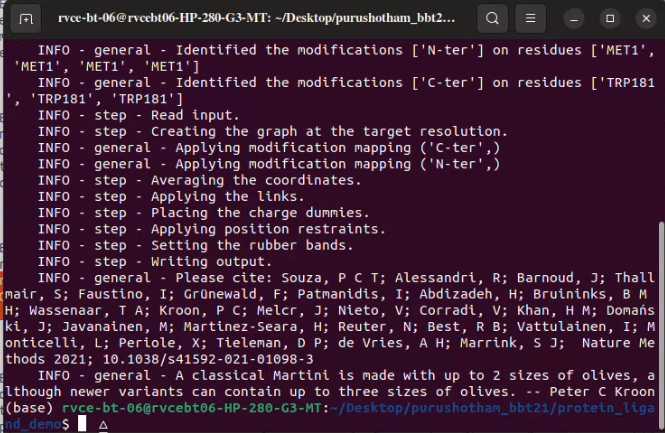
Martinize: Convert All atomic model of AAC2 to Coarse grain model
martinize2 -f 1m4i_singlechain.pdb -o 1m4i_ONLY.top -x 1m4i_CG.pdb -ss CCCCTTCCEEEEGGGCCHHHHHHHHHHHHHHTTTCCCHHHHHHTCSSEEEEEEETTEEEEEEEEEEEEEEETTEEEEEEEEEEEEECGGGTTSSHHHHHHHHHHHHHHHHCSEEEEECCTTTHHHHHHTTCEECCSCEEEEETTEEEECGGGTTTEEEEESSCCCCTTSCEEEECCSSCCC -p backbone -ff martini3001 -elastic -ef 500.0 -el 0.5 -eu 0.8 -scfix -cy auto
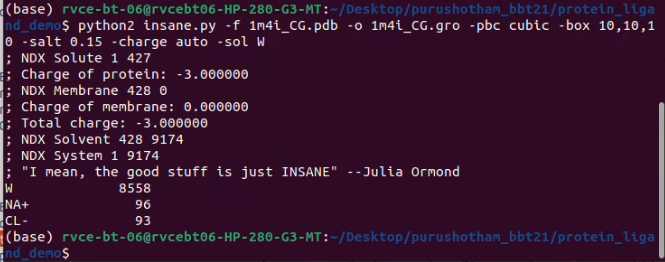
Add water and ions using Insane.py script
python2 insane.py -f 1m4i_CG.pdb -o 1m4i_CG.gro -pbc cubic -box 10,10,10 -salt 0.15 -charge auto -sol W
copy the number of water and ions (without signs) to Topology file (1m4i_ONLY.top), also add required itp files and rename it to 1m4i.top
Insert Ligand molecule
Ligand has been parameterised and included for simulation
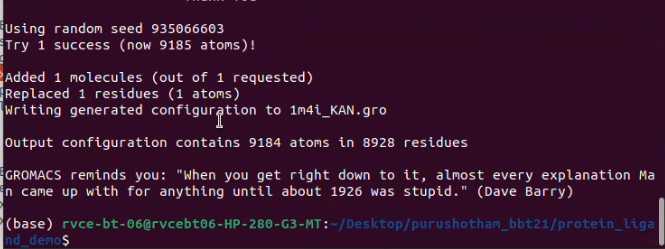
gmx insert-molecules -f 1m4i_CG.gro -nmol 1 -ci KAN.gro -o 1m4i_KAN.gro -replace
replace water by pressing the water selection. After successfull execution, change the number of water molecules and add kanamycin in molecules section and itp file of kanamycin in the topology file.