Multicolour flow cytometry protocol for dogs
Takanori Kitamura, Maciej Parys
Abstract
Although immunotherapy is becoming a standard approach of human cancer treatment, only a minor fraction of patients responds to the therapy. It is therefore required to determine the sub-populations of patients who will respond to immunotherapies along with developing novel strategies to improve efficacy of anti-tumor immune reactions. Development of novel immunotherapies is currently heavily relying on mouse models of cancer. These models are important for better understanding of mechanisms behind tumor immune escape and investigation of novel strategies to overcome it. Nevertheless, the murine models do not necessarily represent the complexity of spontaneously occurring cancers in humans. Dogs spontaneously develop a wide range of cancer types with an intact immune system under similar environment and exposure to humans, which can serve as translational models in cancer immunotherapy research. To date though, there is still relatively limited amount of information regarding immune cell profiles in canine cancers. One possible reason could be the lack of established method to isolate and simultaneously detect different immune cell types in neoplastic tissues. Here we describe a protocol for multi-color flow cytometry to distinguish immune cell types in blood, lymph nodes, and neoplastic tissues from dogs with cancer. Our results demonstrate that a 9-color flow cytometry panel enables to characterize different cell subpopulations including myeloid cells. We also show that the panel allows to detect minor/aberrant subsets within a mixed population of cells in various neoplastic samples including blood, lymph node and solid tumors. To our knowledge, this is the first simultaneous immune cell detection panel applicable for solid tumors in dogs. This multi-color flow cytometry panel has the potential to inform future basic research focusing on immune cell functions in translational canine cancer models.
Steps
Sample collection and processing- blood and lymph node
Blood: Collect 1-2 mL of peripheral blood from the vein of dogs into tubes including EDTA.
LN: Collect fine-needle aspirate, using a 23G needle from the enlarged LN and place it into EDTA tubes with RPMI 1640 supplemented with 10% FBS.
The yield and number of aspirates depend on lymph node size and the operator skills. In our hospital, the usual yield is between 5x106to 1x107 , and each animal has two to three lymph node aspirates performed.
- Place the tubes on ice or at 4ºC until further processing.
Transfer the samples to 15 mL tubes and add 5 mL of 1x RBC lysis buffer
Incubate the tubes at room temperature for 10 minutes.
Add 5 mL of Staining Buffer (PBS including 2% BSA).
Centrifuge the tubes at 350 xg for 5 minutes.
Discard supernatant and resuspend the pellet with 5 mL of Staining Buffer.
Centrifuge the tubes at 350 xg for 5 minutes.
Discard supernatant and resuspend the pellet with 500 mL of Staining Buffer.
Filter the cell suspension through 5 mL round-bottom polystyrene test tubes with cell strainer.
Take 10 uL of cell suspension, mix with 10 uL of trypan blue, and count the cell number.
Adjust the cell density to 2.5x106 cells/mL by Staining Buffer.
- The tubes including cell suspension should be kept on ice or at 4ºC.
Sample collection and processing - tumour samples
Transfer surgically dissected tissues into tubes including 2-4 mL of cold MACS tissue storage solution (Miltenyi).
- Place the tubes on ice or at 4ºC until further processing.
Prepare enzyme mix solution (Tumor Dissociation Kit, Miltenyi) by adding the following components into tissue dissociation tubes (e.g., gentleMACS-C tube); 4.7 mL DMEM, 200 mL Enzyme-H, 20 mL Enzyme-R, and 25 mL Enzyme-A and add it to the minced tumour sample
-
DMEM should not contain Fetal Bovine Serum
-
Although some epitopes can be affected by the dissociation process, there was no indication that enzymes affect the epitopes targeted by antibodies used in this panel. Moreover, these targets are stable during enzymatic treatment according to the manufacturer’s data. In case of modifications to the panel, however, a list of affected epitopes provided by the manufacturer should be referred.
Dissociate the tissue using a dissociator (e.g., gentleMACS Dissociator, h_tumor_01).
Incubate the tubes at 37ºC for 30 minutes under continuous rotation.
Dissociate the tissue by a dissociator.
Incubate the tubes at 37ºC for 30 minutes under continuous rotation.
Dissociate the tissue by a dissociator.
- Tissue dissociation condition depends on tumor type, thus monitoring of the dissociation level is recommended. In many cases, a single dissociation following 30 minutes incubation may be sufficient to dissociate the tumor completely.
Resuspend the samples and filter the cell suspensions via 40mm cell strainer placed on 50mL tubes.
Centrifuge the tubes at 500 xg for 5 minutes.
Filter the cell suspension through 5 mL round-bottom polystyrene test tubes with cell strainer.
Take 10 uL of cell suspension, mix with 10 uL of trypan blue, and count the cell number.
Adjust the cell density to 2.5x106 live cells/mL by Staining Buffer.
- Keep the tubes including cell suspension on ice or at 4ºC.
Sample staining
Aliquot 100 mL of cell suspension to 1.5 mL tubes (2.5x105 live cells/tube)
- At least two tubes (one stained one unstained) are needed.
Add 1 uL of Fc receptor binding inhibitor antibody (eBioscience) to each tube.
Incubate the cells on ice for 30 minutes
Add fluorochrome-conjugated antibodies to each tube.
Antibodies required for this flow cytometry panel, along with the recommended volume of use are listed in Materials section.
Incubate the cells on ice in dark for 30-60 minutes.
Add 0.1mL of a viability dye (Zombie Aqua, Biolegend) to each tube
Incubate the cells at room temperature for 15 minutes in dark
Add 1 mL of Staining Buffer
Centrifuge the tubes at 500 xg for 5 minutes.
Discard supernatant and resuspend the cell pellets with 400 mL of Staining Buffer
Filter the cell suspension through 5 mL round-bottom polystyrene test tubes with cell strainer.
-
Place the tubes on ice or at 4ºC until analysis
-
Prepare single stained Compensation Beads at the same time as sample staining:
i. Mix the beads by vortexing and aliquot one drop of beads into new tubes.
ii. Add 1 uL of a single antibody into each tube.
iii. Place the tubes on ice in dark for 15-30 minutes.
iv. Add 1 mL of Staining Buffer and centrifuge the tubes at 500 xg for 5 minutes
v. Discard supernatant and resuspend the beads with 400 mL of Staining Buffer.
- Appropriate compensation is crucial for multi-colour flow cytometry assays. It should be done routinely to ensure validity of the results.
Flow cytometry
The protocol is validated for LSR Fortessa II (BD Biosciences) which is equipped with 4 lasers and 16 detectors, i.e., 2 detectors of the 488 nm blue laser, 6 detectors of the 405 nm violet laser, 3 detectors of the 640 nm red laser, and 5 detectors of the 561 nm yellow-green laser. Voltage for fluorescence channels is set using an unstained cell sample.
- to identify also rare populations, at least 30 000 CD45+ cells need to be captured on flow cytometer
Data analysis
Data can be analysed using various commercially available programs. Our group usually uses FlowJo, as it is the most user friendly, but other programs, such as FCS Express, are also potentially available.
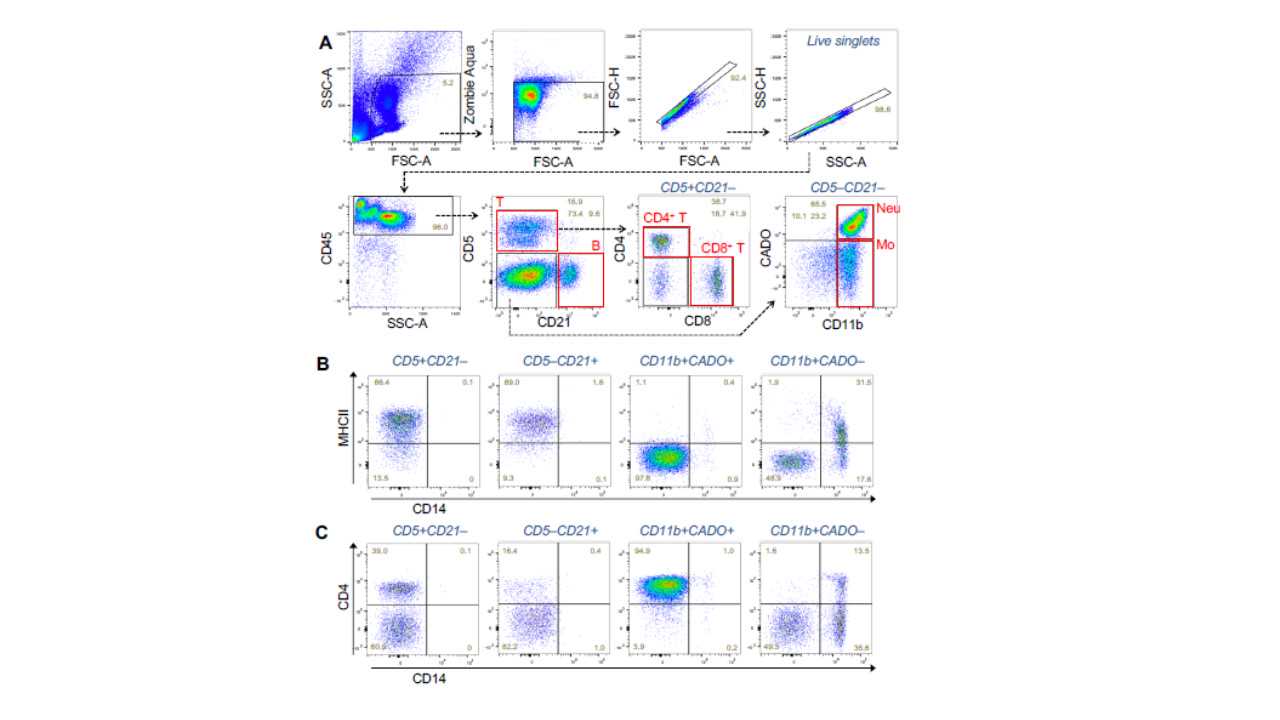
Gating strategy is shown in the figure below