Loading a Radseq Library on Nextseq 500
Angie Chia
Abstract
Protocol to load a Radseq Library on EFGL Nextseq 500
Steps
NextSeq 500 Prep
Power cycling & Manual Wash : Perform a manual wash (Post Run Manual Wash), approximately two hours prior to run/or on the morning of run.
Equipment
| Value | Label |
|---|---|
| NextSeq 500 System | NAME |
| Sequencer | TYPE |
| Illumina | BRAND |
| ********* | SKU |
Power cycle NextSeq 500
- Tap on Home, if not already on Home screen
- Select Manage Instrument -> System Shut down Options
- Select Shut Down
- Wait
0h 5m 0s, before powering back up by pressing on the On button in front of machine - Enter login credentials once Windows startup has completed.
- Wait for machine to initialize NextSeq software, and displays Home screen once ready.
Perform a Post Run Manual Wash (if a run was completed within the past 2-3 days, OK with quick wash instead)
- On Home screen, select Perform Wash -> Manual Wash
- Follow the instructions on NextSeq 500's screen
- When loading wash buffer tray, be sure to fill position 34 with 0.01% ddH20/TWEEN about 3/4 full
- Tap on Start to begin wash
Important: If there is a run in place prior to your wash, the waste container has 1.8% formamide. It needs to be disposed of properly into a waste bottle. Similarly, if reagent tray is in the machine, remove formamide container (location 6) and dispose of it properly (zip-loc bag - contaminated waste trash can). All other liquid/solid waste can be disposed of in sink/trash can.
Thaw Reagent Tray and Flow Cell - It is important that the output of both the reagent tray and flow cell matches i.e. MID/MID or HIGH/HIGH
Retrieve a High-300 output or Mid-300 output Illumina Reagent Tray from the freezer and place in bucket/container of room temperature tap water. Water level should be to the bottom of the reagent tray handle. Let thaw for 1h 0m 0s in water bath.
Retrieve 1 tube of HT1 buffer from the freezer, and place in a beaker of room temperature tap water to thaw as well.
At the 30 minute mark of reagent tray thaw process, retrieve a matching output flow cell (i.e. MID/MID or HIGH/HIGH) from the fridge. Open flow cell packaging, but leave it in its packaging on bench to let it come to room temperature.
NextSeq Library Prep
Begin prepping RAD library for loading, only after reagent tray has been thawing for 40 minutes, and flow cell left on bench for 10 minutes.
Prepare 0.2 N NaOH denature solution. In a new 1.5mL LoBind tube (label tube 0.2 ):
- Add
10µLof 1 N NaOH +40µLof molecular grade lab water - Gently vortex, and briefly spin down.
- Let sit at room temperature.
Add the following components in order into a new 1.5mL LoBind tube (label tube DL ):
5µLof 0.2 N NaOH +5µLof 4nM RAD library - do not pump, as this can introduce bubbles- Vortex gently, spin
300x g(using IEC micromax) - Incubate DL at room temperature for
0h 5m 0s - Add
5µLof 200mM Tris-HCl pH 7.0 to DL - Vortex gently and spin down
To DL tube, add 985µL of HT1 buffer.
If library is not at 4nM , use the equation below to determine the amount of HT1 buffer to add:
Gently vortex DL tube, and spin down. Keep tube in a benchtop cooler.
Library is now at 20pM .
Depending on the output you'll want to achieve for your library, further dilute your library to the desired concentration prior to loading onto reagent tray for sequencing. The table below is an example that our lab uses for Mid/High - 300 cycles output libraries.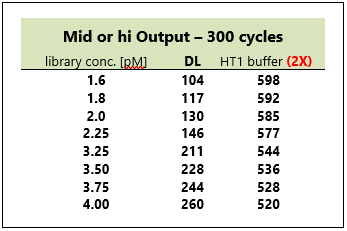
In a new 2.0mL tube (label tube 10 ), add the components accordingly to the table above.
- Gently vortex, and spin down. Tube 10 has a final volume of
1300µL - Keep tube in benchtop cooler till reagent tray is ready to be loaded.
Prep NextSeq reagent tray
Once reagent tray has been thawing for an hour, remove tray from water bath and wipe dry.
- Invert reagent tray gently 5 times to mix reagents, and gently tap tray on bench top to collect all liquid to the bottom.
- Use a 1000µl pipette tip and punch a hole in position 10. Discard tip.
- Load all
1300µLof tube 10 into position 10 of reagent tray.
Loading reagent tray on NextSeq 500
Verify both reagent tray and flow cell output matches. 1. Take a new buffer cartridge
- Inspect the flow cell by making sure brackets on the back are in place, and flushed. If there's any dirt on top of flow cell, gently remove with a Kimwipe.
On Home screen, select Sequence
- "Manual" would been pre-selected, tap on Next
Enter Run Parameters:
- Run Name: Use your library name (e.g. L0012)
- Library ID: Same as Run Name
- Check the "Paired end" box
- Read 1: 150 Index 1: 6 Index 2: 6 Read 2: 150 (index cycles are optional)
- Tap on Next
Follow the instructions on NextSeq 500's screen to load buffer cartridge and reagent tray for sequencing.
Once all pre-run checks are complete on the machine, the run will start automatically. (Typical High-300 Output run takes ~32 hours, and Mid-300 Output takes ~26 hours)

