General genotyping of Myzus persicae using PCR with microsatellite markers and gel electrophoresis
Mariska M. Beekman, Marcel Dicke, Bas J. Zwaan, Eveline Verhulst, Bart Pannebakker
Abstract
This protocol can be used to 'roughly' genotype green peach aphids, Myzus persicae (Sulzer), based on microsatellites using PCR and gel electrophoresis with high percentage agarose gels. Please note this protocol does not result in exact allele information as it uses gel-electrophoresis for allele identification and comparisons instead of capillary-electrophoresis. Therefore, allele sizes are judged by eye and thus results are prone to human error as differentiating between identical and different-length alleles might be a challenge.
Microsatellites are regions in the genome with repeating DNA sequences. This protocol targets microsatellites with di-repeats, meaning they contain repeats of two different bases, for example GAGAGAGAGA. Due to slippage during DNA replications, the number of repeats in a microsatellite region can change over time at a rate higher than that of alternative regions in the genome. The number of repeats an organism has for a certain microsatellite region represents a specific allele. The allele sizes of multiple microsatellite regions can be determined to distinguish an organisms multi-locus genotype (MLG). Because M. persicae reproduces by parthenogenesis, you can use microsatellites to determine which aphids are clone-mates (aphids coming from the same parthenogenic ancestor), as these will have the same MLG.
The microsatellite regions will be targeted with PCR. Alleles with more repeats will result in a larger PCR product. As larger PCR products travel slower through an agarose gel during gel-electrophoresis than smaller PCR products, the pattern of bands seen on the agarose gel will be unique for each unique allele. It is possible to target and amplify multiple microsatellite loci at the same time in multiplex PCR. This will result in more information in a single PCR reaction, but with a trade-off when too many markers are combined as the final result can become blurry or difficult to interpret.
IMPORTANT: This protocol can be used to check if various samples of M. persicae have similar or distinct MLGs. This protocol on its own can not be used to assign a specific genotype to a sample.
Steps
Materials
Chelex and proteinase K based DNA extraction
Pipettes and pipette tips
Eppendorf tubes
Centrifuge
Heat block or water bath
Ultrapure water
Chelex 100 resin (Bio-Rad, Hercules, CA, USA)
Proteinase K (20 mg/mL; Promega, Southampton, UK)
PCR
Pipettes and pipette tips
PCR machine
PCR tubes
Eppendorf tubes
Ultrapure water
Primers (see table below)
QIAGEN Multiplex PCR Master Mix (206143; Qiagen, Venlo, the Netherlands) OR a different DNA polymerase + dNTPs + potentially additional MgCl2
DNA of the aphids to be tested
DNA of the aphid genotype of interest (reference/ positive control)
Primers
| A | B | C | D |
|---|---|---|---|
| Primer | Sequence | Multiplex | Reference |
| M37_F | GTGTGAGTAAGTCGTATTG | 1 | Sloane et al., 2001 |
| M37_R | TTGTATTATGTACCTGTGC | 1 | " |
| M40_F | ACACGCATACAAGAATAGGG | 2 | " |
| M40_R | AGAGGAGGCAGAGGTGAAAC | 2 | " |
| M86_F | TCCACTAAGACCTCAAACAC | 2 | " |
| M86_R | ATTTATTATGTCGTTCCGCC | 2 | " |
| myz2_F | TGGCGAGAGAGAAAGACCTGC | 1 | Wilson et al., 2004 |
| myz2_R | TCGGAAGACAGAGACATCGAGA | 1 | " |
| myz9_F | AACCTCACCTCGTGGAGTTCG | 2 | " |
| myz9_R | CTTGGATGTGTGTGGGGTGC | 2 | " |
Tabel 1: Primers to target different microsatellite loci, M37/M40/M86/myz2/myz9, in the Myzus persciae. Markers with the most information (most alleles and the largest size differences between alleles) are M86, myz2 and myz9. The least information comes from marker M37. Primers in the same multiplex can be combined in a single PCR reaction.
Methods
DNA extractions
You can use any preferred protocol to extract DNA from the aphids. You don’t need a lot or high quality DNA. Extract DNA from single aphids (preferably adult, non-damaged, non-parasitized). We use the DNA extraction protocol described below, which is also published in Beekman and Donner et al. (2022).
Chelex and proteinase K based DNA extraction
Make a 5% (v/v) Chelex 100 resin solution in ultrapure water. For each aphid sample you want to genotype, add 100µL of the 5% Chelex solution in an eppendorf tube and add 2.5µL proteinase K . When extracting DNA from aphids stored in ethanol, make sure to let the ethanol evaporate completely before adding the aphid to the eppendorf tube. Grind up the aphid in the Chelex and proteinase K solution, for instance by using a clean pipette tip. Shortly vortex and spin down the tube. Incubate the tubes for at least 1h 0m 0sat 56°C (can also be done overnight) and subsequently heat to 98°Cfor 0h 8m 0s to inactivate the proteinase K. Cool down on ice. Centrifuge the tubes at high speed for 0h 4m 0s. The DNA will be in the supernatant. Be careful not to take any Chelex beads while pipetting off the supernatant, as Chelex resin will inhibit downstream PCR reactions. Dilute DNA at 33% (v/v) in ultrapure water and use immediate, or store either short-term at 4°C or long-term at -20°C until further use.
PCR reactions
PCR reactions are carried out in 5µL-volumes containing 1x QIAGEN Multiplex PCR Master Mix (containing dNTPs, HotStartTaq DNA polymerase and 3millimolar (mM) MgCl2 as final concentration), 0.2millimolar (mM) of each primer, and 1µL Chelex extracted DNA (which was diluted at 1/3). It is possible to use different DNA polymerases but I don’t know what the final product will look like on gel, it is possible that there is more smear and less clear individual bands. When doing multiplex PCR using a different DNA polymerase, it might also be necessary to increase the [Mg2+] to make sure all primers anneal during PCR.
Which and how many microsatellite markers to target is up to you and depends on how much time,
money and resources you want to spend, as well on how certain you want to be of your results. I would recommend to do a multiplex of M86 and myz9. If you want to be more sure that samples with the same pattern for these loci have the same genotype, you can do an extra PCR for M37 and myz2 multiplexed.
Make a master mix for the PCR reactions containing all ingredients except for the DNA. Always include a negative control in which no DNA is added, as well as at least one positive control for the M. persicae genotype of interest. If the positive control is not included of fails, it will not be possible to say if the collected aphids have the genotype of interest. Even better would be to also include a DNA sample of a M. persicae genotypes which is known to be different from the genotype of interest.
Thermocycling protocol PCR machine
| A | B | C |
|---|---|---|
| temperature | duration | cycles |
| 95 °C | 15 min | |
| 94 °C | 30 s | |
| 57 °C | 60 s | |
| 72 °C | 45 s | |
| 72 °C | 5 min | |
| 12 °C | ∞ |
Table 2: Thermocycling conditions for the PCR for when using the QIAGEN Multiplex PCR Master Mix
Gel electrophoresis
The PCR products will be visualized on a 3.5-4% agarose gel.
Make a 3.5-4% agarose gel. It might be difficult to dissolve the agarose. Stirring with a magnetic stirrer in between rounds of boiling will help. Stain the gel with the stain of your preference. Pour the gel and let it set properly.
Add loading dye to the PCR products and load them onto the gel. Don't load too much PCR product as this makes it difficult to distinguish separate bands. Start by trying out 1.5µL. The gel should run quite long to make sure all bands are properly separated. Exact time will depend on the voltage, and voltage will depend on the size of your gel. The gels from the examples (section 4) were ran for 1.5-2 hours at 90 volt. Running longer will increase the distance between bands, which makes for easier and more precise comparison of genotypes, but it will also result in fainter bands. You can check the gel after 45min and run longer if necessary.
Each row of the gel should contain at least one sample of the aphid genotype of interest so that the
pattern of DNA bands can be easily compared with the pattern of the genotype of interest. At the beginning and end of each row, add a DNA ladder, preferably a 100bp step ladder.
Examples of results
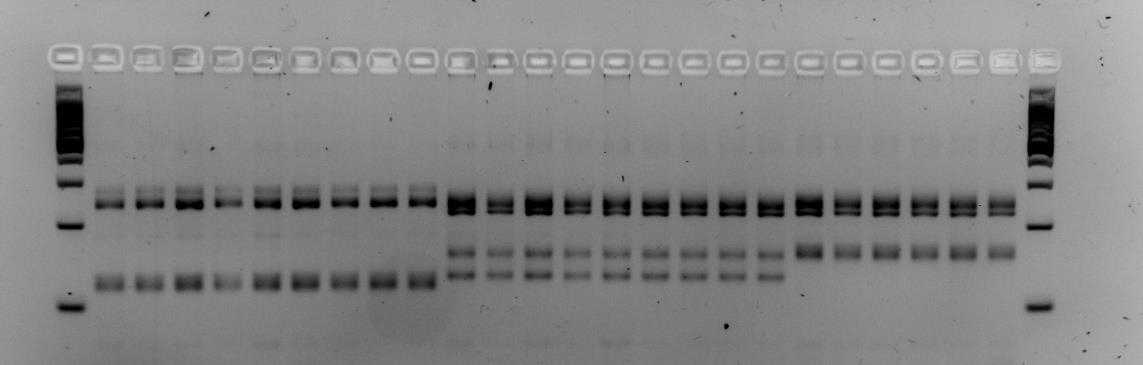
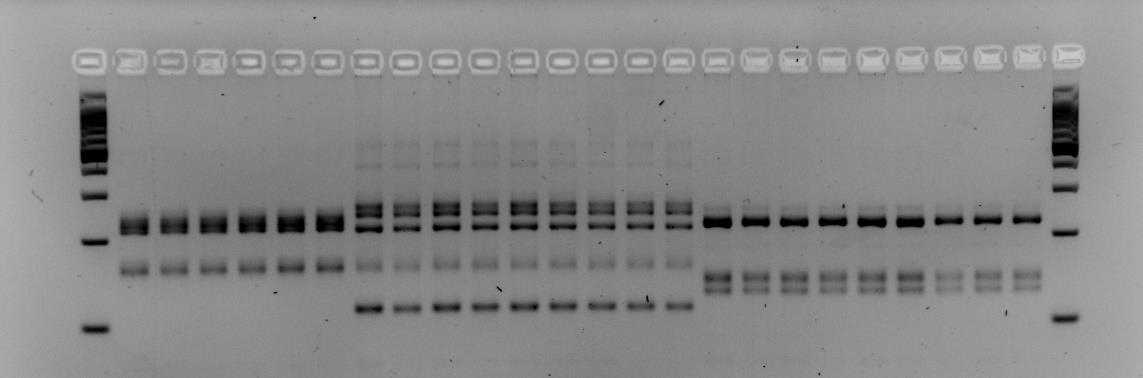
Below is an example of results that were obtained when using primers M86 and myz9 in multiplex. The first and last columns are a 100bp step ladder. Every other column is a single aphid sample. The pattern of the PCR fragments represent the respective genotype of an aphid. Brightness of the bands does not matter but it is the height of the bands that determine whether an aphid has a different genotype. In the examples below we see that both gels contain three different multi-locus genotypes.
!Careful! Make sure the 100, 200 and 300 fragments of the ladder have sufficiently separated (Fig. 1). If not sufficiently separated, it will be impossible to distinguish size differences between the amplicons. Additionally, samples with different patterns are for sure different clones, however, samples with similar patterns could also different clones that by chance have the same (or very similar) length alleles for the loci you tested. To increase the certainty that samples indeed belong to the same clone-mates, include extra microsatellite markers.